Testing Variance Prior for cTWAS
wesleycrouse
2023-06-26
Last updated: 2023-08-18
Checks: 6 1
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ce02b6f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Untracked files:
Untracked: gwas.RData
Untracked: ld_R_info.RData
Untracked: z_snp_pos_ebi-a-GCST004131.RData
Untracked: z_snp_pos_ebi-a-GCST004132.RData
Untracked: z_snp_pos_ebi-a-GCST004133.RData
Untracked: z_snp_pos_scz-2018.RData
Untracked: z_snp_pos_ukb-a-360.RData
Untracked: z_snp_pos_ukb-d-30780_irnt.RData
Unstaged changes:
Modified: analysis/variance_prior_testing.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/variance_prior_testing.Rmd) and HTML
(docs/variance_prior_testing.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | ce02b6f | wesleycrouse | 2023-08-18 | fixing variance prior report |
| Rmd | 1632c6b | wesleycrouse | 2023-07-27 | fixing gamma prior results |
| Rmd | 05b0117 | wesleycrouse | 2023-07-27 | adding results for inv-gamma prior on variance |
analysis_id <- "ukb-d-30660_irnt_Liver"
trait_id <- "ukb-d-30660_irnt"
source("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ctwas_config.R")E-M with Inverse-Gamma Prior on Variance
Likelihood Function
The complete log likelihood for a single group. For completeness, [brackets] denote variables that are conditionally independent.
Dropping conditionally independent variables from the notation:
Gamma prior on the precision
Hyperparameter \(a\) is the shape and \(b\) is the rate.
\(\sigma^{-2} \sim \mathrm{Gamma}(a,b)\)
\(\mathrm{logp}(\sigma^{-2}) = a\mathrm{log}(b) - \mathrm{log}(\Gamma(a)) + (a-1)\mathrm{log}(\sigma^{-2}) -b\sigma^{-2}\)
E-M update for the variance
