Rheumatoid factor (quantile) - Liver
wesleycrouse
2021-09-09
Last updated: 2021-09-09
Checks: 6 1
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 59e5f4d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Unstaged changes:
Modified: analysis/ukb-d-30500_irnt_Liver.Rmd
Modified: analysis/ukb-d-30600_irnt_Liver.Rmd
Modified: analysis/ukb-d-30610_irnt_Liver.Rmd
Modified: analysis/ukb-d-30620_irnt_Liver.Rmd
Modified: analysis/ukb-d-30630_irnt_Liver.Rmd
Modified: analysis/ukb-d-30640_irnt_Liver.Rmd
Modified: analysis/ukb-d-30650_irnt_Liver.Rmd
Modified: analysis/ukb-d-30660_irnt_Liver.Rmd
Modified: analysis/ukb-d-30670_irnt_Liver.Rmd
Modified: analysis/ukb-d-30680_irnt_Liver.Rmd
Modified: analysis/ukb-d-30690_irnt_Liver.Rmd
Modified: analysis/ukb-d-30700_irnt_Liver.Rmd
Modified: analysis/ukb-d-30710_irnt_Liver.Rmd
Modified: analysis/ukb-d-30720_irnt_Liver.Rmd
Modified: analysis/ukb-d-30730_irnt_Liver.Rmd
Modified: analysis/ukb-d-30740_irnt_Liver.Rmd
Modified: analysis/ukb-d-30750_irnt_Liver.Rmd
Modified: analysis/ukb-d-30760_irnt_Liver.Rmd
Modified: analysis/ukb-d-30770_irnt_Liver.Rmd
Modified: analysis/ukb-d-30780_irnt_Liver.Rmd
Modified: analysis/ukb-d-30790_irnt_Liver.Rmd
Modified: analysis/ukb-d-30800_irnt_Liver.Rmd
Modified: analysis/ukb-d-30810_irnt_Liver.Rmd
Modified: analysis/ukb-d-30820_irnt_Liver.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-d-30820_irnt_Liver.Rmd) and HTML (docs/ukb-d-30820_irnt_Liver.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | cbf7408 | wesleycrouse | 2021-09-08 | adding enrichment to reports |
| html | cbf7408 | wesleycrouse | 2021-09-08 | adding enrichment to reports |
| Rmd | 4970e3e | wesleycrouse | 2021-09-08 | updating reports |
| html | 4970e3e | wesleycrouse | 2021-09-08 | updating reports |
| Rmd | 627a4e1 | wesleycrouse | 2021-09-07 | adding heritability |
| Rmd | dfd2b5f | wesleycrouse | 2021-09-07 | regenerating reports |
| html | dfd2b5f | wesleycrouse | 2021-09-07 | regenerating reports |
| Rmd | 61b53b3 | wesleycrouse | 2021-09-06 | updated PVE calculation |
| html | 61b53b3 | wesleycrouse | 2021-09-06 | updated PVE calculation |
| Rmd | 837dd01 | wesleycrouse | 2021-09-01 | adding additional fixedsigma report |
| Rmd | d0a5417 | wesleycrouse | 2021-08-30 | adding new reports to the index |
| Rmd | 0922de7 | wesleycrouse | 2021-08-18 | updating all reports with locus plots |
| html | 0922de7 | wesleycrouse | 2021-08-18 | updating all reports with locus plots |
| html | 1c62980 | wesleycrouse | 2021-08-11 | Updating reports |
| Rmd | 5981e80 | wesleycrouse | 2021-08-11 | Adding more reports |
| html | 5981e80 | wesleycrouse | 2021-08-11 | Adding more reports |
| Rmd | 05a98b7 | wesleycrouse | 2021-08-07 | adding additional results |
| html | 05a98b7 | wesleycrouse | 2021-08-07 | adding additional results |
| html | 03e541c | wesleycrouse | 2021-07-29 | Cleaning up report generation |
| Rmd | 276893d | wesleycrouse | 2021-07-29 | Updating reports |
| html | 276893d | wesleycrouse | 2021-07-29 | Updating reports |
Overview
These are the results of a ctwas analysis of the UK Biobank trait Rheumatoid factor (quantile) using Liver gene weights.
The GWAS was conducted by the Neale Lab, and the biomarker traits we analyzed are discussed here. Summary statistics were obtained from IEU OpenGWAS using GWAS ID: ukb-d-30820_irnt. Results were obtained from from IEU rather than Neale Lab because they are in a standardard format (GWAS VCF). Note that 3 of the 34 biomarker traits were not available from IEU and were excluded from analysis.
The weights are mashr GTEx v8 models on Liver eQTL obtained from PredictDB. We performed a full harmonization of the variants, including recovering strand ambiguous variants. This procedure is discussed in a separate document. (TO-DO: add report that describes harmonization)
LD matrices were computed from a 10% subset of Neale lab subjects. Subjects were matched using the plate and well information from genotyping. We included only biallelic variants with MAF>0.01 in the original Neale Lab GWAS. (TO-DO: add more details [number of subjects, variants, etc])
Weight QC
TO-DO: add enhanced QC reporting (total number of weights, why each variant was missing for all genes)
qclist_all <- list()
qc_files <- paste0(results_dir, "/", list.files(results_dir, pattern="exprqc.Rd"))
