LDL direct (quantile) - Adipose_Subcutaneous
wesleycrouse
2021-09-7
Last updated: 2021-11-15
Checks: 6 1
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 81c11f7. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: analysis/figure/
Unstaged changes:
Modified: analysis/ukb-d-30780_irnt_Adipose_Subcutaneous_known.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-d-30780_irnt_Adipose_Subcutaneous_known.Rmd) and HTML (docs/ukb-d-30780_irnt_Adipose_Subcutaneous_known.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 81c11f7 | wesleycrouse | 2021-11-15 | fixing report |
| Rmd | c123245 | wesleycrouse | 2021-11-15 | LDL adipose sub |
Overview
These are the results of a ctwas analysis of the UK Biobank trait LDL direct using Adipose_Subcutaneous gene weights.
The GWAS was conducted by the Neale Lab, and the biomarker traits we analyzed are discussed here. Summary statistics were obtained from IEU OpenGWAS using GWAS ID: ukb-d-30780_irnt. Results were obtained from from IEU rather than Neale Lab because they are in a standardard format (GWAS VCF). Note that 3 of the 34 biomarker traits were not available from IEU and were excluded from analysis.
The weights are mashr GTEx v8 models on Adipose_Subcutaneous eQTL obtained from PredictDB. We performed a full harmonization of the variants, including recovering strand ambiguous variants. This procedure is discussed in a separate document. (TO-DO: add report that describes harmonization)
LD matrices were computed from a 10% subset of Neale lab subjects. Subjects were matched using the plate and well information from genotyping. We included only biallelic variants with MAF>0.01 in the original Neale Lab GWAS. (TO-DO: add more details [number of subjects, variants, etc])
Weight QC
TO-DO: add enhanced QC reporting (total number of weights, why each variant was missing for all genes)
qclist_all <- list()
qc_files <- paste0(results_dir, "/", list.files(results_dir, pattern="exprqc.Rd"))
for (i in 1:length(qc_files)){
load(qc_files[i])
chr <- unlist(strsplit(rev(unlist(strsplit(qc_files[i], "_")))[1], "[.]"))[1]
qclist_all[[chr]] <- cbind(do.call(rbind, lapply(qclist,unlist)), as.numeric(substring(chr,4)))
}
qclist_all <- data.frame(do.call(rbind, qclist_all))