Take the expectation over \(\beta,\Gamma|\pi^{t},\sigma^{t}\), then take the derivative with respect to \(\sigma^{-2}\) and set it equal to zero. \(\alpha_{j}\) is the PIP of variable \(j\).
\((\sigma^{2})^{t+1} = \frac{0.5\sum_{j}[\alpha_{j}\tau_{j}]+b}{0.5\sum[\alpha_j] + a - 1}\)
Case 31 - Bilirubin with Liver - Independent Variance
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case31")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
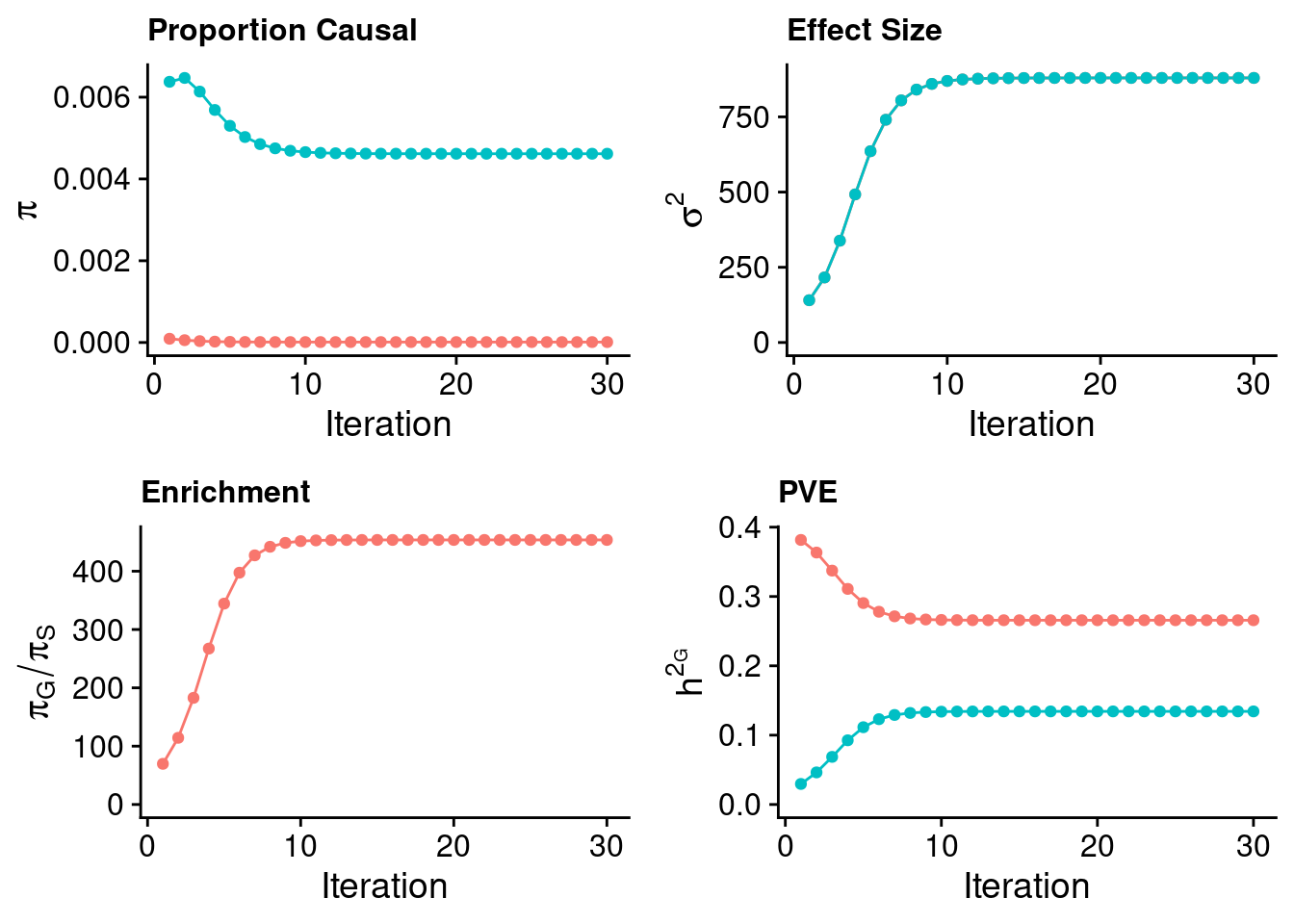
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))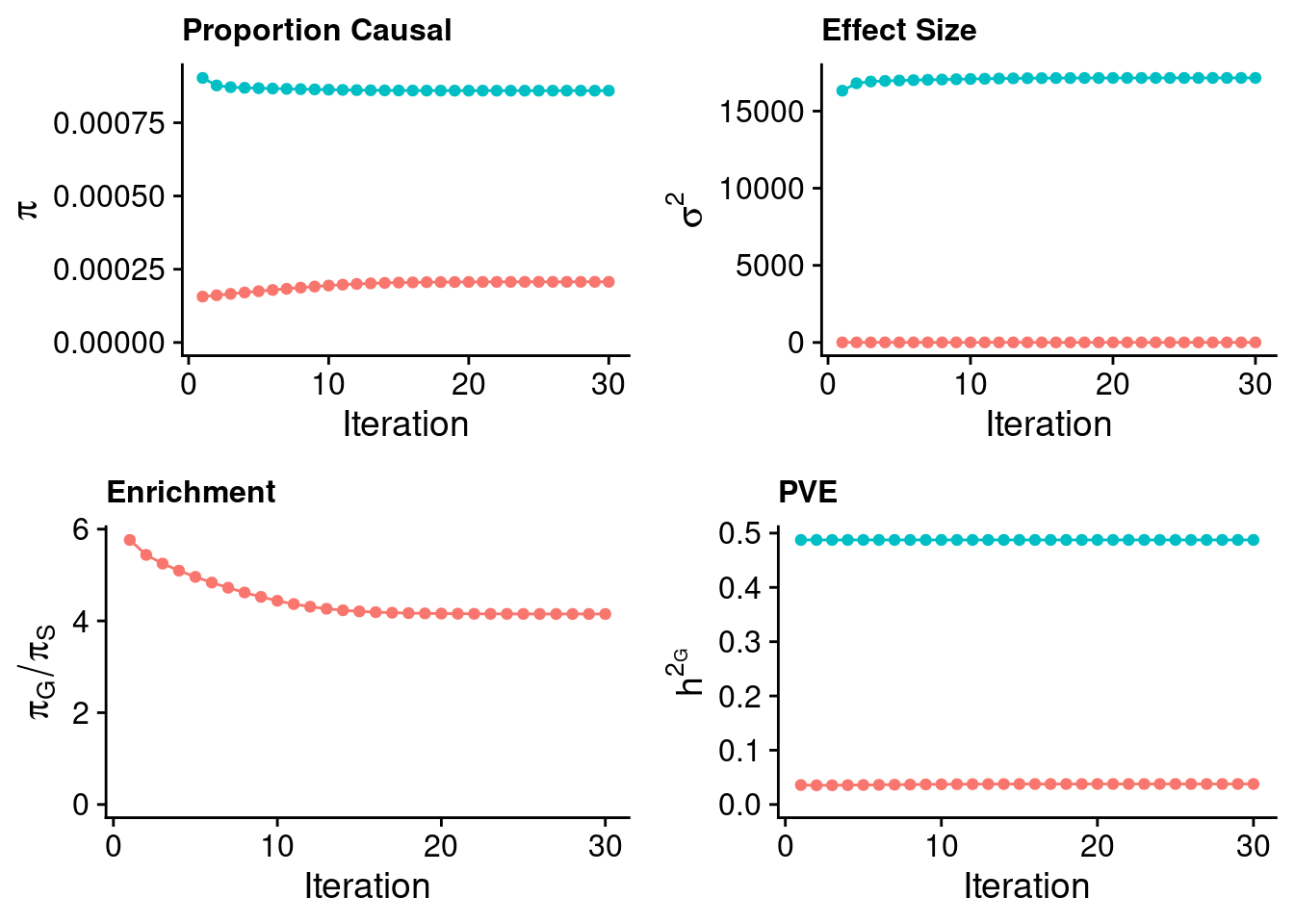
| Version | Author | Date |
|---|---|---|
| ce02b6f | wesleycrouse | 2023-08-18 |
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0008594861 0.0002070674 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
17150.42864 6.14623 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
4.150755 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) gene SNP
0.48745533 0.03778347 #total PVE
sum(estimated_group_pve)[1] 0.5252388#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) gene SNP
0.92806421 0.07193579 Case 35 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=2, b=10
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case35")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))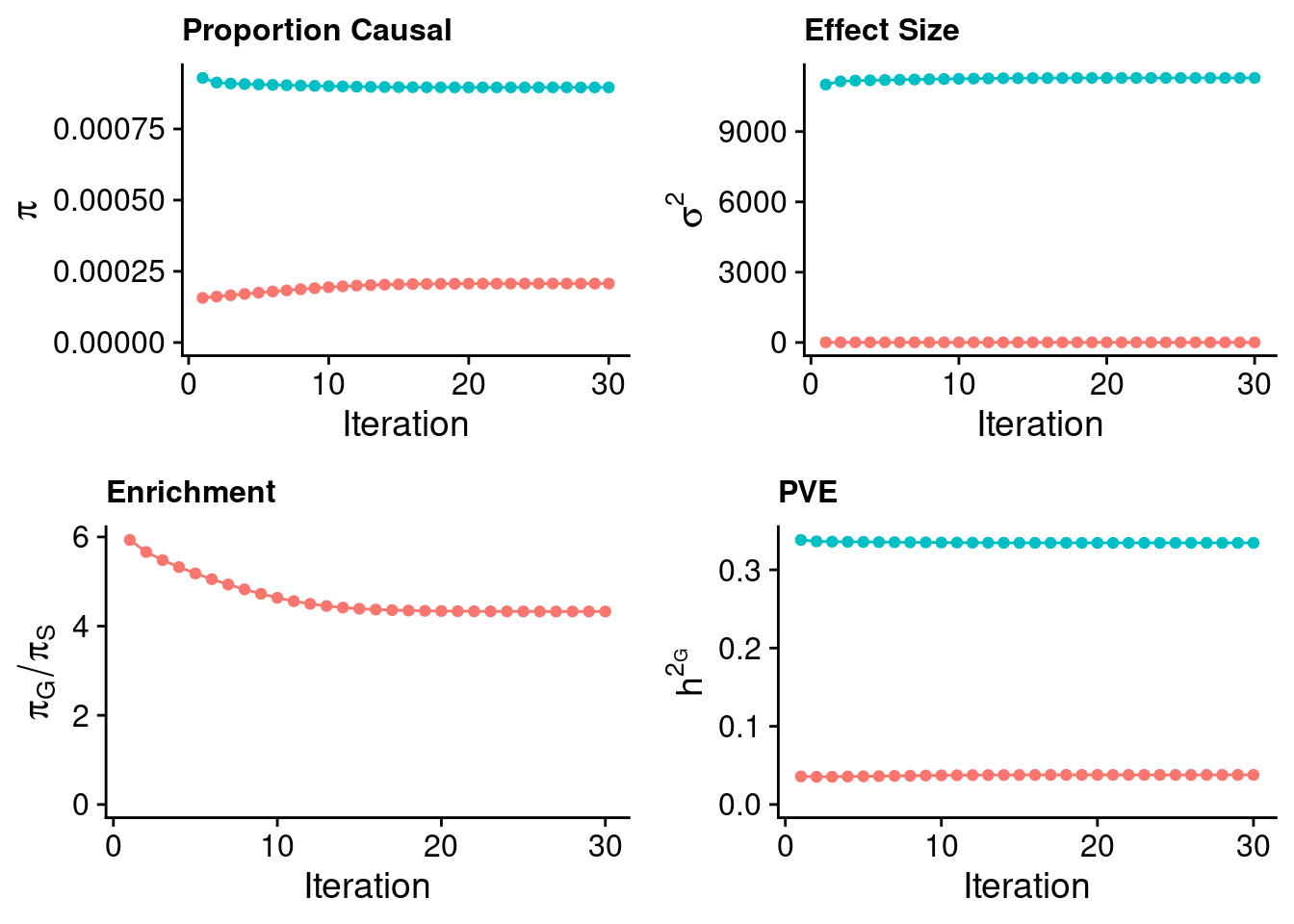
| Version | Author | Date |
|---|---|---|
| ce02b6f | wesleycrouse | 2023-08-18 |
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0008957946 0.0002069582 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
11288.781541 6.160552 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
4.328384 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) gene SNP
0.33440788 0.03785153 #total PVE
sum(estimated_group_pve)[1] 0.3722594#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) gene SNP