for (i in 1:length(qc_files)){
load(qc_files[i])
chr <- unlist(strsplit(rev(unlist(strsplit(qc_files[i], "_")))[1], "[.]"))[1]
qclist_all[[chr]] <- cbind(do.call(rbind, lapply(qclist,unlist)), as.numeric(substring(chr,4)))
}
qclist_all <- data.frame(do.call(rbind, qclist_all))
colnames(qclist_all)[ncol(qclist_all)] <- "chr"
rm(qclist, wgtlist, z_gene_chr)
#number of imputed weights
nrow(qclist_all)[1] 10901#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
1070 768 652 417 494 611 548 408 405 434 634 629 195 365 354
16 17 18 19 20 21 22
526 663 160 859 306 114 289 #proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.8366205Load ctwas results
#load ctwas results
ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#make unique identifier for regions
ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
#compute PVE for each gene/SNP
ctwas_res$PVE = ctwas_res$susie_pip*ctwas_res$mu2/sample_size #check PVE calculation
#separate gene and SNP results
ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
ctwas_gene_res <- data.frame(ctwas_gene_res)
ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
ctwas_snp_res <- data.frame(ctwas_snp_res)
#add gene information to results
sqlite <- RSQLite::dbDriver("SQLite")
db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
query <- function(...) RSQLite::dbGetQuery(db, ...)
gene_info <- query("select gene, genename from extra")
gene_info <- query("select gene, genename, gene_type from extra")
RSQLite::dbDisconnect(db)
ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#add z score to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd")) #for new version, stored after harmonization
z_snp <- readRDS(paste0(results_dir, "/", trait_id, ".RDS")) #for old version, unharmonized
z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,] #subset snps to those included in analysis, note some are duplicated, need to match which allele was used
ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)] #for duplicated snps, this arbitrarily uses the first allele
ctwas_snp_res$z_flag <- as.numeric(ctwas_snp_res$id %in% z_snp$id[duplicated(z_snp$id)]) #mark the unclear z scores, flag=1
#formatting and rounding for tables
ctwas_gene_res$z <- round(ctwas_gene_res$z,2)
ctwas_snp_res$z <- round(ctwas_snp_res$z,2)
ctwas_gene_res$susie_pip <- round(ctwas_gene_res$susie_pip,3)
ctwas_snp_res$susie_pip <- round(ctwas_snp_res$susie_pip,3)
ctwas_gene_res$mu2 <- round(ctwas_gene_res$mu2,2)
ctwas_snp_res$mu2 <- round(ctwas_snp_res$mu2,2)
ctwas_gene_res$PVE <- signif(ctwas_gene_res$PVE, 2)
ctwas_snp_res$PVE <- signif(ctwas_snp_res$PVE, 2)
#merge gene and snp results with added information
ctwas_gene_res$z_flag=NA
ctwas_snp_res$genename=NA
ctwas_snp_res$gene_type=NA
ctwas_res <- rbind(ctwas_gene_res,
ctwas_snp_res[,colnames(ctwas_gene_res)])
#store columns to report
report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos"))]
first_cols <- c("genename", "region_tag")
report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])
report_cols_snps <- c("id", report_cols[-1])
#get number of SNPs from s1 results; adjust for thin
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)Check convergence of parameters
library(ggplot2)
library(cowplot)
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
df <- data.frame(niter = rep(1:ncol(group_prior_rec), 2),
value = c(group_prior_rec[1,], group_prior_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_rec)))
df$group <- as.factor(df$group)
df$value[df$group=="SNP"] <- df$value[df$group=="SNP"]*thin #adjust parameter to account for thin argument
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Prior mean") +
theme_cowplot()
df <- data.frame(niter = rep(1:ncol(group_prior_var_rec), 2),
value = c(group_prior_var_rec[1,], group_prior_var_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_var_rec)))
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Prior variance") +
theme_cowplot()
plot_grid(p_pi, p_sigma2)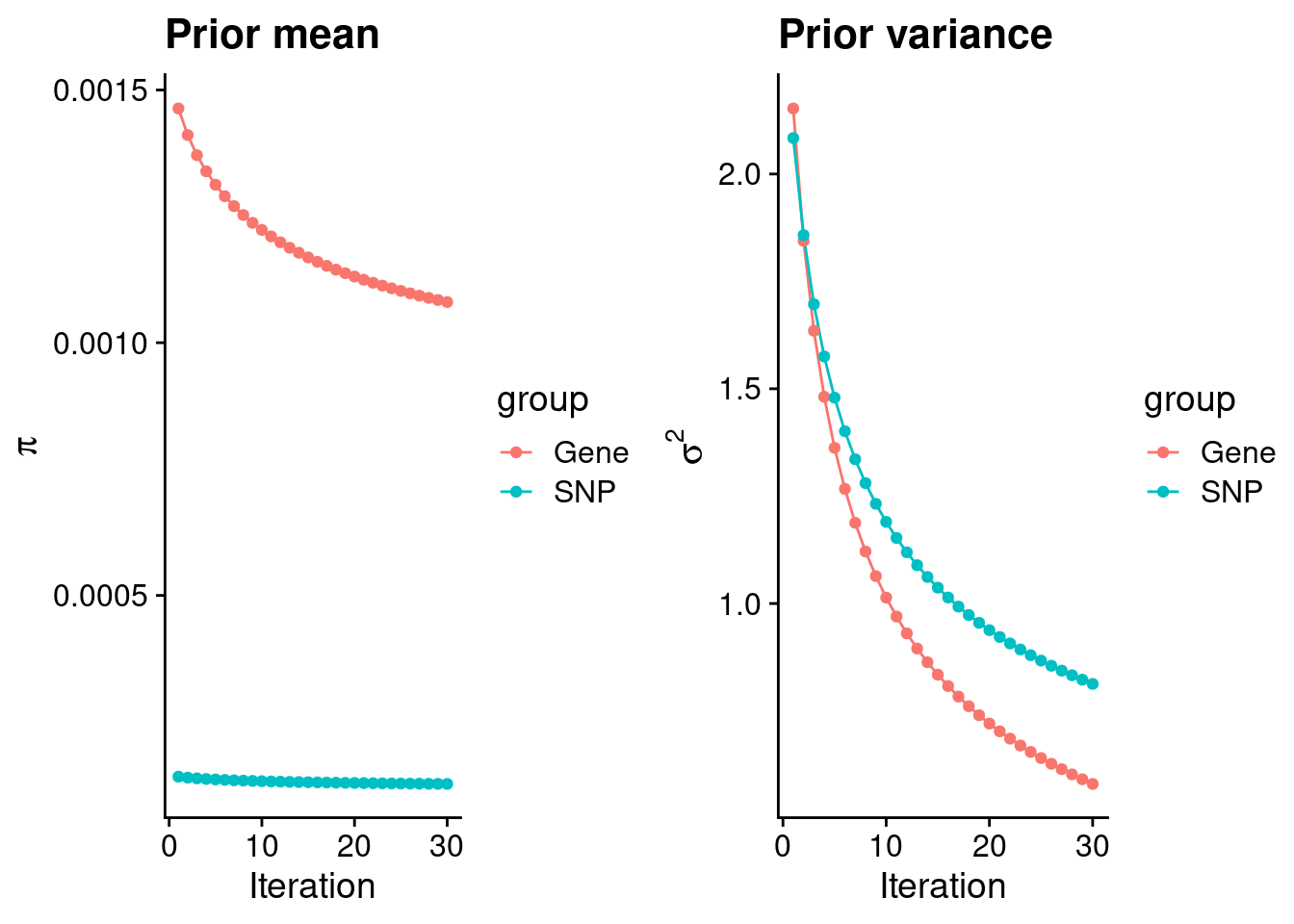
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0010804922 0.0001269809 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
0.5798947 0.8127999 #report sample size
print(sample_size)[1] 30565#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 10901 8697330#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.0002234666 0.0293686203 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.00518315 0.64644319Genes with highest PIPs