colnames(qclist_all)[ncol(qclist_all)] <- "chr"
rm(qclist, wgtlist, z_gene_chr)
#number of imputed weights
nrow(qclist_all)[1] 12961#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
1278 918 741 500 602 740 638 485 494 502 777 731 256 426 444
16 17 18 19 20 21 22
610 799 198 977 380 144 321 #proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.7639843Load ctwas results
Check convergence of parameters
library(ggplot2)
library(cowplot)
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
df <- data.frame(niter = rep(1:ncol(group_prior_rec), 2),
value = c(group_prior_rec[1,], group_prior_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_rec)))
df$group <- as.factor(df$group)
df$value[df$group=="SNP"] <- df$value[df$group=="SNP"]*thin #adjust parameter to account for thin argument
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Prior mean") +
theme_cowplot()
df <- data.frame(niter = rep(1:ncol(group_prior_var_rec), 2),
value = c(group_prior_var_rec[1,], group_prior_var_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_var_rec)))
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Prior variance") +
theme_cowplot()
plot_grid(p_pi, p_sigma2)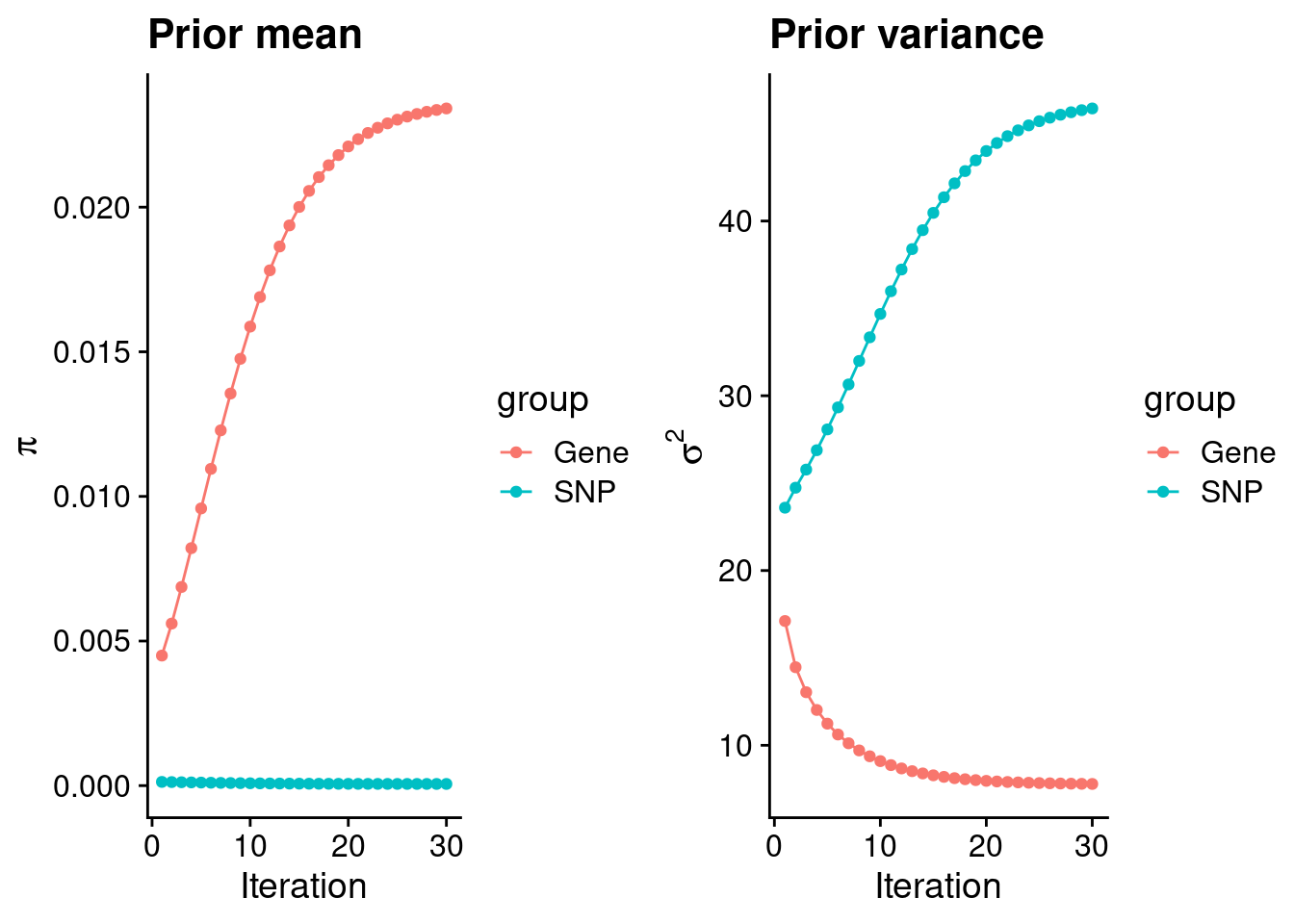
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0234130881 0.0000584326 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
7.788427 46.440462 #report sample size
print(sample_size)[1] 343621#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 12961 8696600#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.00687808 0.06867862 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.07281326 1.20053323Genes with highest PIPs
#distribution of PIPs
hist(ctwas_gene_res$susie_pip, xlim=c(0,1), main="Distribution of Gene PIPs")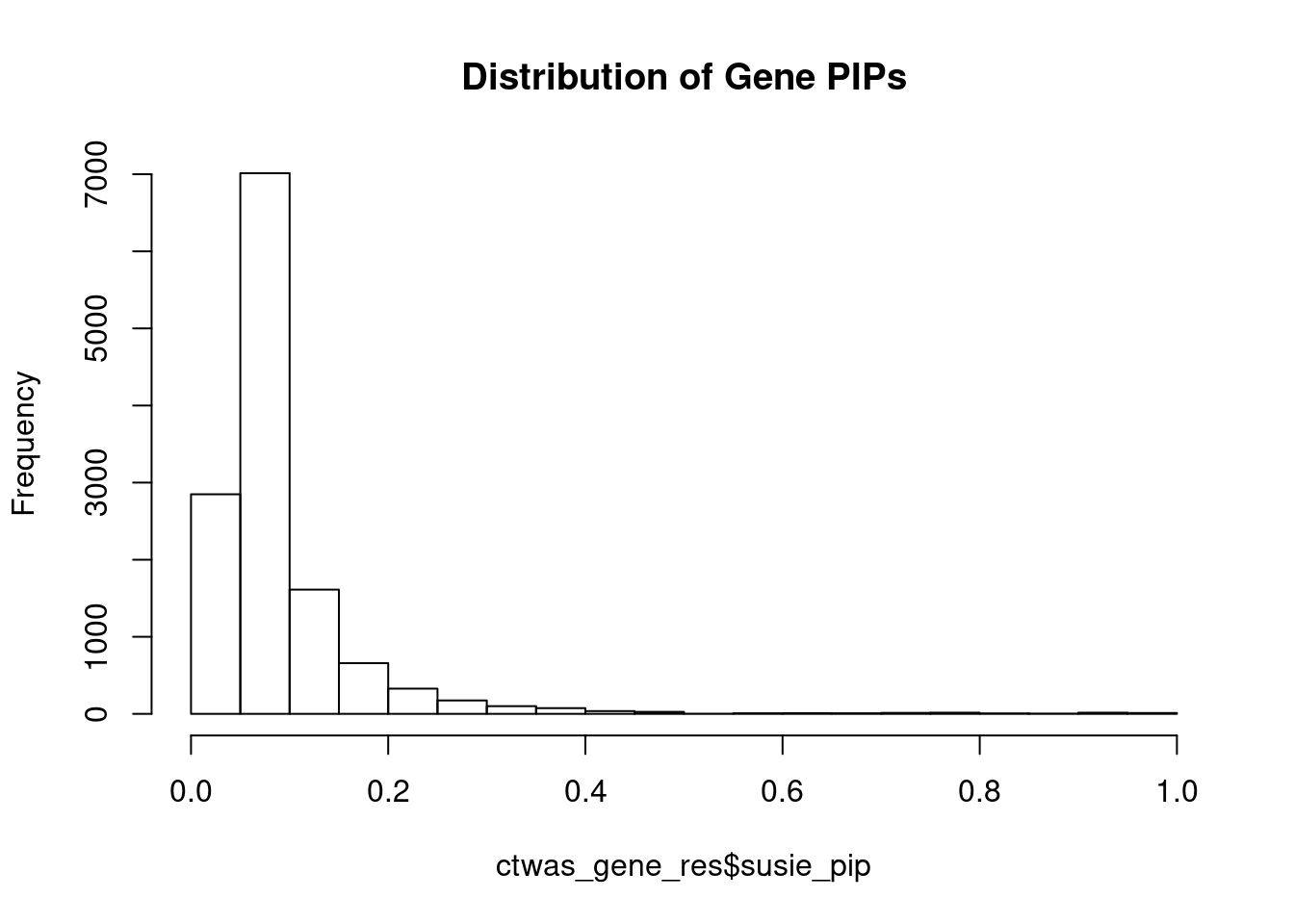
#genes with PIP>0.8 or 20 highest PIPs
head(ctwas_gene_res[order(-ctwas_gene_res$susie_pip),report_cols], max(sum(ctwas_gene_res$susie_pip>0.8), 20)) genename region_tag susie_pip mu2 PVE z
5923 ABCA8 17_39 0.9982625 28.49746 8.278872e-05 5.086840
2199 FCGRT 19_34 0.9967913 10378.34712 3.010598e-02 -4.165895
7045 ZDHHC7 16_49 0.9967008 24.23491 7.029534e-05 -4.891663
7879 ACP6 1_73 0.9932775 21.77810 6.295220e-05 4.613976