0.8983195 0.1016805 Case 36 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=5, b=40
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case36")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))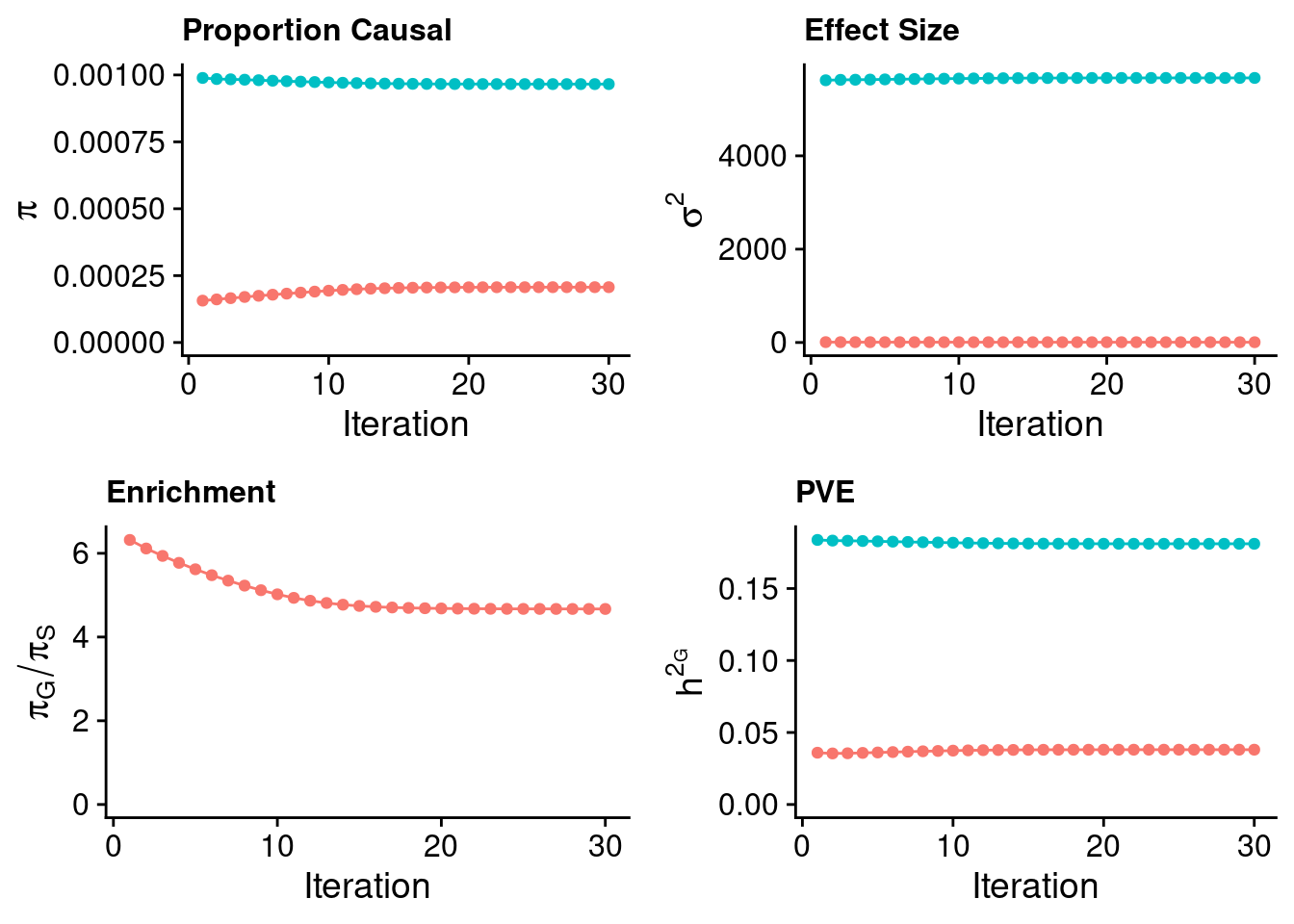
| Version | Author | Date |
|---|---|---|
| ce02b6f | wesleycrouse | 2023-08-18 |
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0009658282 0.0002067951 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
5669.185717 6.200549 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
4.67046 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) gene SNP
0.18106793 0.03806725 #total PVE
sum(estimated_group_pve)[1] 0.2191352#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) gene SNP
0.8262842 0.1737158 Case 37 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=41, b=400
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case37")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))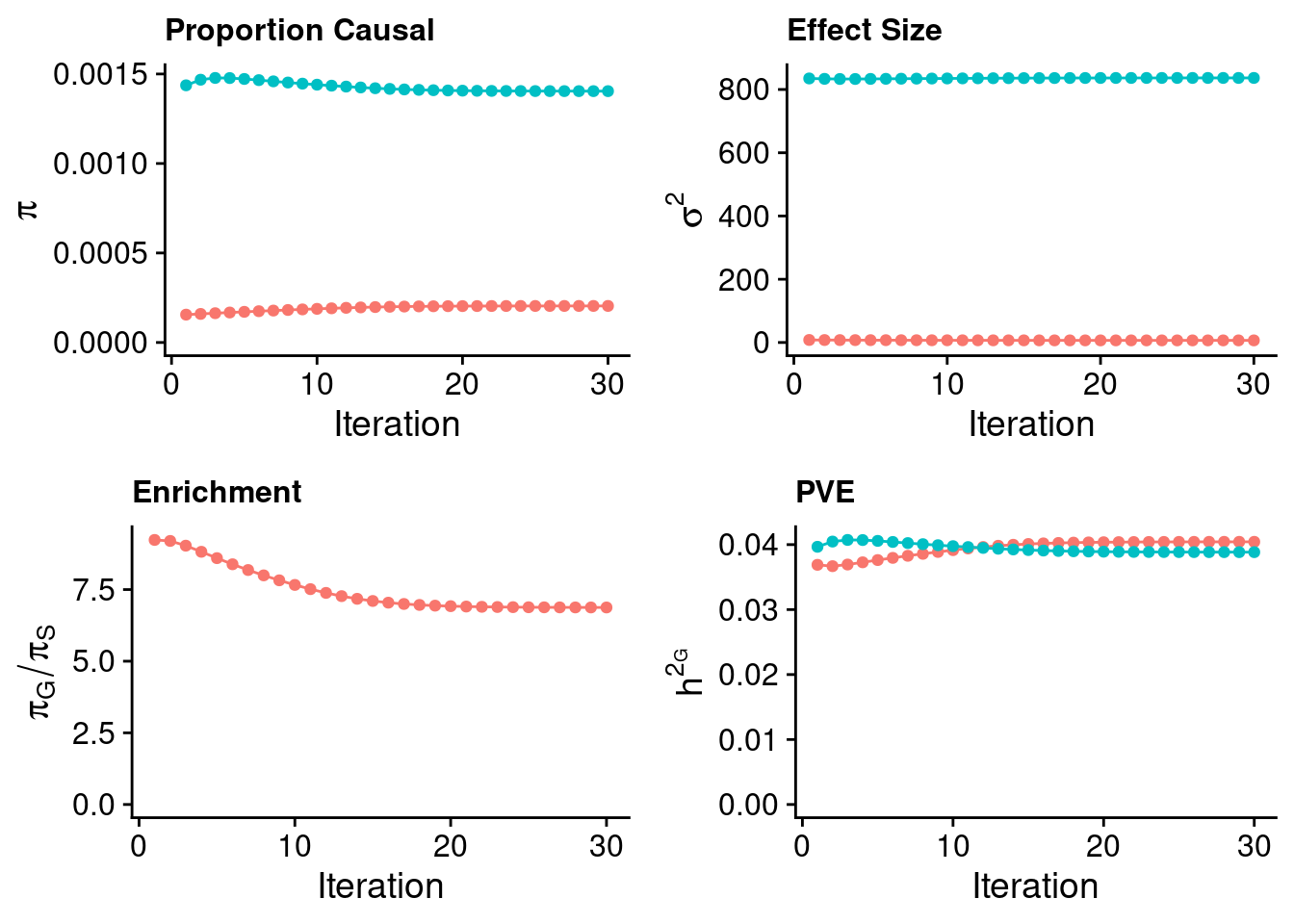
| Version | Author | Date |
|---|---|---|
| ce02b6f | wesleycrouse | 2023-08-18 |
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0014040979 0.0002042366 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
836.537388 6.667974 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
6.874859 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) SNP gene
0.04043046 0.03884219 #total PVE
sum(estimated_group_pve)[1] 0.07927265#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) SNP gene
0.5100177 0.4899823 Case 39 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=401, b=4000
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case39")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))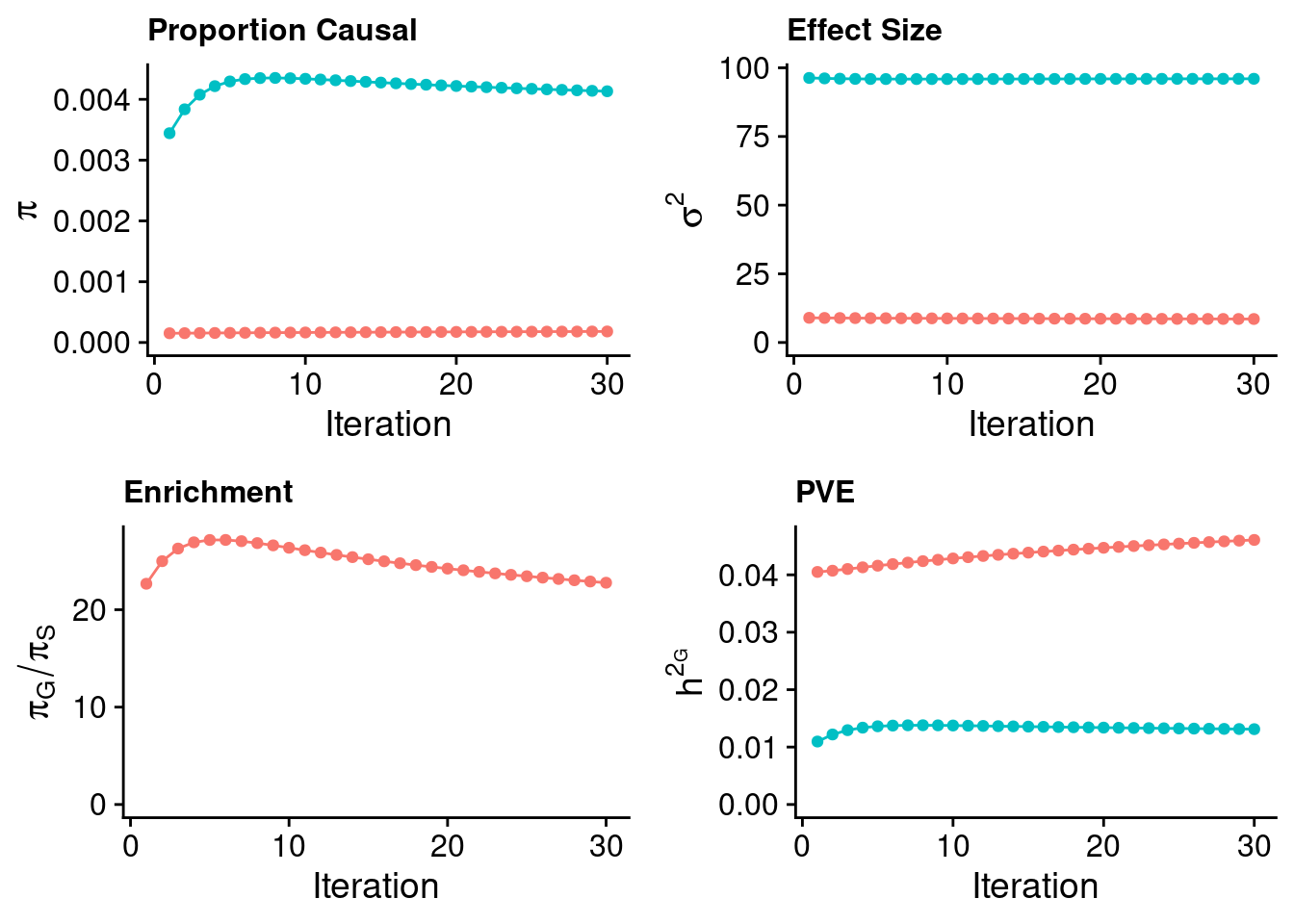