#distribution of PIPs
hist(ctwas_gene_res$susie_pip, xlim=c(0,1), main="Distribution of Gene PIPs")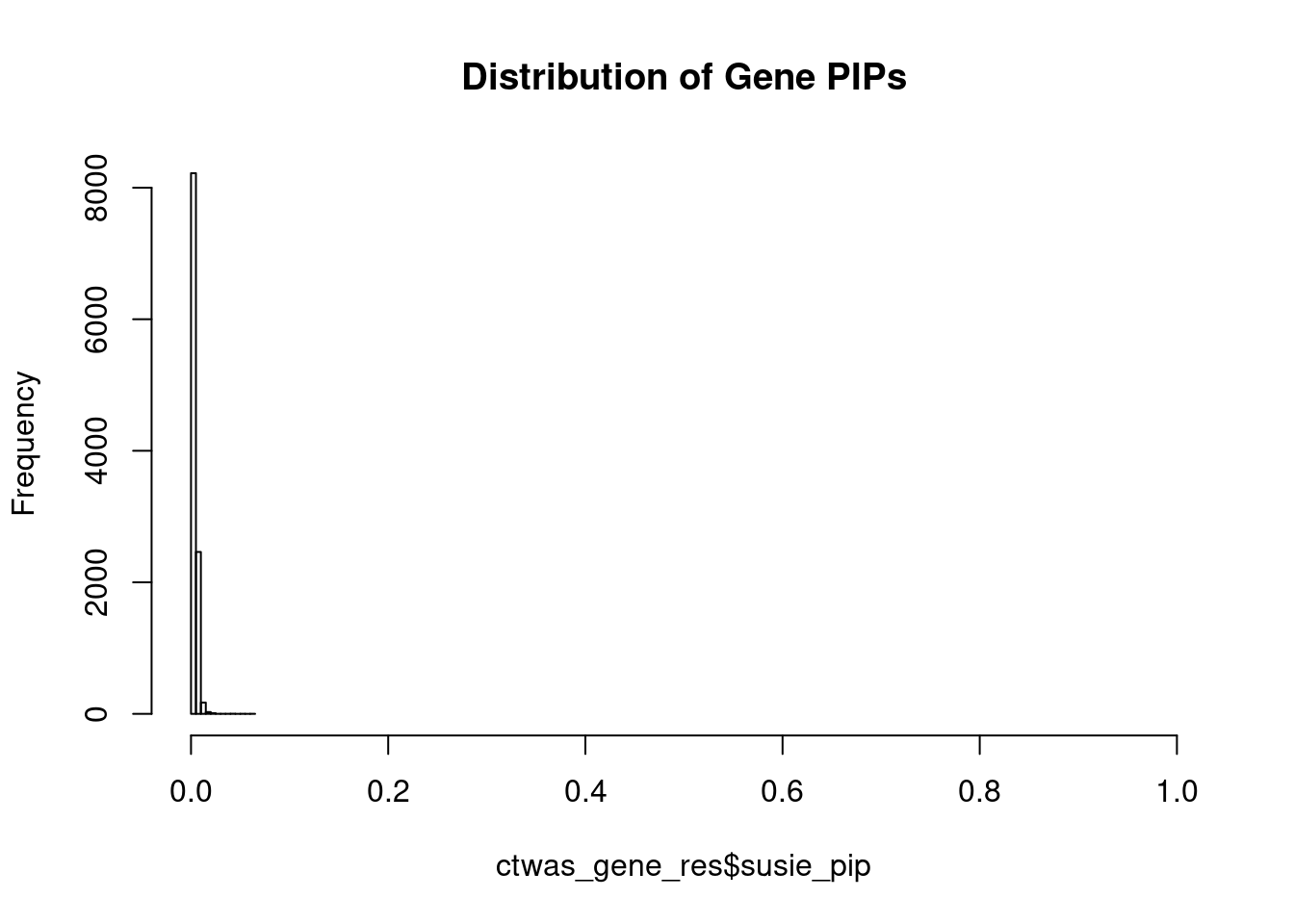
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#genes with PIP>0.8 or 20 highest PIPs
head(ctwas_gene_res[order(-ctwas_gene_res$susie_pip),report_cols], max(sum(ctwas_gene_res$susie_pip>0.8), 20)) genename region_tag susie_pip mu2 PVE z
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
9822 TLR5 1_114 0.040 10.31 1.4e-05 -3.61
6785 PCSK7 11_70 0.034 9.45 1.1e-05 -3.47
11232 GOLGA8N 15_9 0.026 8.51 7.3e-06 -3.27
9533 TMEM173 5_82 0.024 7.92 6.3e-06 -3.12
2669 HECA 6_92 0.023 8.47 6.3e-06 3.39
1299 COMT 22_4 0.023 9.25 6.8e-06 3.41
10471 ARHGAP11A 15_9 0.022 7.83 5.6e-06 3.11
8253 NDUFA3 19_37 0.022 7.90 5.7e-06 -3.08
8438 FGGY 1_37 0.021 7.66 5.3e-06 -3.19
9536 KCNH7 2_98 0.021 8.14 5.7e-06 -3.55
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
3238 HDHD3 9_58 0.021 8.11 5.4e-06 3.23
5249 ZCCHC14 16_52 0.021 7.68 5.3e-06 3.00
5368 SIK1 21_22 0.021 7.88 5.5e-06 -3.40
11299 AC007879.2 2_122 0.020 7.14 4.6e-06 2.95
875 CAMSAP3 19_7 0.020 7.63 5.0e-06 2.99Genes with largest effect sizes
#plot PIP vs effect size
plot(ctwas_gene_res$susie_pip, ctwas_gene_res$mu2, xlab="PIP", ylab="mu^2", main="Gene PIPs vs Effect Size")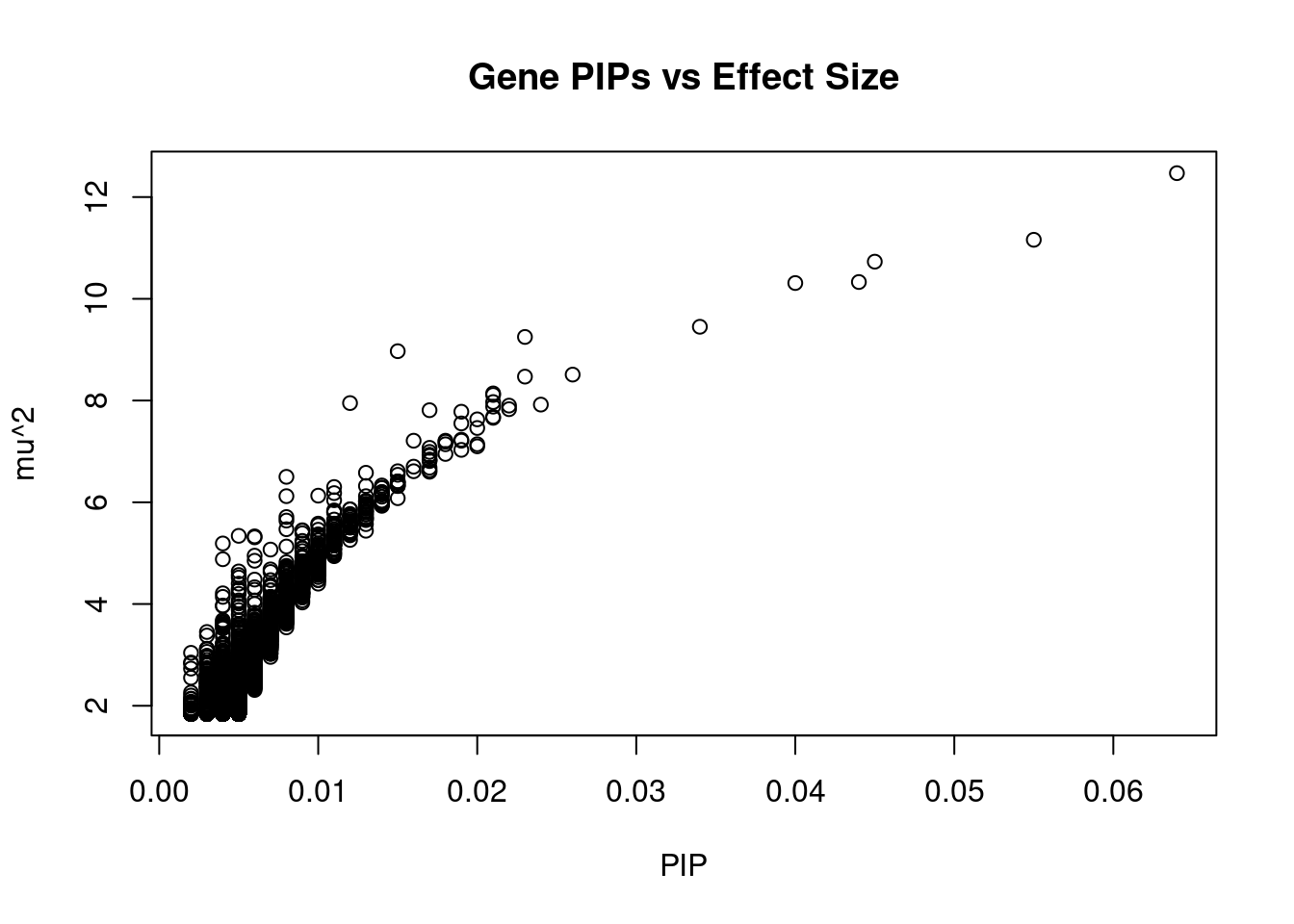
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#genes with 20 largest effect sizes
head(ctwas_gene_res[order(-ctwas_gene_res$mu2),report_cols],20) genename region_tag susie_pip mu2 PVE z
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
9822 TLR5 1_114 0.040 10.31 1.4e-05 -3.61
6785 PCSK7 11_70 0.034 9.45 1.1e-05 -3.47
1299 COMT 22_4 0.023 9.25 6.8e-06 3.41
10600 PBX2 6_26 0.015 8.97 4.5e-06 3.68
11232 GOLGA8N 15_9 0.026 8.51 7.3e-06 -3.27
2669 HECA 6_92 0.023 8.47 6.3e-06 3.39
9536 KCNH7 2_98 0.021 8.14 5.7e-06 -3.55
3238 HDHD3 9_58 0.021 8.11 5.4e-06 3.23
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
10645 PSORS1C2 6_25 0.012 7.95 3.2e-06 -3.42
9533 TMEM173 5_82 0.024 7.92 6.3e-06 -3.12
8253 NDUFA3 19_37 0.022 7.90 5.7e-06 -3.08
5368 SIK1 21_22 0.021 7.88 5.5e-06 -3.40
10471 ARHGAP11A 15_9 0.022 7.83 5.6e-06 3.11
6417 RBM45 2_108 0.017 7.81 4.4e-06 -3.32
4766 SLC31A1 9_58 0.019 7.78 4.8e-06 -3.28Genes with highest PVE
#genes with 20 highest pve
head(ctwas_gene_res[order(-ctwas_gene_res$PVE),report_cols],20) genename region_tag susie_pip mu2 PVE z
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
9822 TLR5 1_114 0.040 10.31 1.4e-05 -3.61
6785 PCSK7 11_70 0.034 9.45 1.1e-05 -3.47
11232 GOLGA8N 15_9 0.026 8.51 7.3e-06 -3.27
1299 COMT 22_4 0.023 9.25 6.8e-06 3.41
9533 TMEM173 5_82 0.024 7.92 6.3e-06 -3.12
2669 HECA 6_92 0.023 8.47 6.3e-06 3.39
9536 KCNH7 2_98 0.021 8.14 5.7e-06 -3.55
8253 NDUFA3 19_37 0.022 7.90 5.7e-06 -3.08
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
10471 ARHGAP11A 15_9 0.022 7.83 5.6e-06 3.11
5368 SIK1 21_22 0.021 7.88 5.5e-06 -3.40
3238 HDHD3 9_58 0.021 8.11 5.4e-06 3.23
8438 FGGY 1_37 0.021 7.66 5.3e-06 -3.19
5249 ZCCHC14 16_52 0.021 7.68 5.3e-06 3.00
875 CAMSAP3 19_7 0.020 7.63 5.0e-06 2.99
4766 SLC31A1 9_58 0.019 7.78 4.8e-06 -3.28Genes with largest z scores
#genes with 20 largest z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
10600 PBX2 6_26 0.015 8.97 4.5e-06 3.68
9822 TLR5 1_114 0.040 10.31 1.4e-05 -3.61
9536 KCNH7 2_98 0.021 8.14 5.7e-06 -3.55
6785 PCSK7 11_70 0.034 9.45 1.1e-05 -3.47
10645 PSORS1C2 6_25 0.012 7.95 3.2e-06 -3.42
1299 COMT 22_4 0.023 9.25 6.8e-06 3.41
5368 SIK1 21_22 0.021 7.88 5.5e-06 -3.40
2669 HECA 6_92 0.023 8.47 6.3e-06 3.39
6417 RBM45 2_108 0.017 7.81 4.4e-06 -3.32
4766 SLC31A1 9_58 0.019 7.78 4.8e-06 -3.28
11232 GOLGA8N 15_9 0.026 8.51 7.3e-06 -3.27
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
3238 HDHD3 9_58 0.021 8.11 5.4e-06 3.23
8438 FGGY 1_37 0.021 7.66 5.3e-06 -3.19
9533 TMEM173 5_82 0.024 7.92 6.3e-06 -3.12
10471 ARHGAP11A 15_9 0.022 7.83 5.6e-06 3.11Comparing z scores and PIPs
#set nominal signifiance threshold for z scores
alpha <- 0.05
#bonferroni adjusted threshold for z scores
sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#Q-Q plot for z scores
obs_z <- ctwas_gene_res$z[order(ctwas_gene_res$z)]
exp_z <- qnorm((1:nrow(ctwas_gene_res))/nrow(ctwas_gene_res))