7623 PKN3 9_66 0.9854511 42.73347 1.225529e-04 -6.885356
4395 POR 7_48 0.9843314 32.69729 9.366414e-05 6.026895
2313 PLPPR2 19_10 0.9790947 28.36151 8.081172e-05 3.965665
3730 C10orf88 10_77 0.9771524 31.39957 8.929071e-05 -6.762952
5600 FAM117B 2_120 0.9695151 38.09714 1.074898e-04 7.877231
3647 CCDC92 12_75 0.9604840 26.34034 7.362610e-05 -5.343007
10255 KLHDC7A 1_13 0.9544827 18.66743 5.185288e-05 4.124187
10447 FKRP 19_33 0.9482623 20.61788 5.689744e-05 4.425174
1279 SRRT 7_62 0.9450584 25.66844 7.059571e-05 4.744831
632 SPHK2 19_33 0.9412590 38.80542 1.062972e-04 -8.721460
14551 AC007950.2 15_29 0.9412296 30.76191 8.426148e-05 5.555780
5327 IL1RN 2_67 0.9410797 20.79413 5.694919e-05 4.455379
1003 TPD52 8_57 0.9370510 18.68606 5.095669e-05 -4.121885
10272 SPTY2D1 11_13 0.9335133 28.13983 7.644734e-05 -5.557123
3633 CCND2 12_4 0.9318759 19.26549 5.224666e-05 -4.128258
1526 CWF19L1 10_64 0.9256794 28.62700 7.711817e-05 5.813958
3674 KDSR 18_35 0.9199769 18.92509 5.066818e-05 -4.119957
161 MPND 19_5 0.9197113 19.83813 5.309731e-05 -4.322018
7441 TMED4 7_32 0.9191981 36.87829 9.865071e-05 7.688274
8904 PDHB 3_40 0.9140947 24.02176 6.390228e-05 3.361078
4022 ACVR1C 2_94 0.9057641 19.38221 5.109033e-05 -4.185879
9348 KCNK3 2_16 0.9044393 20.54412 5.407385e-05 -4.772296
9036 PCSK9 1_34 0.8982431 105.00205 2.744808e-04 17.210869
1559 SCD 10_64 0.8966611 18.64373 4.864984e-05 -4.541468
11865 C2CD4A 15_28 0.8713009 21.59986 5.476958e-05 4.535165
13258 LINC01184 5_78 0.8593862 18.23008 4.559291e-05 -3.918127
6023 NTN5 19_33 0.8492136 47.56594 1.175529e-04 10.033010
7079 UBASH3B 11_74 0.8475160 23.99612 5.918467e-05 4.906621
5607 PARP9 3_76 0.8452748 22.78261 5.604304e-05 3.744644
6967 PELO 5_30 0.8402073 61.55834 1.505198e-04 8.522224
1237 CPNE3 8_62 0.8147843 18.04139 4.277922e-05 3.753687
3724 GPAM 10_70 0.8129956 19.45034 4.601885e-05 4.000718
num_eqtl
5923 2
2199 2
7045 3
7879 4
7623 2
4395 2
2313 1
3730 1
5600 2
3647 5
10255 1
10447 5
1279 2
632 1
14551 1
5327 1
1003 2
10272 1
3633 2
1526 2
3674 2
161 2
7441 2
8904 2
4022 1
9348 1
9036 1
1559 1
11865 1
13258 2
6023 2
7079 1
5607 1
6967 1
1237 3
3724 1Genes with largest effect sizes
#plot PIP vs effect size