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0041325068 0.0001814764 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
95.997710 8.552013 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
22.7716 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) SNP gene
0.04607546 0.01311884 #total PVE
sum(estimated_group_pve)[1] 0.0591943#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) SNP gene
0.7783766 0.2216234 Case 40 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=4001, b=40000
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case40")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))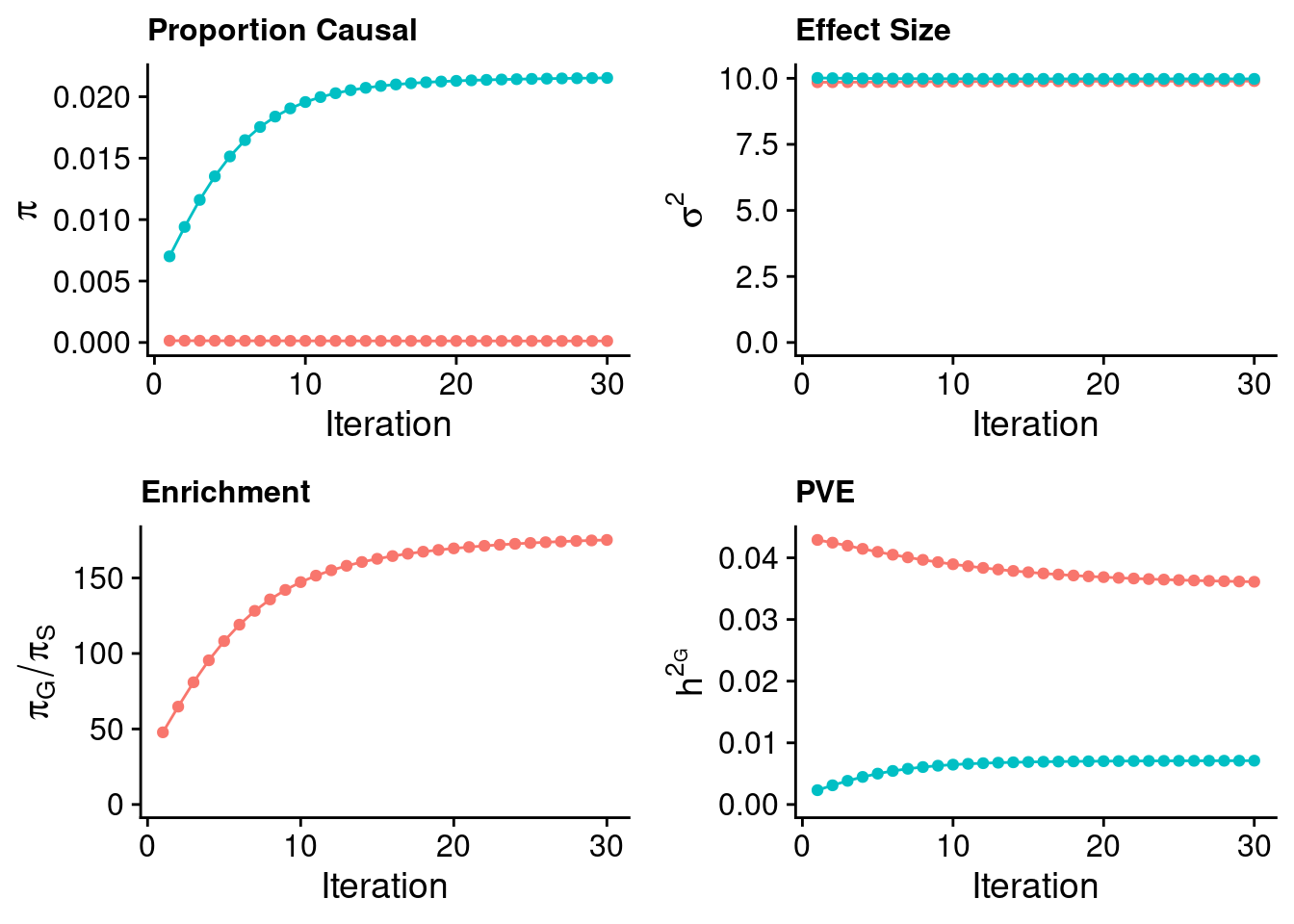
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0215283290 0.0001229033 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
9.978117 9.896473 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
175.1647 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) SNP gene
0.036109819 0.007103623 #total PVE
sum(estimated_group_pve)[1] 0.04321344#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) SNP gene
0.8356154 0.1643846 Case 38 - Bilirubin with Liver - Inv-Gamma prior on variance of SNPs and genes; a=1000001, b=10000000
Load ctwas results
results_dir <- paste0("/project2/mstephens/wcrouse/ctwas_multigroup_testing/ukb-d-30780_irnt/multigroup_case38")
weight <- "/project2/compbio/predictdb/mashr_models/mashr_Liver.db"
#load information for all genes
gene_info <- data.frame(gene=as.character(), genename=as.character(), gene_type=as.character(), weight=as.character())
for (i in 1:length(weight)){
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, weight[i])
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info_current <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