plot(exp_z, obs_z, xlab="Expected z", ylab="Observed z", main="Gene z score Q-Q plot")
abline(a=0,b=1)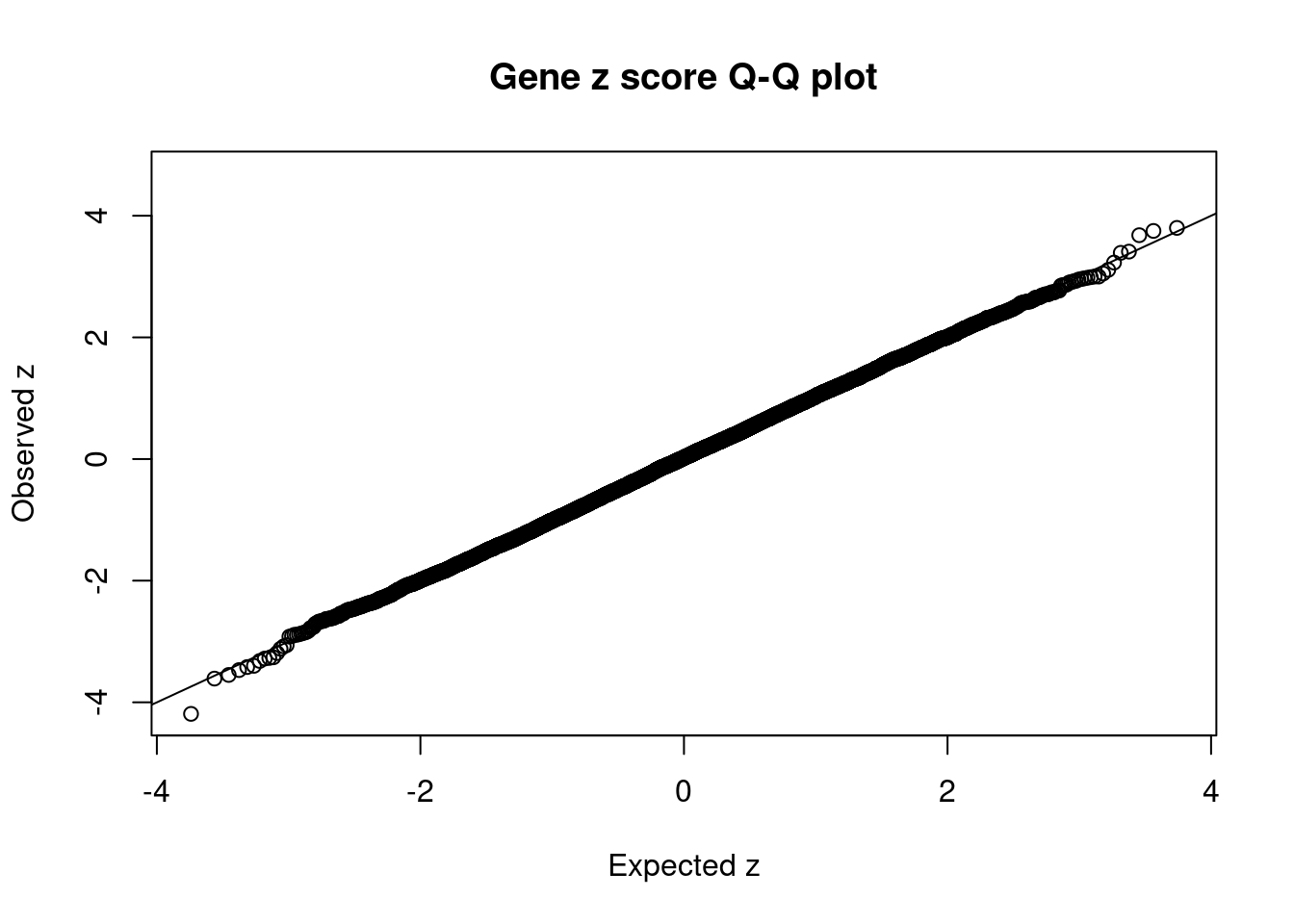
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#plot z score vs PIP
plot(abs(ctwas_gene_res$z), ctwas_gene_res$susie_pip, xlab="abs(z)", ylab="PIP")
abline(v=sig_thresh, col="red", lty=2)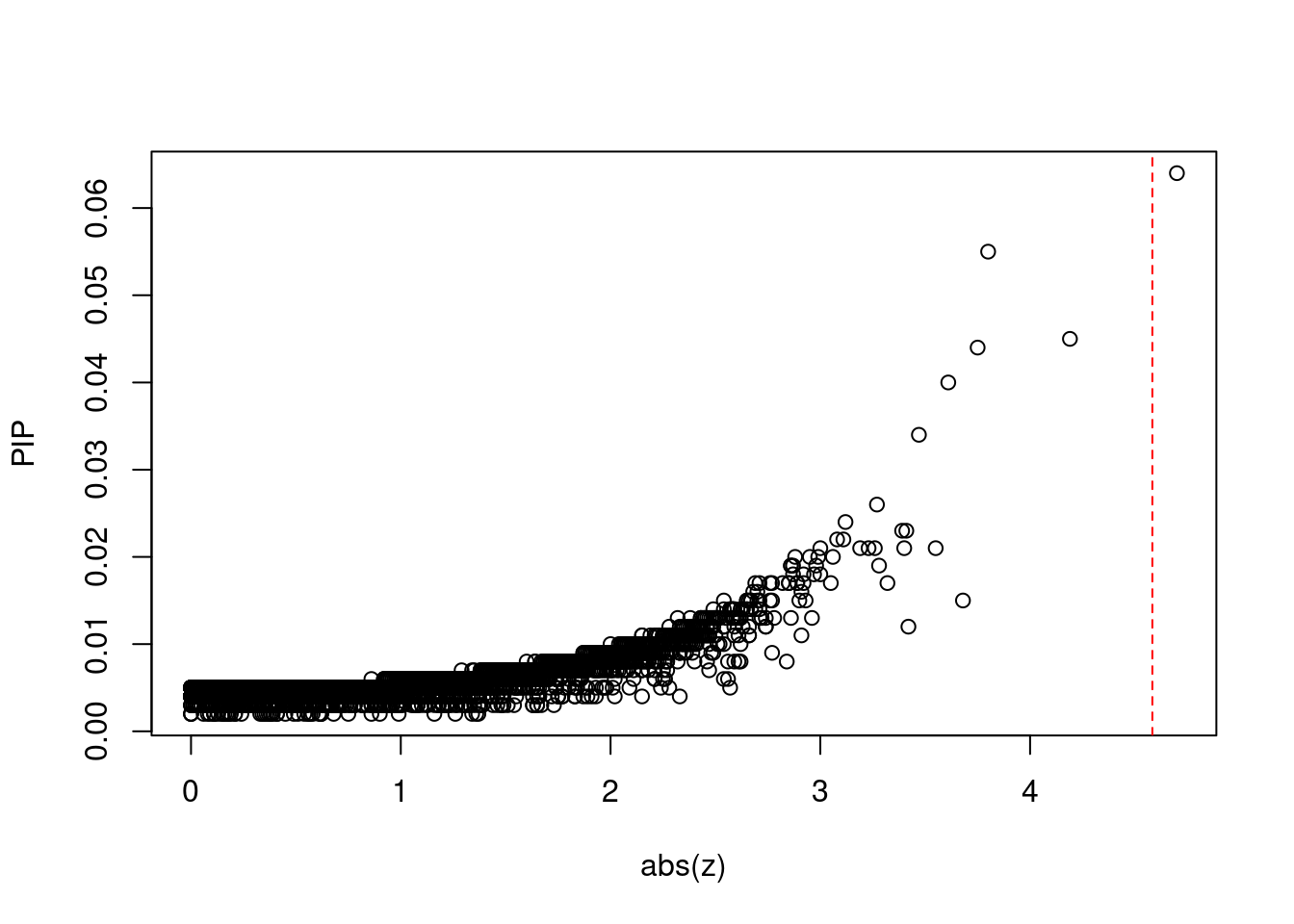
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#proportion of significant z scores
mean(abs(ctwas_gene_res$z) > sig_thresh)[1] 9.17347e-05#genes with most significant z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
10600 PBX2 6_26 0.015 8.97 4.5e-06 3.68
9822 TLR5 1_114 0.040 10.31 1.4e-05 -3.61
9536 KCNH7 2_98 0.021 8.14 5.7e-06 -3.55
6785 PCSK7 11_70 0.034 9.45 1.1e-05 -3.47
10645 PSORS1C2 6_25 0.012 7.95 3.2e-06 -3.42
1299 COMT 22_4 0.023 9.25 6.8e-06 3.41
5368 SIK1 21_22 0.021 7.88 5.5e-06 -3.40
2669 HECA 6_92 0.023 8.47 6.3e-06 3.39
6417 RBM45 2_108 0.017 7.81 4.4e-06 -3.32
4766 SLC31A1 9_58 0.019 7.78 4.8e-06 -3.28
11232 GOLGA8N 15_9 0.026 8.51 7.3e-06 -3.27
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
3238 HDHD3 9_58 0.021 8.11 5.4e-06 3.23
8438 FGGY 1_37 0.021 7.66 5.3e-06 -3.19
9533 TMEM173 5_82 0.024 7.92 6.3e-06 -3.12
10471 ARHGAP11A 15_9 0.022 7.83 5.6e-06 3.11Locus plots for genes and SNPs
ctwas_gene_res_sortz <- ctwas_gene_res[order(-abs(ctwas_gene_res$z)),]
n_plots <- 5
for (region_tag_plot in head(unique(ctwas_gene_res_sortz$region_tag), n_plots)){
ctwas_res_region <- ctwas_res[ctwas_res$region_tag==region_tag_plot,]
start <- min(ctwas_res_region$pos)
end <- max(ctwas_res_region$pos)
ctwas_res_region <- ctwas_res_region[order(ctwas_res_region$pos),]
ctwas_res_region_gene <- ctwas_res_region[ctwas_res_region$type=="gene",]
ctwas_res_region_snp <- ctwas_res_region[ctwas_res_region$type=="SNP",]
#region name
print(paste0("Region: ", region_tag_plot))
#table of genes in region
print(ctwas_res_region_gene[,report_cols])
par(mfrow=c(4,1))
#gene z scores
plot(ctwas_res_region_gene$pos, abs(ctwas_res_region_gene$z), xlab="Position", ylab="abs(gene_z)", xlim=c(start,end),
ylim=c(0,max(sig_thresh, abs(ctwas_res_region_gene$z))),
main=paste0("Region: ", region_tag_plot))
abline(h=sig_thresh,col="red",lty=2)
#significance threshold for SNPs
alpha_snp <- 5*10^(-8)
sig_thresh_snp <- qnorm(1-alpha_snp/2, lower=T)
#snp z scores
plot(ctwas_res_region_snp$pos, abs(ctwas_res_region_snp$z), xlab="Position", ylab="abs(snp_z)",xlim=c(start,end),
ylim=c(0,max(sig_thresh_snp, max(abs(ctwas_res_region_snp$z)))))
abline(h=sig_thresh_snp,col="purple",lty=2)
#gene pips
plot(ctwas_res_region_gene$pos, ctwas_res_region_gene$susie_pip, xlab="Position", ylab="Gene PIP", xlim=c(start,end), ylim=c(0,1))
abline(h=0.8,col="blue",lty=2)
#snp pips
plot(ctwas_res_region_snp$pos, ctwas_res_region_snp$susie_pip, xlab="Position", ylab="SNP PIP", xlim=c(start,end), ylim=c(0,1))
abline(h=0.8,col="blue",lty=2)
}[1] "Region: 19_14"
genename region_tag susie_pip mu2 PVE z
3865 KLF2 19_14 0.004 2.51 3.6e-07 -1.03
3864 EPS15L1 19_14 0.004 1.84 2.2e-07 -0.01
1970 C19orf44 19_14 0.004 1.87 2.2e-07 0.23
1087 CHERP 19_14 0.004 2.39 3.3e-07 0.94
12176 CTD-3222D19.12 19_14 0.004 2.39 3.3e-07 0.94
3863 SLC35E1 19_14 0.004 2.01 2.5e-07 -0.51
12158 CTC-429P9.2 19_14 0.005 2.69 4.0e-07 1.16
10885 SMIM7 19_14 0.009 5.19 1.5e-06 2.33
779 TMEM38A 19_14 0.008 4.69 1.2e-06 -2.10
3866 F2RL3 19_14 0.005 2.99 4.9e-07 1.32
6732 CPAMD8 19_14 0.004 1.84 2.2e-07 0.10
4167 HAUS8 19_14 0.006 3.40 6.2e-07 1.51
1362 MYO9B 19_14 0.004 1.89 2.3e-07 -0.31
452 USE1 19_14 0.009 5.04 1.4e-06 -2.25
1361 OCEL1 19_14 0.008 4.75 1.2e-06 2.18
6733 ANKLE1 19_14 0.004 2.24 3.0e-07 0.83
4064 MRPL34 19_14 0.011 5.80 2.0e-06 -2.59
4063 DDA1 19_14 0.005 2.66 4.0e-07 1.18
3842 ABHD8 19_14 0.004 2.42 3.4e-07 -1.01
4057 GTPBP3 19_14 0.004 1.97 2.4e-07 -0.44
821 ANO8 19_14 0.004 1.84 2.2e-07 0.11
4058 PLVAP 19_14 0.004 1.87 2.2e-07 -0.20
4059 BST2 19_14 0.005 2.88 4.6e-07 -1.27
12691 BISPR 19_14 0.004 2.07 2.6e-07 0.58
5354 MVB12A 19_14 0.004 1.93 2.4e-07 0.39
9870 TMEM221 19_14 0.004 2.30 3.1e-07 0.84
7769 FAM129C 19_14 0.004 2.46 3.5e-07 -0.97
4060 SLC27A1 19_14 0.004 2.33 3.2e-07 -0.87
4065 PGLS 19_14 0.004 2.13 2.7e-07 -0.64
4062 COLGALT1 19_14 0.004 1.86 2.2e-07 0.19
4078 FCHO1 19_14 0.004 2.11 2.7e-07 0.65
9121 B3GNT3 19_14 0.004 1.85 2.2e-07 0.14