plot(ctwas_gene_res$susie_pip, ctwas_gene_res$mu2, xlab="PIP", ylab="mu^2", main="Gene PIPs vs Effect Size")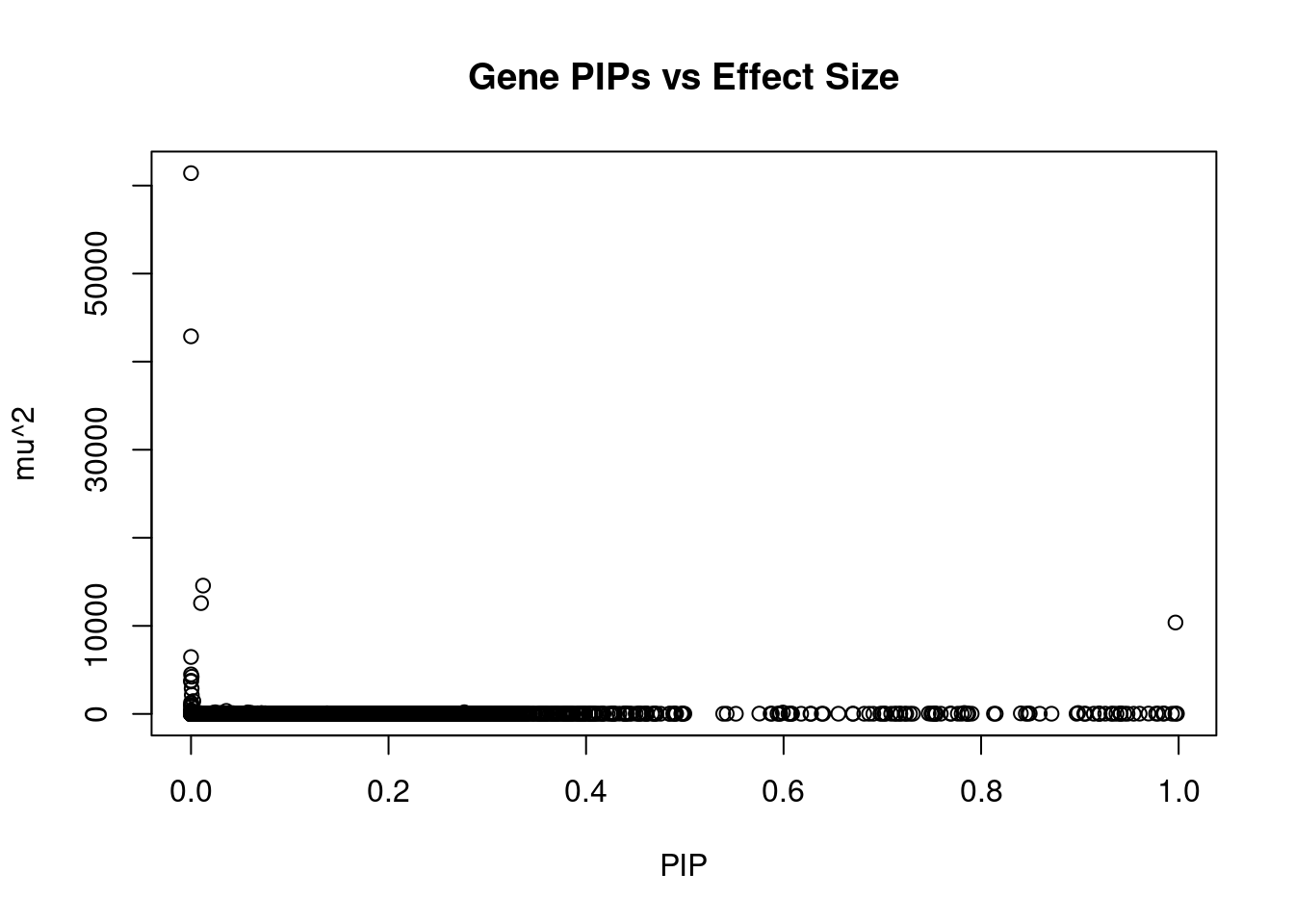
#genes with 20 largest effect sizes
head(ctwas_gene_res[order(-ctwas_gene_res$mu2),report_cols],20) genename region_tag susie_pip mu2 PVE z
75 KMT2E 7_65 0.000000e+00 61406.4269 0.000000e+00 -3.0023371
5123 SRPK2 7_65 0.000000e+00 42876.5960 0.000000e+00 -2.5902427
6042 RPS11 19_34 1.218825e-02 14569.1716 5.167690e-04 -2.4990920
1412 FLT3LG 19_34 1.013001e-02 12581.6580 3.709097e-04 2.4043507
2199 FCGRT 19_34 9.967913e-01 10378.3471 3.010598e-02 -4.1658948
13638 RTEL1 20_38 0.000000e+00 6459.8182 0.000000e+00 -3.9462192
8994 USP39 2_54 2.710276e-12 4489.2019 3.540813e-14 4.0835994
2528 CCNJ 10_61 6.651905e-04 4224.3905 8.177686e-06 -4.6393182
1879 GMEB2 20_38 0.000000e+00 3769.8594 0.000000e+00 2.2095747
8997 VAMP5 2_54 6.218803e-12 3677.0268 6.654630e-14 4.6225955
4296 PRRG2 19_34 6.346413e-04 2889.6595 5.336977e-06 2.0148130
4298 PRR12 19_34 7.826670e-04 2154.4303 4.907155e-06 1.9257259
4297 SCAF1 19_34 2.374574e-03 1461.2603 1.009796e-05 2.1876224
3611 VAMP8 2_54 2.393565e-09 1286.7765 8.963315e-12 -0.5259735
2206 SLC17A7 19_34 4.120128e-04 1175.7166 1.409723e-06 -1.7087222
4571 APOE 19_31 0.000000e+00 999.9678 0.000000e+00 37.8075498
9035 CPT1C 19_34 4.669232e-04 920.8332 1.251258e-06 0.8492599
13190 TNFRSF6B 20_38 0.000000e+00 822.9465 0.000000e+00 2.2944943
12031 LIME1 20_38 0.000000e+00 421.6586 0.000000e+00 -2.9819676
3883 POLK 5_44 3.571822e-02 345.0503 3.586679e-05 23.9950413
num_eqtl
75 1
5123 2
6042 1
1412 1
2199 2
13638 3
8994 1
2528 1
1879 1
8997 2
4296 2
4298 1
4297 1
3611 2
2206 2
4571 2
9035 2
13190 3
12031 1
3883 1Genes with highest PVE
#genes with 20 highest pve
head(ctwas_gene_res[order(-ctwas_gene_res$PVE),report_cols],20) genename region_tag susie_pip mu2 PVE z
2199 FCGRT 19_34 0.99679127 10378.34712 3.010598e-02 -4.165895
6042 RPS11 19_34 0.01218825 14569.17164 5.167690e-04 -2.499092
1412 FLT3LG 19_34 0.01013001 12581.65797 3.709097e-04 2.404351
1287 CETP 16_30 0.59931467 157.61639 2.749012e-04 13.824230
9036 PCSK9 1_34 0.89824307 105.00205 2.744808e-04 17.210869
3267 NRBP1 2_16 0.78298651 106.73558 2.432113e-04 5.571352