gene_info_current$weight <- weight[i]
gene_info <- rbind(gene_info, gene_info_current)
}
gene_info$group <- sapply(1:nrow(gene_info), function(x){paste0(unlist(strsplit(tools::file_path_sans_ext(rev(unlist(strsplit(gene_info$weight[x], "/")))[1]), "_"))[-1], collapse="_")})
gene_info$gene_id <- paste(gene_info$gene, gene_info$group, sep="|")
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#load z scores for SNPs and collect sample size
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
sample_size <- z_snp$ss
sample_size <- as.numeric(names(which.max(table(sample_size))))
# #compute PVE for each gene/SNP
# ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type != "SNP", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# ctwas_gene_res$gene_id <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x, split="[|]"))[1]})
# ctwas_gene_res$group <- ctwas_gene_res$type
# ctwas_gene_res$alt_name <- paste0(ctwas_gene_res$gene_id, "|", ctwas_gene_res$group)
#
# ctwas_gene_res$genename <- NA
# ctwas_gene_res$gene_type <- NA
#
# group_list <- unique(ctwas_gene_res$group)
#
# for (j in 1:length(group_list)){
# print(j)
# group <- group_list[j]
#
# res_group_indx <- which(ctwas_gene_res$group==group)
# gene_info_group <- gene_info[gene_info$group==group,,drop=F]
#
# ctwas_gene_res[res_group_indx,c("genename", "gene_type")] <- gene_info_group[sapply(ctwas_gene_res$alt_name[res_group_indx], match, gene_info_group$gene_id), c("genename", "gene_type")]
# }
#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
#ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
#ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