11583 INSL3 19_14 0.004 2.17 2.8e-07 -0.71
2053 JAK3 19_14 0.004 2.50 3.6e-07 -0.99
2054 ARRDC2 19_14 0.004 1.93 2.4e-07 0.46
1346 IL12RB1 19_14 0.004 2.20 2.9e-07 -0.82
1359 MAST3 19_14 0.004 1.94 2.4e-07 0.55
2055 PIK3R2 19_14 0.004 2.16 2.8e-07 -0.86
10964 IFI30 19_14 0.064 12.47 2.6e-05 4.70
11755 MPV17L2 19_14 0.011 6.04 2.2e-06 2.91
2056 RAB3A 19_14 0.006 3.54 6.7e-07 -1.63
2057 PDE4C 19_14 0.004 1.84 2.2e-07 -0.17
4084 KIAA1683 19_14 0.004 2.51 3.6e-07 -0.96
4085 JUND 19_14 0.006 3.48 6.5e-07 -1.47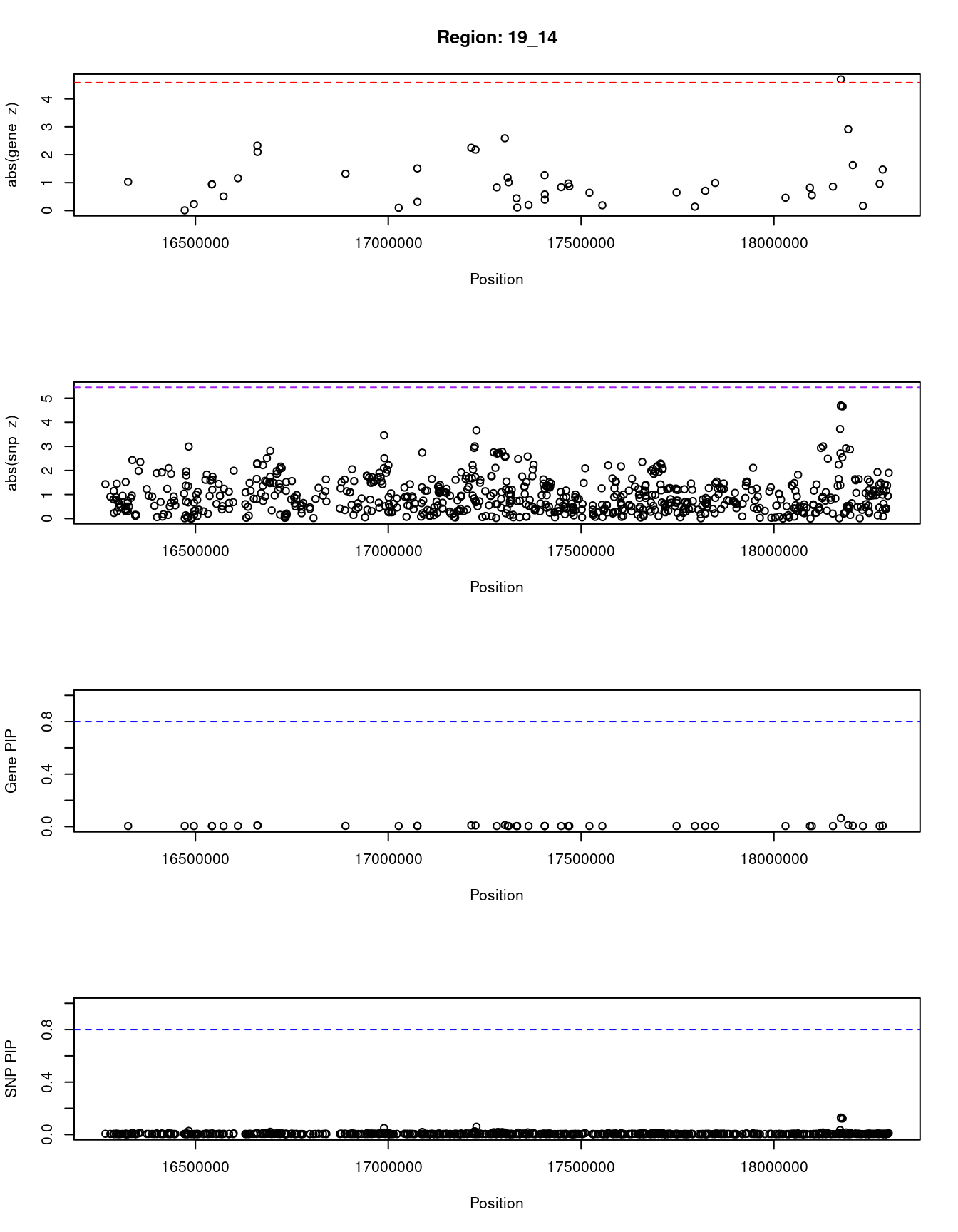
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
[1] "Region: 6_20"
genename region_tag susie_pip mu2 PVE z
5755 SLC17A4 6_20 0.004 1.98 2.7e-07 0.46
3641 SLC17A1 6_20 0.004 1.91 2.6e-07 -0.38
3640 SLC17A3 6_20 0.007 3.88 9.0e-07 -2.00
2660 SLC17A2 6_20 0.021 7.97 5.6e-06 -3.26
2662 TRIM38 6_20 0.045 10.73 1.6e-05 -4.19
12334 U91328.19 6_20 0.004 2.12 3.0e-07 -0.69
12379 U91328.22 6_20 0.004 1.89 2.5e-07 -0.18
9850 HIST1H1C 6_20 0.004 1.86 2.5e-07 0.10
167 HFE 6_20 0.004 1.99 2.7e-07 -0.53
9169 HIST1H2AC 6_20 0.005 2.87 5.0e-07 -1.57
6602 HIST1H2BD 6_20 0.006 3.43 7.0e-07 -1.86
12482 HIST1H2BE 6_20 0.004 1.91 2.6e-07 -0.36
12602 HIST1H2BF 6_20 0.004 1.84 2.4e-07 -0.15
12502 HIST1H3E 6_20 0.004 1.98 2.7e-07 -0.52
12544 HIST1H2BH 6_20 0.005 2.48 3.9e-07 0.93
6604 HIST1H4H 6_20 0.004 1.86 2.5e-07 -0.05
9736 BTN3A2 6_20 0.005 2.69 4.5e-07 -1.65
3635 BTN2A2 6_20 0.004 1.85 2.4e-07 -0.12
298 BTN3A1 6_20 0.004 2.13 3.0e-07 0.59
2597 BTN3A3 6_20 0.004 1.86 2.5e-07 0.26
2702 BTN2A1 6_20 0.004 2.02 2.8e-07 -0.61
9373 HMGN4 6_20 0.005 2.28 3.4e-07 -0.78
5765 ABT1 6_20 0.005 2.31 3.5e-07 -0.92
9231 ZNF322 6_20 0.005 2.75 4.7e-07 1.30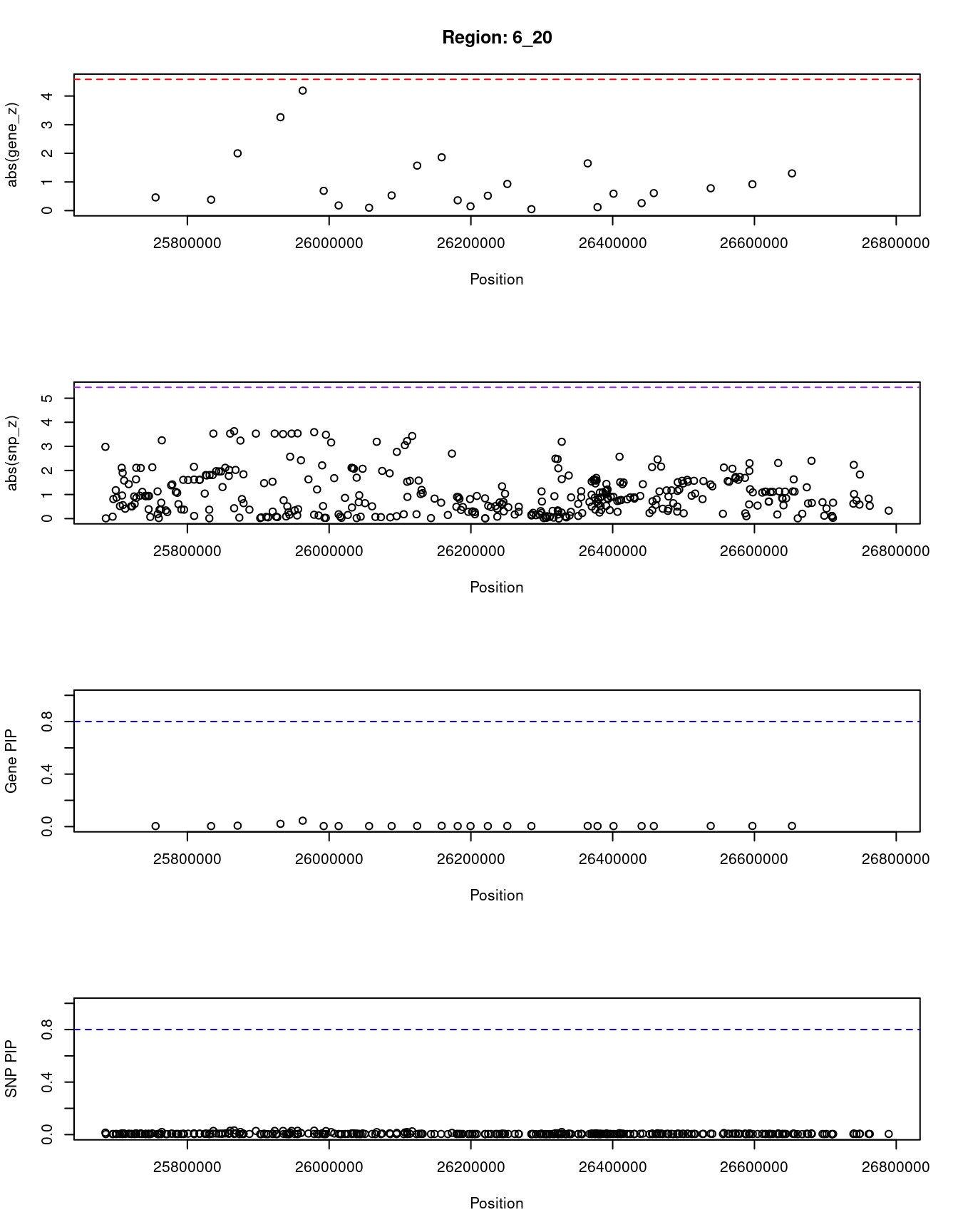
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
[1] "Region: 17_29"
genename region_tag susie_pip mu2 PVE z
563 NGFR 17_29 0.005 2.05 3.1e-07 0.57
3384 SPOP 17_29 0.004 1.89 2.8e-07 0.29
3386 SLC35B1 17_29 0.004 1.88 2.7e-07 0.25
69 PDK2 17_29 0.005 2.03 3.1e-07 0.55
2352 PPP1R9B 17_29 0.007 3.29 7.0e-07 -1.47
2354 SGCA 17_29 0.004 1.84 2.7e-07 -0.09
7721 TMEM92 17_29 0.055 11.16 2.0e-05 3.80
234 XYLT2 17_29 0.004 1.84 2.7e-07 0.06
2355 MRPL27 17_29 0.004 1.86 2.7e-07 0.21
7723 ACSF2 17_29 0.005 2.02 3.1e-07 0.54
4724 CHAD 17_29 0.004 1.85 2.7e-07 0.16
12555 RP11-94C24.13 17_29 0.005 1.91 2.8e-07 0.34
4720 RSAD1 17_29 0.008 4.06 1.1e-06 1.88
420 EPN3 17_29 0.007 3.31 7.2e-07 -1.52
82 SPATA20 17_29 0.005 2.00 3.0e-07 -0.53
2359 ABCC3 17_29 0.008 3.85 9.6e-07 1.73
6377 ANKRD40 17_29 0.005 2.26 3.7e-07 -0.81
5278 TOB1 17_29 0.006 3.17 6.6e-07 -1.45
12017 RP11-700H6.4 17_29 0.007 3.73 9.0e-07 -1.71
11567 RP11-700H6.1 17_29 0.004 1.86 2.7e-07 -0.22
131 SPAG9 17_29 0.004 1.86 2.7e-07 -0.19
11398 NME1 17_29 0.006 2.97 5.8e-07 1.33
11504 NME2 17_29 0.008 4.15 1.1e-06 -1.88
181 UTP18 17_29 0.005 2.00 3.0e-07 -0.49
180 MBTD1 17_29 0.004 1.84 2.7e-07 -0.03
12127 LINC02073 17_29 0.005 2.03 3.1e-07 -0.56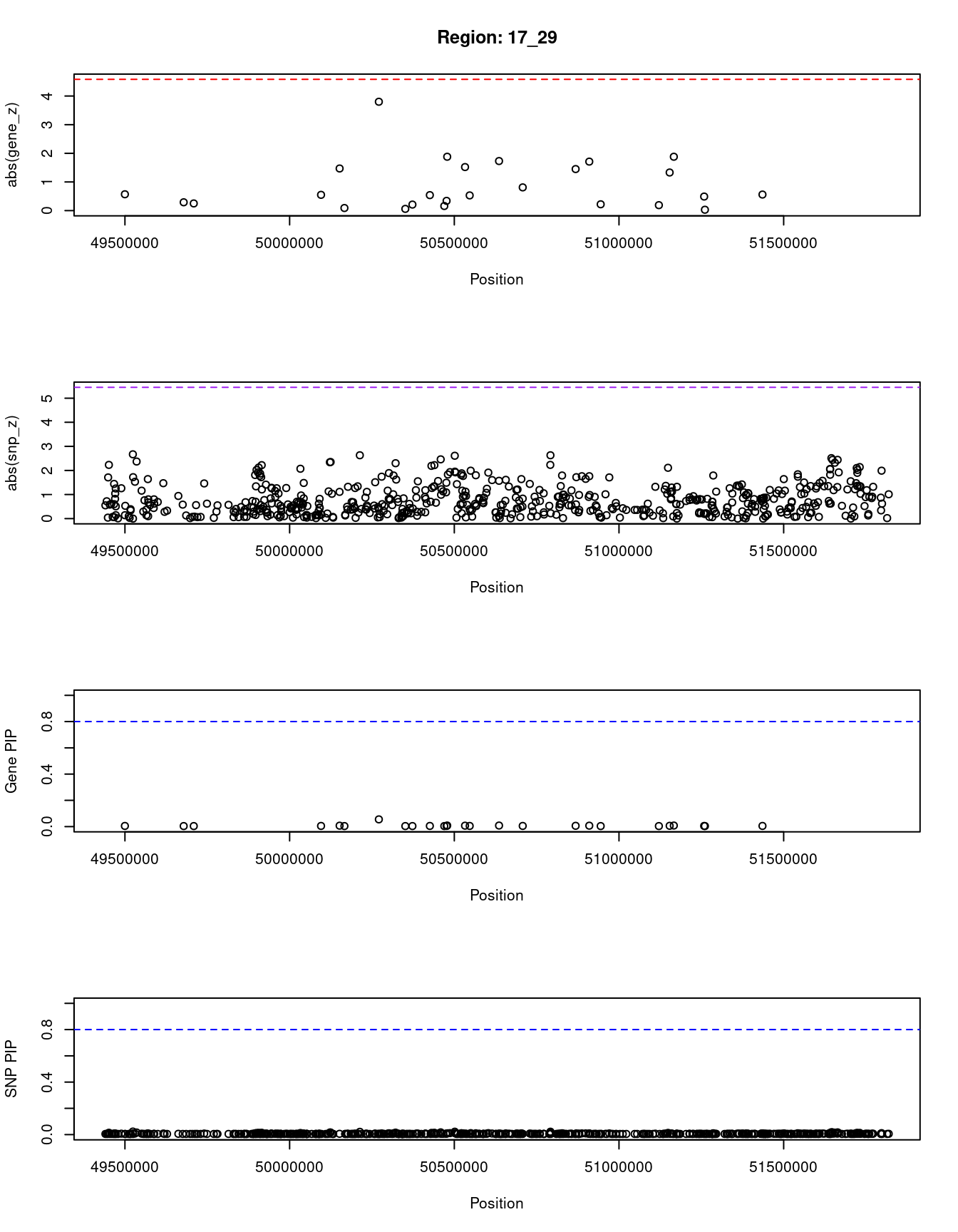
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
[1] "Region: 18_12"
genename region_tag susie_pip mu2 PVE z
4477 CABLES1 18_12 0.004 1.84 2.6e-07 -0.04
4476 TMEM241 18_12 0.004 1.85 2.7e-07 0.15
1708 RIOK3 18_12 0.044 10.33 1.5e-05 3.75
5304 C18orf8 18_12 0.006 3.25 6.8e-07 -1.47
5306 NPC1 18_12 0.004 1.84 2.6e-07 0.04
454 LAMA3 18_12 0.005 2.02 3.1e-07 0.53
7914 TTC39C 18_12 0.008 4.18 1.1e-06 -1.87
6311 CABYR 18_12 0.005 2.23 3.6e-07 -0.78
12078 RP11-799B12.4 18_12 0.005 2.38 4.0e-07 -0.90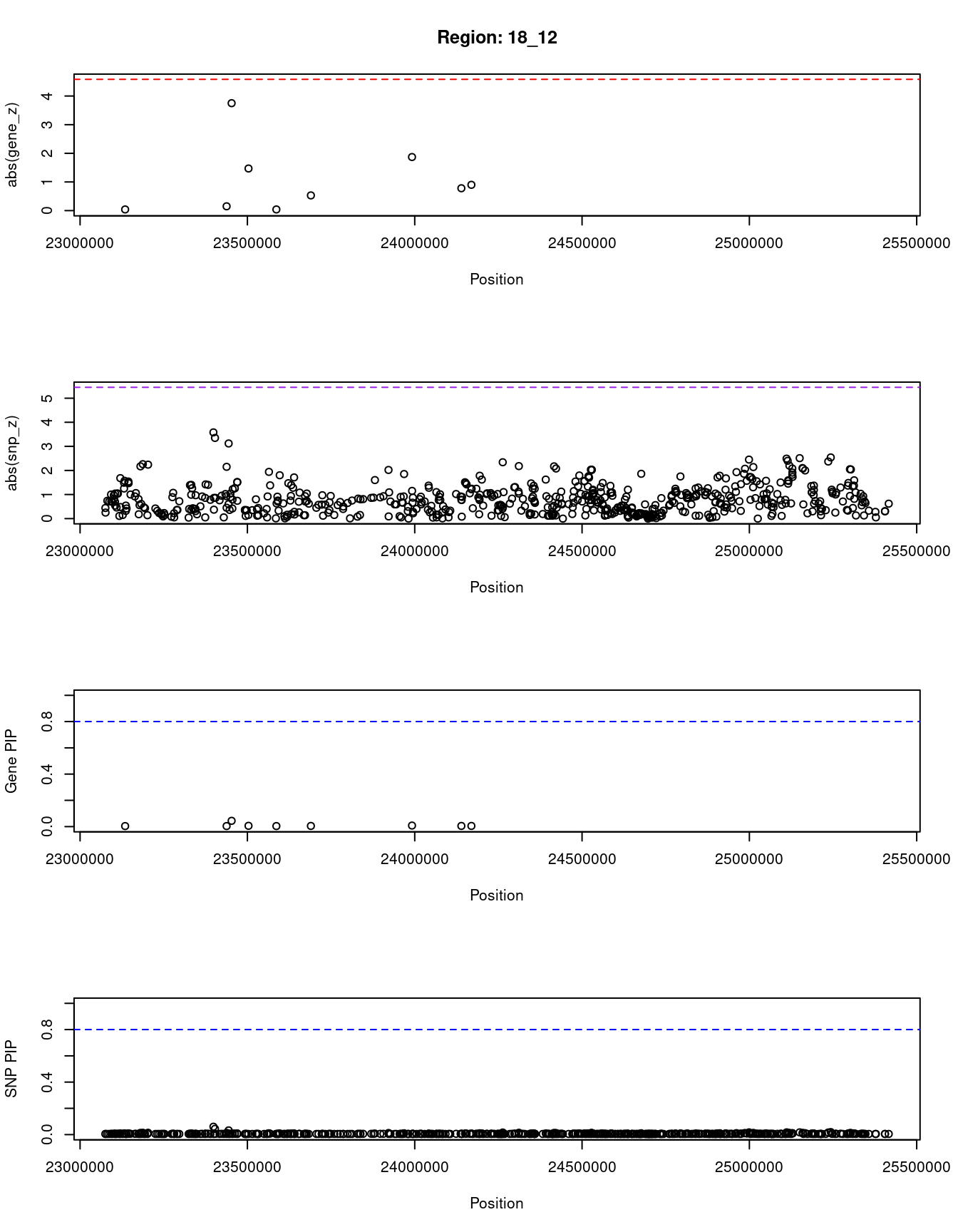
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
[1] "Region: 6_26"
genename region_tag susie_pip mu2 PVE z
10632 BAG6 6_26 0.003 2.36 1.9e-07 -0.81
10634 AIF1 6_26 0.003 3.38 3.7e-07 1.65
10633 PRRC2A 6_26 0.002 1.84 1.3e-07 0.18
10631 APOM 6_26 0.004 4.21 5.7e-07 2.02
10630 C6orf47 6_26 0.004 3.65 4.3e-07 1.77
10629 CSNK2B 6_26 0.002 1.86 1.3e-07 -0.19
11414 LY6G5B 6_26 0.006 5.31 9.8e-07 2.54
10628 LY6G5C 6_26 0.006 5.33 9.9e-07 2.56
10627 ABHD16A 6_26 0.003 3.11 3.1e-07 1.67
10626 MPIG6B 6_26 0.002 1.88 1.4e-07 -0.38
10849 DDAH2 6_26 0.003 2.57 2.2e-07 1.20
10625 MSH5 6_26 0.003 3.12 3.2e-07 -1.54
10848 CLIC1 6_26 0.005 4.57 6.9e-07 -2.24
10623 VWA7 6_26 0.004 3.96 5.0e-07 -1.91
10622 LSM2 6_26 0.003 3.45 3.8e-07 1.63
10621 HSPA1L 6_26 0.002 2.00 1.5e-07 0.54