4991 PSRC1 1_67 0.27672417 192.00911 1.546284e-04 -22.968124
6967 PELO 5_30 0.84020734 61.55834 1.505198e-04 8.522224
7623 PKN3 9_66 0.98545107 42.73347 1.225529e-04 -6.885356
6023 NTN5 19_33 0.84921360 47.56594 1.175529e-04 10.033010
5600 FAM117B 2_120 0.96951505 38.09714 1.074898e-04 7.877231
632 SPHK2 19_33 0.94125900 38.80542 1.062972e-04 -8.721460
5576 DNAJC13 3_82 0.74690937 48.23841 1.048531e-04 -4.919872
7441 TMED4 7_32 0.91919811 36.87829 9.865071e-05 7.688274
4395 POR 7_48 0.98433137 32.69729 9.366414e-05 6.026895
3730 C10orf88 10_77 0.97715244 31.39957 8.929071e-05 -6.762952
14551 AC007950.2 15_29 0.94122959 30.76191 8.426148e-05 5.555780
5923 ABCA8 17_39 0.99826247 28.49746 8.278872e-05 5.086840
2313 PLPPR2 19_10 0.97909467 28.36151 8.081172e-05 3.965665
13216 TMEM199 17_17 0.76873569 35.77300 8.002997e-05 6.692808
num_eqtl
2199 2
6042 1
1412 1
1287 2
9036 1
3267 1
4991 2
6967 1
7623 2
6023 2
5600 2
632 1
5576 2
7441 2
4395 2
3730 1
14551 1
5923 2
2313 1
13216 2Genes with largest z scores
#genes with 20 largest z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
4571 APOE 19_31 0.000000e+00 999.96778 0.000000e+00 37.80755
12880 AC067959.1 2_13 4.072320e-11 330.71746 3.919398e-14 -25.52145
3883 POLK 5_44 3.571822e-02 345.05025 3.586679e-05 23.99504
4991 PSRC1 1_67 2.767242e-01 192.00911 1.546284e-04 -22.96812
9036 PCSK9 1_34 8.982431e-01 105.00205 2.744808e-04 17.21087
11376 ANKDD1B 5_44 2.216674e-02 122.31840 7.890672e-06 15.52949
1211 APOB 2_13 2.837784e-10 181.86756 1.501948e-13 -15.03583
2354 ATP13A1 19_15 5.739104e-02 175.75452 2.935424e-05 -14.74773
8740 GATAD2A 19_15 2.463813e-02 187.72384 1.346008e-05 -14.43456
6024 GEMIN7 19_31 0.000000e+00 157.43955 0.000000e+00 14.13454
1287 CETP 16_30 5.993147e-01 157.61639 2.749012e-04 13.82423
6093 CELSR2 1_67 8.522725e-03 109.89040 2.725577e-06 13.72575
4570 NECTIN2 19_31 0.000000e+00 55.57768 0.000000e+00 -13.59762
8941 FEN1 11_34 6.056125e-02 130.93608 2.307674e-05 12.59183
4644 YIPF2 19_9 6.639134e-14 177.08337 3.421444e-17 11.69253
268 NPC1L1 7_32 4.099715e-02 82.55662 9.849767e-06 10.76193
4377 SMARCA4 19_9 0.000000e+00 101.79287 0.000000e+00 10.69172
7824 USP1 1_39 7.150444e-02 94.23231 1.960890e-05 10.57358