# #merge gene and snp results with added information
# ctwas_snp_res$gene_id=NA
# ctwas_snp_res$group="SNP"
# ctwas_snp_res$alt_name=NA
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
# #store columns to report
# report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos", "alt_name", "gene_id"))]
# first_cols <- c("genename", "group", "region_tag")
# report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])Check convergence of parameters
library(ggplot2)
library(cowplot)
load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- structure(c(n_snps, nrow(z_gene)), names=c("SNP","gene"))
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#estimated enrichment of genes (all iterations)
estimated_enrichment_all <- t(sapply(rownames(estimated_group_prior_all)[rownames(estimated_group_prior_all)!="SNP"], function(x){estimated_group_prior_all[rownames(estimated_group_prior_all)==x,]/estimated_group_prior_all[rownames(estimated_group_prior_all)=="SNP"]}))
title_size <- 12
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_all), nrow(estimated_group_prior_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_all), function(x){estimated_group_prior_all[x,]})),
group = rep(rownames(estimated_group_prior_all), each=ncol(estimated_group_prior_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Proportion Causal") +
theme_cowplot()
p_pi <- p_pi + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_prior_var_all), nrow(estimated_group_prior_var_all)),
value = unlist(lapply(1:nrow(estimated_group_prior_var_all), function(x){estimated_group_prior_var_all[x,]})),
group = rep(rownames(estimated_group_prior_var_all), each=ncol(estimated_group_prior_var_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Effect Size") +
theme_cowplot()
p_sigma2 <- p_sigma2 + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_group_pve_all), nrow(estimated_group_pve_all)),