10619 C6orf48 6_26 0.002 1.85 1.3e-07 -0.11
10618 SLC44A4 6_26 0.003 2.72 2.5e-07 1.20
10616 EHMT2 6_26 0.002 2.02 1.5e-07 -0.57
10612 SKIV2L 6_26 0.002 2.05 1.6e-07 0.49
10610 STK19 6_26 0.002 1.93 1.4e-07 -0.38
10611 DXO 6_26 0.003 2.70 2.4e-07 1.39
11541 C4A 6_26 0.008 6.50 1.7e-06 -2.84
11216 CYP21A2 6_26 0.004 3.63 4.2e-07 -1.83
11038 C4B 6_26 0.005 4.64 7.1e-07 -2.28
10844 ATF6B 6_26 0.003 2.60 2.3e-07 1.27
7949 TNXB 6_26 0.003 2.83 2.6e-07 -1.31
10606 FKBPL 6_26 0.003 2.46 2.1e-07 -1.00
11007 PPT2 6_26 0.002 2.13 1.6e-07 0.75
10605 PRRT1 6_26 0.003 2.59 2.3e-07 -1.29
11441 EGFL8 6_26 0.003 2.81 2.6e-07 1.46
10603 AGPAT1 6_26 0.003 2.60 2.3e-07 1.22
10601 AGER 6_26 0.003 2.85 2.7e-07 1.33
10602 RNF5 6_26 0.003 2.37 2.0e-07 -0.96
10600 PBX2 6_26 0.015 8.97 4.5e-06 3.68
10599 NOTCH4 6_26 0.003 2.80 2.6e-07 -1.24
10597 HLA-DRA 6_26 0.002 2.20 1.7e-07 -0.99
10402 HLA-DRB5 6_26 0.003 2.35 1.9e-07 0.98
10023 HLA-DRB1 6_26 0.003 2.61 2.3e-07 1.18
10137 HLA-DQA1 6_26 0.002 2.08 1.6e-07 0.62
11366 HLA-DQA2 6_26 0.002 1.84 1.3e-07 0.15
9089 HLA-DQB1 6_26 0.003 2.98 2.9e-07 -1.73
11231 HLA-DQB2 6_26 0.003 2.80 2.6e-07 1.63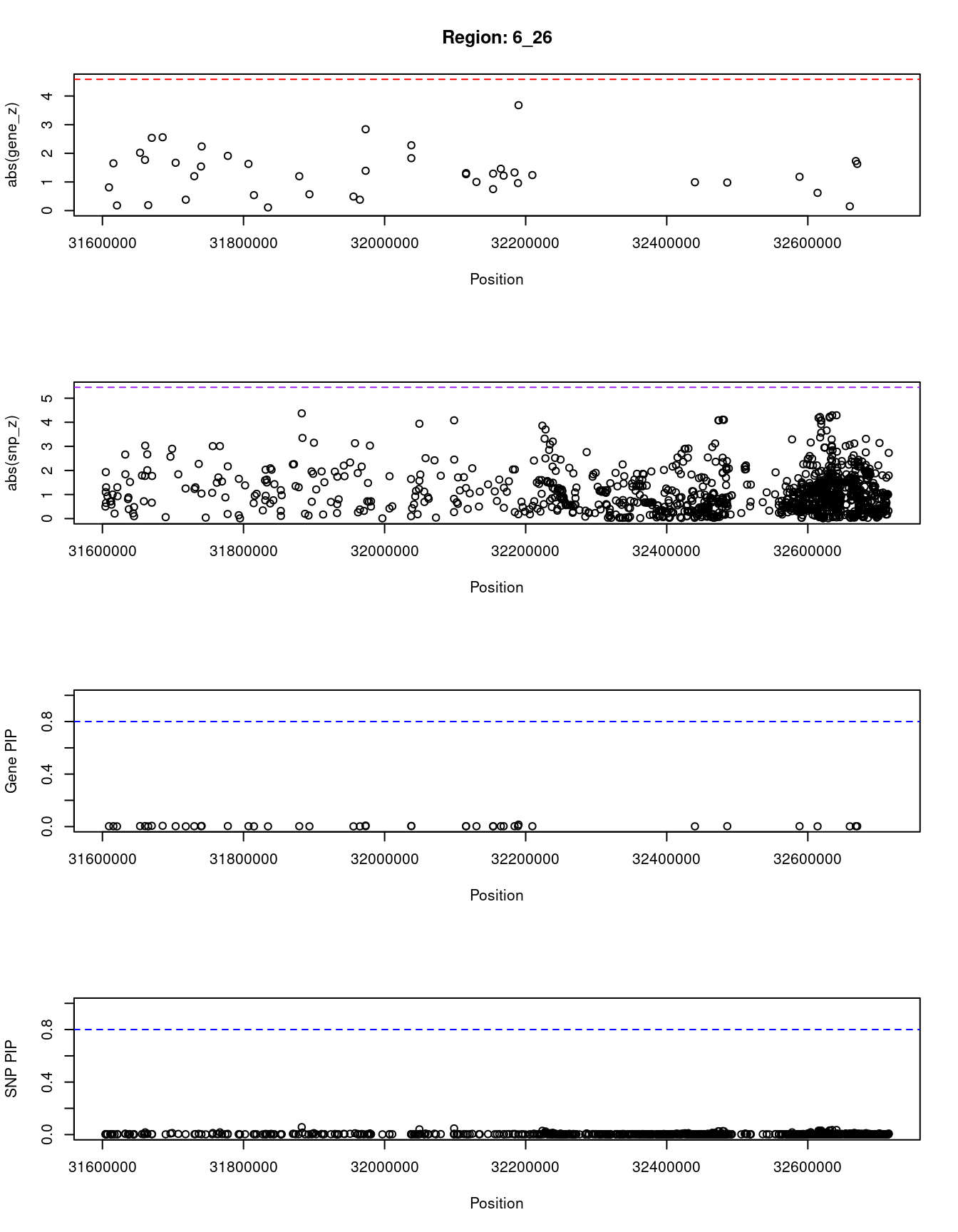
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
SNPs with highest PIPs
#snps with PIP>0.8 or 20 highest PIPs
head(ctwas_snp_res[order(-ctwas_snp_res$susie_pip),report_cols_snps],
max(sum(ctwas_snp_res$susie_pip>0.8), 20)) id region_tag susie_pip mu2 PVE z
832971 rs11557577 20_13 0.234 20.24 1.6e-04 -4.76
221280 rs117111501 4_41 0.213 20.86 1.5e-04 4.73
838805 rs6017234 20_27 0.207 19.88 1.3e-04 -4.60
242736 rs7660796 4_80 0.162 18.50 9.8e-05 4.38
505868 rs75155685 9_51 0.161 18.72 9.9e-05 4.68
607479 rs10772815 12_7 0.145 17.87 8.5e-05 -4.27
608967 rs56198451 12_11 0.138 17.83 8.1e-05 4.45
188130 rs190072323 3_97 0.133 17.41 7.6e-05 4.08
208954 rs35945249 4_18 0.132 17.32 7.5e-05 -4.23
780163 rs1675273 17_44 0.130 17.60 7.5e-05 4.21
813014 rs273265 19_14 0.130 18.12 7.7e-05 -4.70
824212 rs1966573 19_36 0.128 17.88 7.5e-05 -4.20
650317 rs562115589 13_7 0.126 17.54 7.3e-05 -4.20
521053 rs17144212 10_8 0.125 17.25 7.0e-05 -4.14
12749 rs116039993 1_26 0.123 17.03 6.8e-05 4.03
505859 rs118013517 9_51 0.123 17.42 7.0e-05 4.52
813015 rs2241089 19_14 0.123 17.85 7.2e-05 -4.67
813017 rs4808757 19_14 0.123 17.82 7.1e-05 -4.66
3511 rs79089220 1_8 0.121 17.22 6.8e-05 4.08
856603 rs553932 21_22 0.119 17.07 6.6e-05 -4.33SNPs with largest effect sizes
#plot PIP vs effect size
plot(ctwas_snp_res$susie_pip, ctwas_snp_res$mu2, xlab="PIP", ylab="mu^2", main="SNP PIPs vs Effect Size")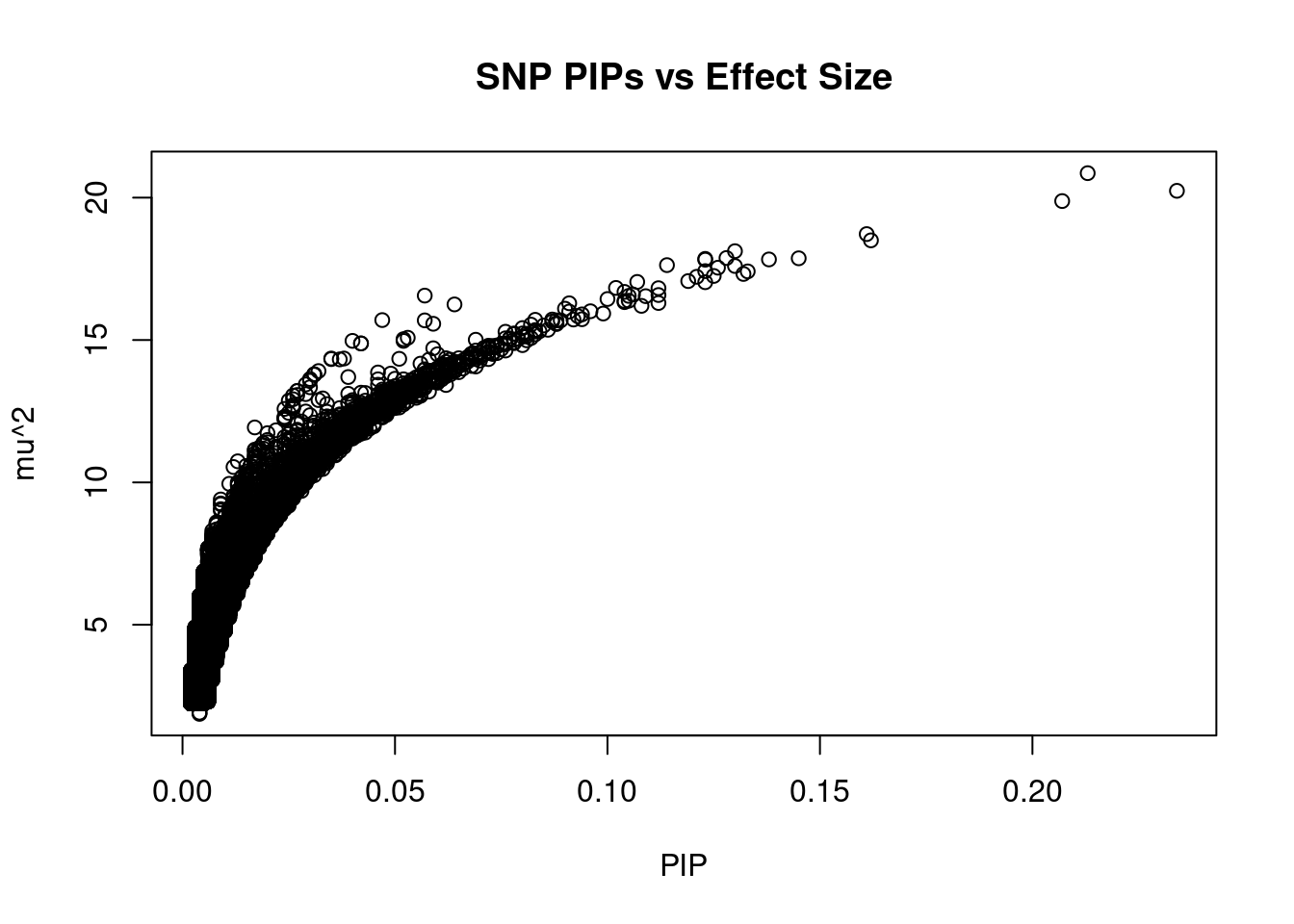
| Version | Author | Date |
|---|---|---|
| dfd2b5f | wesleycrouse | 2021-09-07 |
#SNPs with 50 largest effect sizes
head(ctwas_snp_res[order(-ctwas_snp_res$mu2),report_cols_snps],50) id region_tag susie_pip mu2 PVE z
221280 rs117111501 4_41 0.213 20.86 1.5e-04 4.73
832971 rs11557577 20_13 0.234 20.24 1.6e-04 -4.76
838805 rs6017234 20_27 0.207 19.88 1.3e-04 -4.60
505868 rs75155685 9_51 0.161 18.72 9.9e-05 4.68
242736 rs7660796 4_80 0.162 18.50 9.8e-05 4.38
813014 rs273265 19_14 0.130 18.12 7.7e-05 -4.70
824212 rs1966573 19_36 0.128 17.88 7.5e-05 -4.20
607479 rs10772815 12_7 0.145 17.87 8.5e-05 -4.27
813015 rs2241089 19_14 0.123 17.85 7.2e-05 -4.67
608967 rs56198451 12_11 0.138 17.83 8.1e-05 4.45
813017 rs4808757 19_14 0.123 17.82 7.1e-05 -4.66
527922 rs117340939 10_20 0.114 17.63 6.6e-05 -4.13
780163 rs1675273 17_44 0.130 17.60 7.5e-05 4.21
650317 rs562115589 13_7 0.126 17.54 7.3e-05 -4.20
505859 rs118013517 9_51 0.123 17.42 7.0e-05 4.52
188130 rs190072323 3_97 0.133 17.41 7.6e-05 4.08
208954 rs35945249 4_18 0.132 17.32 7.5e-05 -4.23
521053 rs17144212 10_8 0.125 17.25 7.0e-05 -4.14
3511 rs79089220 1_8 0.121 17.22 6.8e-05 4.08
856603 rs553932 21_22 0.119 17.07 6.6e-05 -4.33
97634 rs200418470 2_58 0.107 17.04 6.0e-05 4.19
12749 rs116039993 1_26 0.123 17.03 6.8e-05 4.03
97636 rs76082620 2_58 0.102 16.83 5.6e-05 4.16
608971 rs16907395 12_11 0.112 16.82 6.2e-05 4.31
160726 rs9311895 3_43 0.104 16.69 5.7e-05 -3.97
297670 rs115431743 5_62 0.106 16.60 5.8e-05 -4.10
704311 rs77066728 14_50 0.112 16.58 6.1e-05 3.99
334851 rs74434374 6_26 0.057 16.56 3.1e-05 -4.37
297679 rs116673434 5_62 0.105 16.55 5.7e-05 -4.10
709342 rs117521738 15_5 0.109 16.54 5.9e-05 -3.98
380830 rs12198594 6_112 0.100 16.44 5.4e-05 3.99