6033 TIMM29 19_9 0.000000e+00 104.96417 0.000000e+00 10.16969
4552 MAU2 19_15 6.748590e-02 74.31016 1.459424e-05 10.16871
num_eqtl
4571 2
12880 1
3883 1
4991 2
9036 1
11376 1
1211 2
2354 3
8740 2
6024 2
1287 2
6093 2
4570 1
8941 1
4644 2
268 1
4377 2
7824 1
6033 1
4552 2Comparing z scores and PIPs
#set nominal signifiance threshold for z scores
alpha <- 0.05
#bonferroni adjusted threshold for z scores
sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#Q-Q plot for z scores
obs_z <- ctwas_gene_res$z[order(ctwas_gene_res$z)]
exp_z <- qnorm((1:nrow(ctwas_gene_res))/nrow(ctwas_gene_res))
plot(exp_z, obs_z, xlab="Expected z", ylab="Observed z", main="Gene z score Q-Q plot")
abline(a=0,b=1)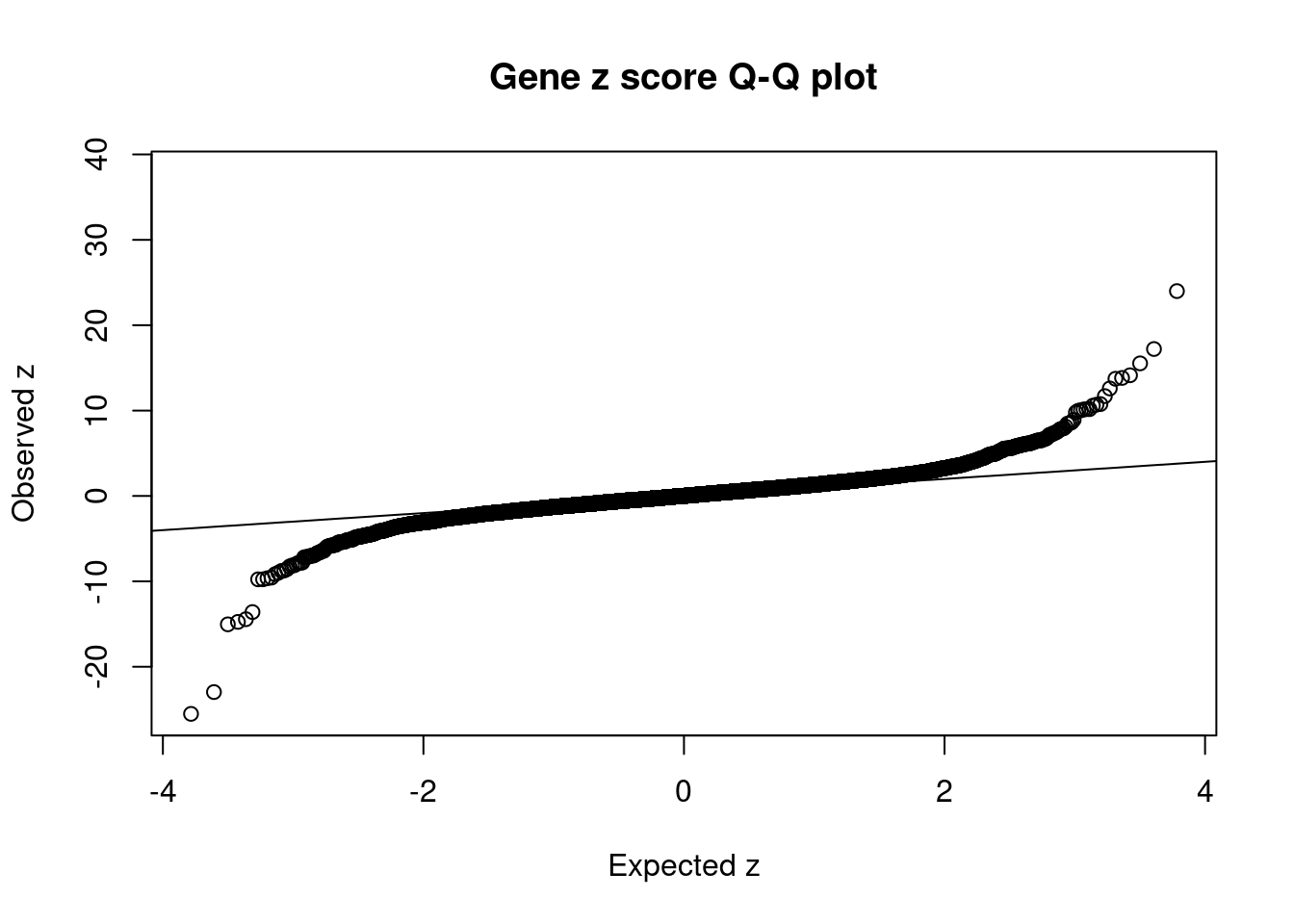
#plot z score vs PIP
plot(abs(ctwas_gene_res$z), ctwas_gene_res$susie_pip, xlab="abs(z)", ylab="PIP")
abline(v=sig_thresh, col="red", lty=2)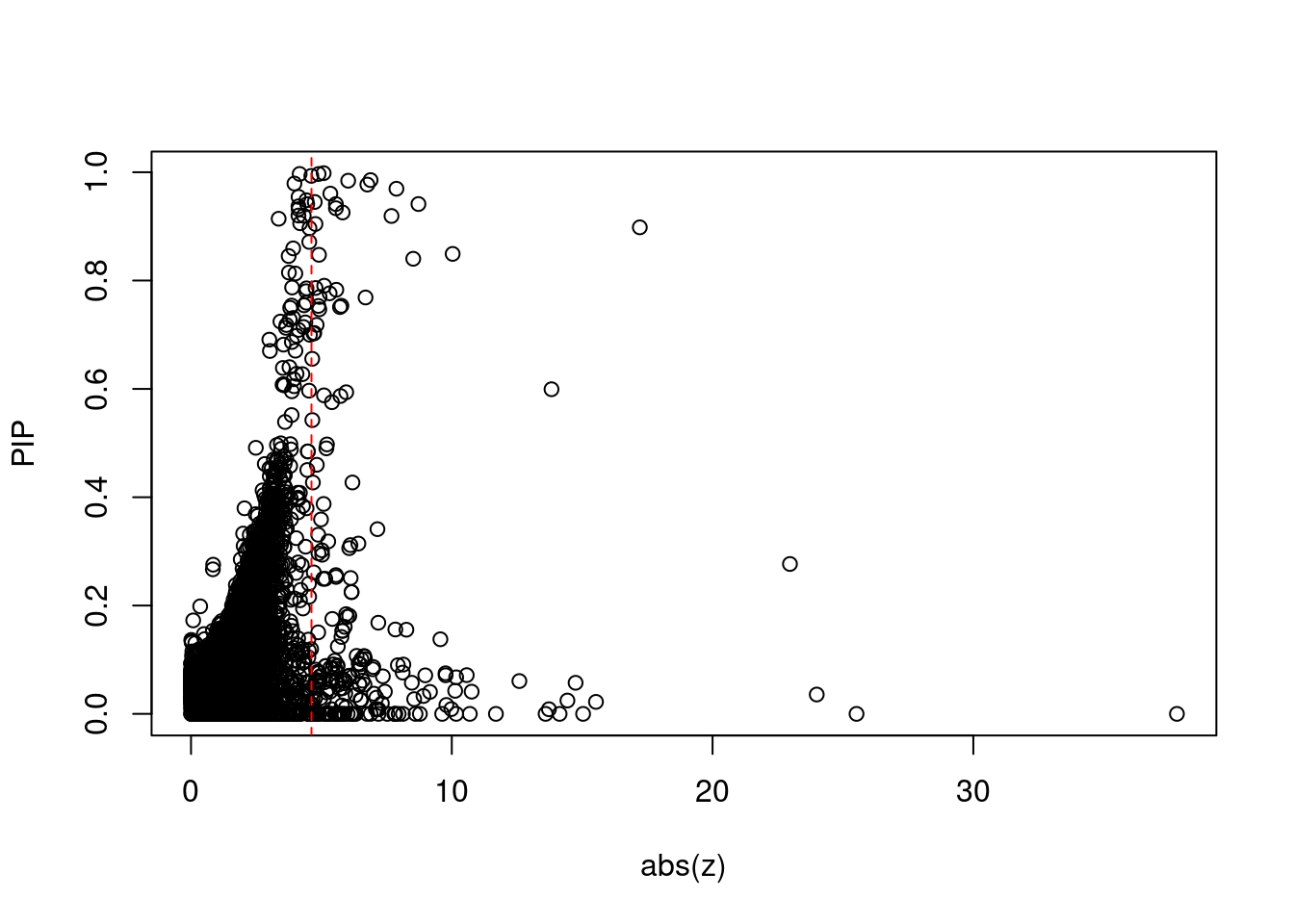
#proportion of significant z scores
mean(abs(ctwas_gene_res$z) > sig_thresh)[1] 0.01751408#genes with most significant z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
4571 APOE 19_31 0.000000e+00 999.96778 0.000000e+00 37.80755
12880 AC067959.1 2_13 4.072320e-11 330.71746 3.919398e-14 -25.52145
3883 POLK 5_44 3.571822e-02 345.05025 3.586679e-05 23.99504
4991 PSRC1 1_67 2.767242e-01 192.00911 1.546284e-04 -22.96812
9036 PCSK9 1_34 8.982431e-01 105.00205 2.744808e-04 17.21087
11376 ANKDD1B 5_44 2.216674e-02 122.31840 7.890672e-06 15.52949
1211 APOB 2_13 2.837784e-10 181.86756 1.501948e-13 -15.03583
2354 ATP13A1 19_15 5.739104e-02 175.75452 2.935424e-05 -14.74773
8740 GATAD2A 19_15 2.463813e-02 187.72384 1.346008e-05 -14.43456
6024 GEMIN7 19_31 0.000000e+00 157.43955 0.000000e+00 14.13454
1287 CETP 16_30 5.993147e-01 157.61639 2.749012e-04 13.82423
6093 CELSR2 1_67 8.522725e-03 109.89040 2.725577e-06 13.72575
4570 NECTIN2 19_31 0.000000e+00 55.57768 0.000000e+00 -13.59762
8941 FEN1 11_34 6.056125e-02 130.93608 2.307674e-05 12.59183
4644 YIPF2 19_9 6.639134e-14 177.08337 3.421444e-17 11.69253
268 NPC1L1 7_32 4.099715e-02 82.55662 9.849767e-06 10.76193
4377 SMARCA4 19_9 0.000000e+00 101.79287 0.000000e+00 10.69172
7824 USP1 1_39 7.150444e-02 94.23231 1.960890e-05 10.57358