value = unlist(lapply(1:nrow(estimated_group_pve_all), function(x){estimated_group_pve_all[x,]})),
group = rep(rownames(estimated_group_pve_all), each=ncol(estimated_group_pve_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_pve <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(h^2[G])) +
ggtitle("PVE") +
theme_cowplot()
p_pve <- p_pve + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
df <- data.frame(niter = rep(1:ncol(estimated_enrichment_all), nrow(estimated_enrichment_all)),
value = unlist(lapply(1:nrow(estimated_enrichment_all), function(x){estimated_enrichment_all[x,]})),
group = rep(rownames(estimated_enrichment_all), each=ncol(estimated_enrichment_all)))
groupnames_for_plots <- sapply(as.character(unique(df$group)), function(x){paste(sapply(unlist(strsplit(x, "_")), substr, start=1, stop=3), collapse="_")})
df$group <- groupnames_for_plots[df$group]
df$group <- as.factor(df$group)
p_enrich <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi[G]/pi[S])) +
ggtitle("Enrichment") +
theme_cowplot()
p_enrich <- p_enrich + theme(plot.title=element_text(size=title_size)) +
expand_limits(y=0) +
guides(color = guide_legend(title = "Group")) + theme (legend.title = element_text(size=12, face="bold"))
plot_grid(p_pi + theme(legend.position = "none"),
p_sigma2 + theme(legend.position = "none"),
p_enrich + theme(legend.position = "none"),
p_pve+ theme(legend.position = "none"))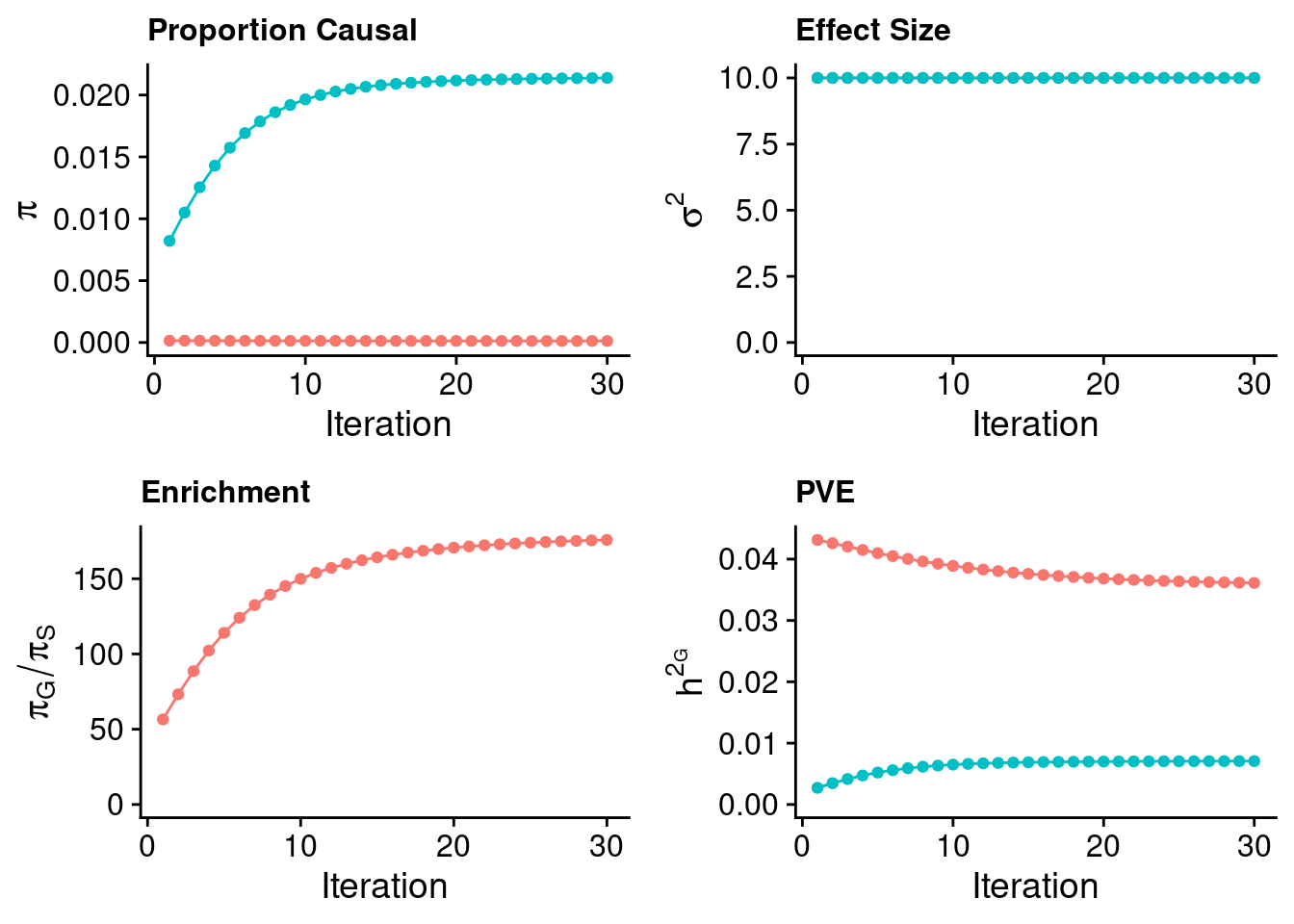
#p_pi
#p_sigma2 + theme(legend.position = "none")
#p_enrich + theme(legend.position = "none")
#p_pve + theme(legend.position = "none")
####################
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
-sort(-estimated_group_prior) gene SNP