367943 rs34181568 6_88 0.105 16.38 5.6e-05 -3.90
520762 rs484392 10_8 0.104 16.37 5.5e-05 3.92
801441 rs8084578 18_38 0.104 16.33 5.6e-05 -4.01
814319 rs78119665 19_17 0.112 16.30 6.0e-05 -4.04
510103 rs10981735 9_58 0.091 16.29 4.8e-05 4.11
289169 rs56203338 5_45 0.064 16.25 3.4e-05 -3.88
1530 rs12082538 1_4 0.108 16.20 5.7e-05 -3.92
627833 rs150360326 12_46 0.090 16.10 4.8e-05 -3.87
735252 rs3785217 16_8 0.096 16.01 5.0e-05 3.90
175280 rs9867405 3_71 0.091 16.00 4.7e-05 4.13
262078 rs28684807 4_117 0.099 15.93 5.2e-05 3.83
414896 rs1734886 7_63 0.094 15.90 4.9e-05 4.11
166197 rs12636636 3_54 0.093 15.84 4.8e-05 3.98
52775 rs17187009 1_105 0.094 15.73 4.9e-05 3.81
57714 rs74751225 1_114 0.087 15.72 4.5e-05 3.82
675062 rs150602177 13_53 0.092 15.72 4.7e-05 -3.84
560583 rs4387284 10_83 0.083 15.71 4.3e-05 -3.92
334909 rs79594290 6_26 0.047 15.70 2.4e-05 -4.08
119153 rs77191462 2_105 0.089 15.69 4.6e-05 -3.78SNPs with highest PVE
#SNPs with 50 highest pve
head(ctwas_snp_res[order(-ctwas_snp_res$PVE),report_cols_snps],50) id region_tag susie_pip mu2 PVE z
832971 rs11557577 20_13 0.234 20.24 1.6e-04 -4.76
221280 rs117111501 4_41 0.213 20.86 1.5e-04 4.73
838805 rs6017234 20_27 0.207 19.88 1.3e-04 -4.60
505868 rs75155685 9_51 0.161 18.72 9.9e-05 4.68
242736 rs7660796 4_80 0.162 18.50 9.8e-05 4.38
607479 rs10772815 12_7 0.145 17.87 8.5e-05 -4.27
608967 rs56198451 12_11 0.138 17.83 8.1e-05 4.45
813014 rs273265 19_14 0.130 18.12 7.7e-05 -4.70
188130 rs190072323 3_97 0.133 17.41 7.6e-05 4.08
208954 rs35945249 4_18 0.132 17.32 7.5e-05 -4.23
780163 rs1675273 17_44 0.130 17.60 7.5e-05 4.21
824212 rs1966573 19_36 0.128 17.88 7.5e-05 -4.20
650317 rs562115589 13_7 0.126 17.54 7.3e-05 -4.20
813015 rs2241089 19_14 0.123 17.85 7.2e-05 -4.67
813017 rs4808757 19_14 0.123 17.82 7.1e-05 -4.66
505859 rs118013517 9_51 0.123 17.42 7.0e-05 4.52
521053 rs17144212 10_8 0.125 17.25 7.0e-05 -4.14
3511 rs79089220 1_8 0.121 17.22 6.8e-05 4.08
12749 rs116039993 1_26 0.123 17.03 6.8e-05 4.03
527922 rs117340939 10_20 0.114 17.63 6.6e-05 -4.13
856603 rs553932 21_22 0.119 17.07 6.6e-05 -4.33
608971 rs16907395 12_11 0.112 16.82 6.2e-05 4.31
704311 rs77066728 14_50 0.112 16.58 6.1e-05 3.99
97634 rs200418470 2_58 0.107 17.04 6.0e-05 4.19
814319 rs78119665 19_17 0.112 16.30 6.0e-05 -4.04
709342 rs117521738 15_5 0.109 16.54 5.9e-05 -3.98
297670 rs115431743 5_62 0.106 16.60 5.8e-05 -4.10
1530 rs12082538 1_4 0.108 16.20 5.7e-05 -3.92
160726 rs9311895 3_43 0.104 16.69 5.7e-05 -3.97
297679 rs116673434 5_62 0.105 16.55 5.7e-05 -4.10
97636 rs76082620 2_58 0.102 16.83 5.6e-05 4.16
367943 rs34181568 6_88 0.105 16.38 5.6e-05 -3.90
801441 rs8084578 18_38 0.104 16.33 5.6e-05 -4.01
520762 rs484392 10_8 0.104 16.37 5.5e-05 3.92
380830 rs12198594 6_112 0.100 16.44 5.4e-05 3.99
262078 rs28684807 4_117 0.099 15.93 5.2e-05 3.83
735252 rs3785217 16_8 0.096 16.01 5.0e-05 3.90
52775 rs17187009 1_105 0.094 15.73 4.9e-05 3.81
414896 rs1734886 7_63 0.094 15.90 4.9e-05 4.11
166197 rs12636636 3_54 0.093 15.84 4.8e-05 3.98
510103 rs10981735 9_58 0.091 16.29 4.8e-05 4.11
627833 rs150360326 12_46 0.090 16.10 4.8e-05 -3.87
175280 rs9867405 3_71 0.091 16.00 4.7e-05 4.13
675062 rs150602177 13_53 0.092 15.72 4.7e-05 -3.84
119153 rs77191462 2_105 0.089 15.69 4.6e-05 -3.78
57714 rs74751225 1_114 0.087 15.72 4.5e-05 3.82
608965 rs147901559 12_11 0.087 15.65 4.5e-05 4.16
645383 rs264505 12_79 0.088 15.59 4.5e-05 3.80
693111 rs11622348 14_28 0.088 15.56 4.5e-05 3.78
781250 rs9890434 17_46 0.088 15.69 4.5e-05 4.41SNPs with largest z scores
#SNPs with 50 largest z scores
head(ctwas_snp_res[order(-abs(ctwas_snp_res$z)),report_cols_snps],50) id region_tag susie_pip mu2 PVE z
832971 rs11557577 20_13 0.234 20.24 1.6e-04 -4.76
221280 rs117111501 4_41 0.213 20.86 1.5e-04 4.73
813014 rs273265 19_14 0.130 18.12 7.7e-05 -4.70
505868 rs75155685 9_51 0.161 18.72 9.9e-05 4.68
813015 rs2241089 19_14 0.123 17.85 7.2e-05 -4.67
813017 rs4808757 19_14 0.123 17.82 7.1e-05 -4.66
838805 rs6017234 20_27 0.207 19.88 1.3e-04 -4.60
505859 rs118013517 9_51 0.123 17.42 7.0e-05 4.52
608967 rs56198451 12_11 0.138 17.83 8.1e-05 4.45
781250 rs9890434 17_46 0.088 15.69 4.5e-05 4.41
242736 rs7660796 4_80 0.162 18.50 9.8e-05 4.38
781249 rs74002714 17_46 0.085 15.51 4.3e-05 4.38
334851 rs74434374 6_26 0.057 16.56 3.1e-05 -4.37
781247 rs74002712 17_46 0.080 15.23 4.0e-05 4.34