6033 TIMM29 19_9 0.000000e+00 104.96417 0.000000e+00 10.16969
4552 MAU2 19_15 6.748590e-02 74.31016 1.459424e-05 10.16871
num_eqtl
4571 2
12880 1
3883 1
4991 2
9036 1
11376 1
1211 2
2354 3
8740 2
6024 2
1287 2
6093 2
4570 1
8941 1
4644 2
268 1
4377 2
7824 1
6033 1
4552 2Locus plots for genes and SNPs
ctwas_gene_res_sortz <- ctwas_gene_res[order(-abs(ctwas_gene_res$z)),]
report_cols_region <- report_cols[!(report_cols %in% c("num_eqtl"))]
n_plots <- 5
for (region_tag_plot in head(unique(ctwas_gene_res_sortz$region_tag), n_plots)){
ctwas_res_region <- ctwas_res[ctwas_res$region_tag==region_tag_plot,]
start <- min(ctwas_res_region$pos)
end <- max(ctwas_res_region$pos)
ctwas_res_region <- ctwas_res_region[order(ctwas_res_region$pos),]
ctwas_res_region_gene <- ctwas_res_region[ctwas_res_region$type=="gene",]
ctwas_res_region_snp <- ctwas_res_region[ctwas_res_region$type=="SNP",]
#region name
print(paste0("Region: ", region_tag_plot))
#table of genes in region
print(ctwas_res_region_gene[,report_cols_region])
par(mfrow=c(4,1))
#gene z scores
plot(ctwas_res_region_gene$pos, abs(ctwas_res_region_gene$z), xlab="Position", ylab="abs(gene_z)", xlim=c(start,end),
ylim=c(0,max(sig_thresh, abs(ctwas_res_region_gene$z))),
main=paste0("Region: ", region_tag_plot))
abline(h=sig_thresh,col="red",lty=2)
#significance threshold for SNPs
alpha_snp <- 5*10^(-8)
sig_thresh_snp <- qnorm(1-alpha_snp/2, lower=T)
#snp z scores
plot(ctwas_res_region_snp$pos, abs(ctwas_res_region_snp$z), xlab="Position", ylab="abs(snp_z)",xlim=c(start,end),
ylim=c(0,max(sig_thresh_snp, max(abs(ctwas_res_region_snp$z)))))
abline(h=sig_thresh_snp,col="purple",lty=2)
#gene pips
plot(ctwas_res_region_gene$pos, ctwas_res_region_gene$susie_pip, xlab="Position", ylab="Gene PIP", xlim=c(start,end), ylim=c(0,1))
abline(h=0.8,col="blue",lty=2)
#snp pips
plot(ctwas_res_region_snp$pos, ctwas_res_region_snp$susie_pip, xlab="Position", ylab="SNP PIP", xlim=c(start,end), ylim=c(0,1))
abline(h=0.8,col="blue",lty=2)
}[1] "Region: 19_31"
genename region_tag susie_pip mu2 PVE z
7552 ZNF233 19_31 0 98.228951 0 -9.63193736
7553 ZNF235 19_31 0 8.185909 0 -0.97877558
624 ZNF112 19_31 0 72.068605 0 7.18938487
14021 ZNF285 19_31 0 10.013342 0 -1.26709542
14628 ZNF229 19_31 0 8.630054 0 1.23011935