0.0213809018 0.0001216473 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
-sort(-estimated_group_prior_var) gene SNP
9.999914 9.999574 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[,ncol(group_prior_var_rec)]
-sort(-estimated_enrichment) gene
175.7614 #report sample size
print(sample_size)[1] 292933#report group size
print(group_size) SNP gene
8696600 9687 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
-sort(-estimated_group_pve) SNP gene
0.036113143 0.007070388 #total PVE
sum(estimated_group_pve)[1] 0.04318353#attributable PVE
-sort(-estimated_group_pve/sum(estimated_group_pve)) SNP gene
0.8362712 0.1637288
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /software/R-4.1.0-no-openblas-el7-x86_64/lib64/R/lib/libRblas.so
LAPACK: /software/R-4.1.0-no-openblas-el7-x86_64/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.35 bslib_0.4.1 colorspace_2.0-3
[5] vctrs_0.5.1 generics_0.1.3 htmltools_0.5.4 yaml_2.3.6
[9] utf8_1.2.2 blob_1.2.3 rlang_1.0.6 jquerylib_0.1.4
[13] later_1.3.0 pillar_1.8.1 withr_2.5.0 glue_1.6.2
[17] DBI_1.1.3 bit64_4.0.5 lifecycle_1.0.3 stringr_1.5.0
[21] munsell_0.5.0 gtable_0.3.1 evaluate_0.18 memoise_2.0.1
[25] labeling_0.4.2 knitr_1.41 callr_3.7.3 fastmap_1.1.0
[29] httpuv_1.6.6 ps_1.7.2 fansi_1.0.3 highr_0.9
[33] Rcpp_1.0.9 promises_1.2.0.1 scales_1.2.1 cachem_1.0.6
[37] jsonlite_1.8.4 farver_2.1.1 fs_1.5.2 bit_4.0.5
[41] digest_0.6.31 stringi_1.7.8 processx_3.8.0 dplyr_1.0.10
[45] getPass_0.2-2 rprojroot_2.0.3 grid_4.1.0 cli_3.4.1
[49] tools_4.1.0 magrittr_2.0.3 sass_0.4.4 tibble_3.1.8
[53] RSQLite_2.2.19 whisker_0.4.1 pkgconfig_2.0.3 data.table_1.14.6
[57] assertthat_0.2.1 rmarkdown_2.18 httr_1.4.4 rstudioapi_0.14
[61] R6_2.5.1 git2r_0.30.1 compiler_4.1.0