856603 rs553932 21_22 0.119 17.07 6.6e-05 -4.33
781243 rs9912120 17_46 0.077 15.05 3.8e-05 4.32
608971 rs16907395 12_11 0.112 16.82 6.2e-05 4.31
335654 rs183736834 6_26 0.035 14.35 1.7e-05 -4.29
335693 rs35686990 6_26 0.035 14.33 1.6e-05 -4.29
781244 rs74002710 17_46 0.073 14.79 3.5e-05 4.28
607479 rs10772815 12_7 0.145 17.87 8.5e-05 -4.27
781238 rs112313398 17_46 0.072 14.75 3.5e-05 4.27
208954 rs35945249 4_18 0.132 17.32 7.5e-05 -4.23
335619 rs192408808 6_26 0.032 13.91 1.5e-05 -4.23
781245 rs112225092 17_46 0.071 14.72 3.4e-05 4.23
335526 rs73730378 6_26 0.031 13.80 1.4e-05 -4.22
335625 rs76434237 6_26 0.031 13.79 1.4e-05 -4.22
780163 rs1675273 17_44 0.130 17.60 7.5e-05 4.21
650317 rs562115589 13_7 0.126 17.54 7.3e-05 -4.20
824212 rs1966573 19_36 0.128 17.88 7.5e-05 -4.20
97634 rs200418470 2_58 0.107 17.04 6.0e-05 4.19
335620 rs536856134 6_26 0.030 13.63 1.3e-05 -4.19
335511 rs199640993 6_26 0.030 13.57 1.3e-05 -4.18
97636 rs76082620 2_58 0.102 16.83 5.6e-05 4.16
608965 rs147901559 12_11 0.087 15.65 4.5e-05 4.16
521053 rs17144212 10_8 0.125 17.25 7.0e-05 -4.14
175280 rs9867405 3_71 0.091 16.00 4.7e-05 4.13
527922 rs117340939 10_20 0.114 17.63 6.6e-05 -4.13
335277 rs9469134 6_26 0.027 13.07 1.1e-05 -4.11
414896 rs1734886 7_63 0.094 15.90 4.9e-05 4.11
510103 rs10981735 9_58 0.091 16.29 4.8e-05 4.11
297670 rs115431743 5_62 0.106 16.60 5.8e-05 -4.10
297679 rs116673434 5_62 0.105 16.55 5.7e-05 -4.10
309462 rs252232 5_84 0.065 14.36 3.0e-05 4.10
335284 rs7748472 6_26 0.026 13.05 1.1e-05 -4.10
3511 rs79089220 1_8 0.121 17.22 6.8e-05 4.08
188130 rs190072323 3_97 0.133 17.41 7.6e-05 4.08
334909 rs79594290 6_26 0.047 15.70 2.4e-05 -4.08
335262 rs9461755 6_26 0.026 12.92 1.1e-05 -4.08
335264 rs2071805 6_26 0.025 12.87 1.1e-05 -4.08Gene set enrichment for genes with PIP>0.8
#GO enrichment analysis
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
#number of genes for gene set enrichment
length(genes)[1] 0if (length(genes)>0){
GO_enrichment <- enrichr(genes, dbs)
for (db in dbs){
print(db)
df <- GO_enrichment[[db]]
df <- df[df$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(df)
}
#DisGeNET enrichment
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <-disease_enrichment(entities=genes, vocabulary = "HGNC",
database = "CURATED" )
df <- res_enrich@qresult[1:10, c("Description", "FDR", "Ratio", "BgRatio")]
print(df)
#WebGestalt enrichment
library(WebGestaltR)
background <- ctwas_gene_res$genename
#listGeneSet()
databases <- c("pathway_KEGG", "disease_GLAD4U", "disease_OMIM")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
print(enrichResult[,c("description", "size", "overlap", "FDR", "database", "userId")])
}
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] enrichR_3.0 cowplot_1.0.0 ggplot2_3.3.3
loaded via a namespace (and not attached):
[1] Biobase_2.44.0 httr_1.4.1
[3] bit64_4.0.5 assertthat_0.2.1
[5] stats4_3.6.1 blob_1.2.1
[7] BSgenome_1.52.0 GenomeInfoDbData_1.2.1
[9] Rsamtools_2.0.0 yaml_2.2.0
[11] progress_1.2.2 pillar_1.6.1
[13] RSQLite_2.2.7 lattice_0.20-38
[15] glue_1.4.2 digest_0.6.20
[17] GenomicRanges_1.36.0 promises_1.0.1
[19] XVector_0.24.0 colorspace_1.4-1
[21] htmltools_0.3.6 httpuv_1.5.1
[23] Matrix_1.2-18 XML_3.98-1.20
[25] pkgconfig_2.0.3 biomaRt_2.40.1
[27] zlibbioc_1.30.0 purrr_0.3.4
[29] scales_1.1.0 whisker_0.3-2
[31] later_0.8.0 BiocParallel_1.18.0
[33] git2r_0.26.1 tibble_3.1.2
[35] farver_2.1.0 generics_0.0.2
[37] IRanges_2.18.1 ellipsis_0.3.2
[39] withr_2.4.1 cachem_1.0.5
[41] SummarizedExperiment_1.14.1 GenomicFeatures_1.36.3
[43] BiocGenerics_0.30.0 magrittr_2.0.1
[45] crayon_1.4.1 memoise_2.0.0
[47] evaluate_0.14 fs_1.3.1
[49] fansi_0.5.0 tools_3.6.1
[51] data.table_1.14.0 prettyunits_1.0.2
[53] hms_1.1.0 lifecycle_1.0.0
[55] matrixStats_0.57.0 stringr_1.4.0
[57] S4Vectors_0.22.1 munsell_0.5.0
[59] DelayedArray_0.10.0 AnnotationDbi_1.46.0
[61] Biostrings_2.52.0 compiler_3.6.1
[63] GenomeInfoDb_1.20.0 rlang_0.4.11
[65] grid_3.6.1 RCurl_1.98-1.1
[67] rjson_0.2.20 VariantAnnotation_1.30.1
[69] labeling_0.3 bitops_1.0-6
[71] rmarkdown_1.13 gtable_0.3.0
[73] curl_3.3 DBI_1.1.1
[75] R6_2.5.0 GenomicAlignments_1.20.1
[77] dplyr_1.0.7 knitr_1.23
[79] rtracklayer_1.44.0 utf8_1.2.1
[81] fastmap_1.1.0 bit_4.0.4
[83] workflowr_1.6.2 rprojroot_2.0.2
[85] stringi_1.4.3 parallel_3.6.1
[87] Rcpp_1.0.6 vctrs_0.3.8
[89] tidyselect_1.1.0 xfun_0.8