8729 ZNF180 19_31 0 25.642421 0 1.41443321
11089 CEACAM19 19_31 0 13.759203 0 3.64086985
898 PVR 19_31 0 30.112592 0 -3.01973306
4570 NECTIN2 19_31 0 55.577675 0 -13.59762224
4572 TOMM40 19_31 0 56.000201 0 5.53230466
4571 APOE 19_31 0 999.967785 0 37.80754977
4573 APOC1 19_31 0 296.620171 0 7.53014709
12950 APOC2 19_31 0 99.467017 0 7.82468739
2196 CLPTM1 19_31 0 48.802068 0 -2.57517256
9263 ZNF296 19_31 0 65.538646 0 5.25423092
123 MARK4 19_31 0 20.312700 0 -2.24637681
6024 GEMIN7 19_31 0 157.439551 0 14.13454209
2198 PPP1R37 19_31 0 35.405859 0 -2.37518201
131 TRAPPC6A 19_31 0 16.600235 0 1.92140959
11388 BLOC1S3 19_31 0 8.975124 0 2.30141189
2203 ERCC2 19_31 0 13.615522 0 1.53401174
3560 CD3EAP 19_31 0 16.730266 0 0.05565582
229 ERCC1 19_31 0 13.134793 0 -2.19401016
12385 PPM1N 19_31 0 20.227762 0 -1.22009641
4224 RTN2 19_31 0 6.803450 0 2.07007103
4226 VASP 19_31 0 23.808738 0 1.26035740
4222 OPA3 19_31 0 11.581430 0 1.32335471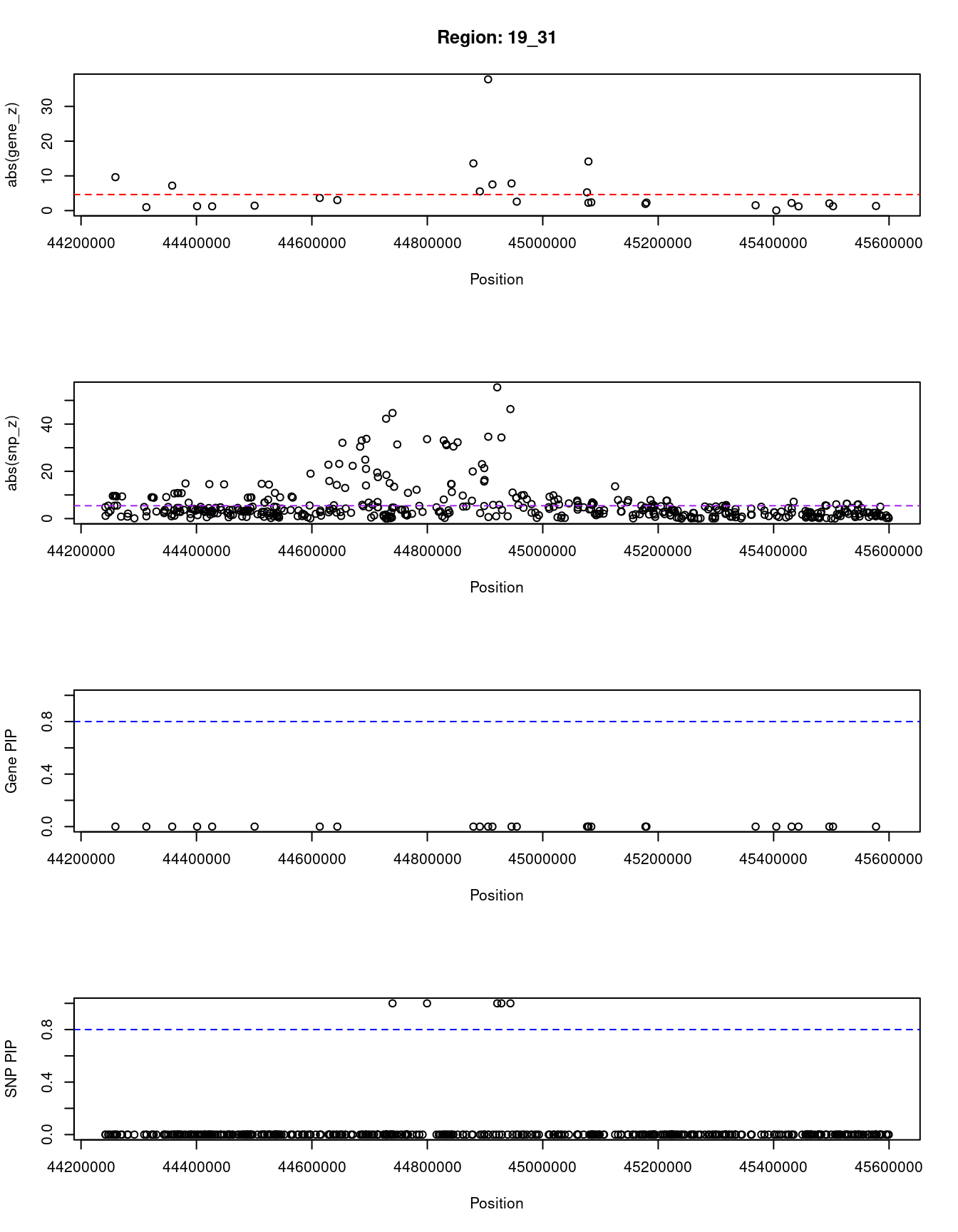
[1] "Region: 2_13"
genename region_tag susie_pip mu2 PVE z
1211 APOB 2_13 2.837784e-10 181.8676 1.501948e-13 -15.03583
12880 AC067959.1 2_13 4.072320e-11 330.7175 3.919398e-14 -25.52145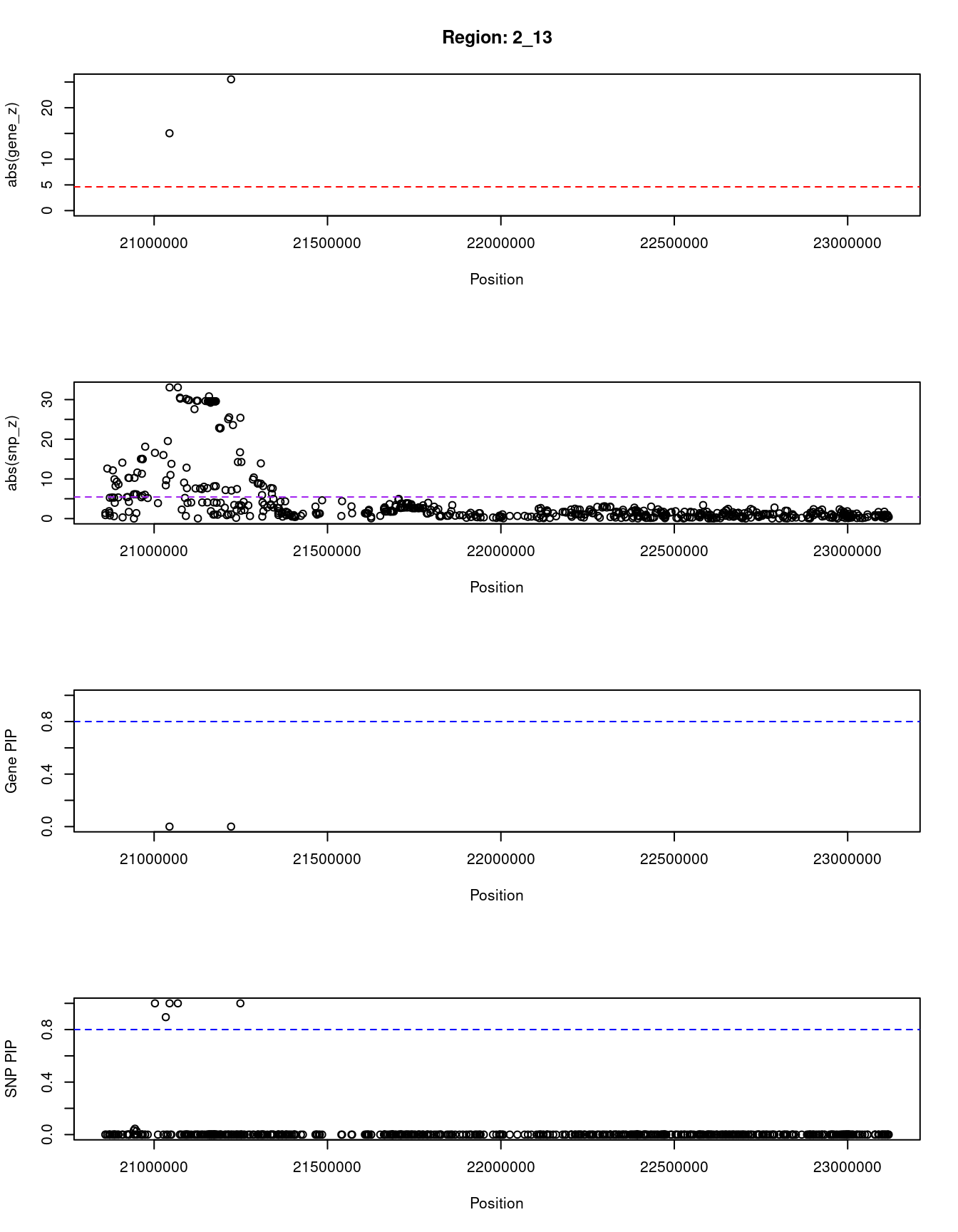
[1] "Region: 5_44"
genename region_tag susie_pip mu2 PVE
492 HEXB 5_44 0.01736765 5.064390 2.559696e-07
9392 ENC1 5_44 0.01622141 4.498465 2.123603e-07
8217 NSA2 5_44 0.02290740 8.434046 5.622534e-07
8218 GFM2 5_44 0.01691796 6.530220 3.215111e-07
11922 FAM169A 5_44 0.01713930 4.950113 2.469043e-07
10025 GCNT4 5_44 0.01663010 4.796782 2.321482e-07
13386 LINC01336 5_44 0.02389685 13.675282 9.510367e-07
3883 POLK 5_44 0.03571822 345.050253 3.586679e-05
14207 CTC-366B18.4 5_44 0.09103018 54.870482 1.453598e-05
11376 ANKDD1B 5_44 0.02216674 122.318403 7.890672e-06
6933 POC5 5_44 0.03730029 38.519190 4.181284e-06
3884 SV2C 5_44 0.27496756 31.503093 2.520896e-05
13509 AC113404.1 5_44 0.02546824 11.766943 8.721332e-07
6412 IQGAP2 5_44 0.02909680 11.478390 9.719557e-07
z
492 -0.2060482
9392 0.2472305
8217 -1.5148382
8218 -1.7610624
11922 0.1096504
10025 -0.4189438
13386 -1.5053404
3883 23.9950413
14207 -8.1427000
11376 15.5294946
6933 -7.0119331
3884 4.2474479
13509 2.3250769
6412 -2.1788866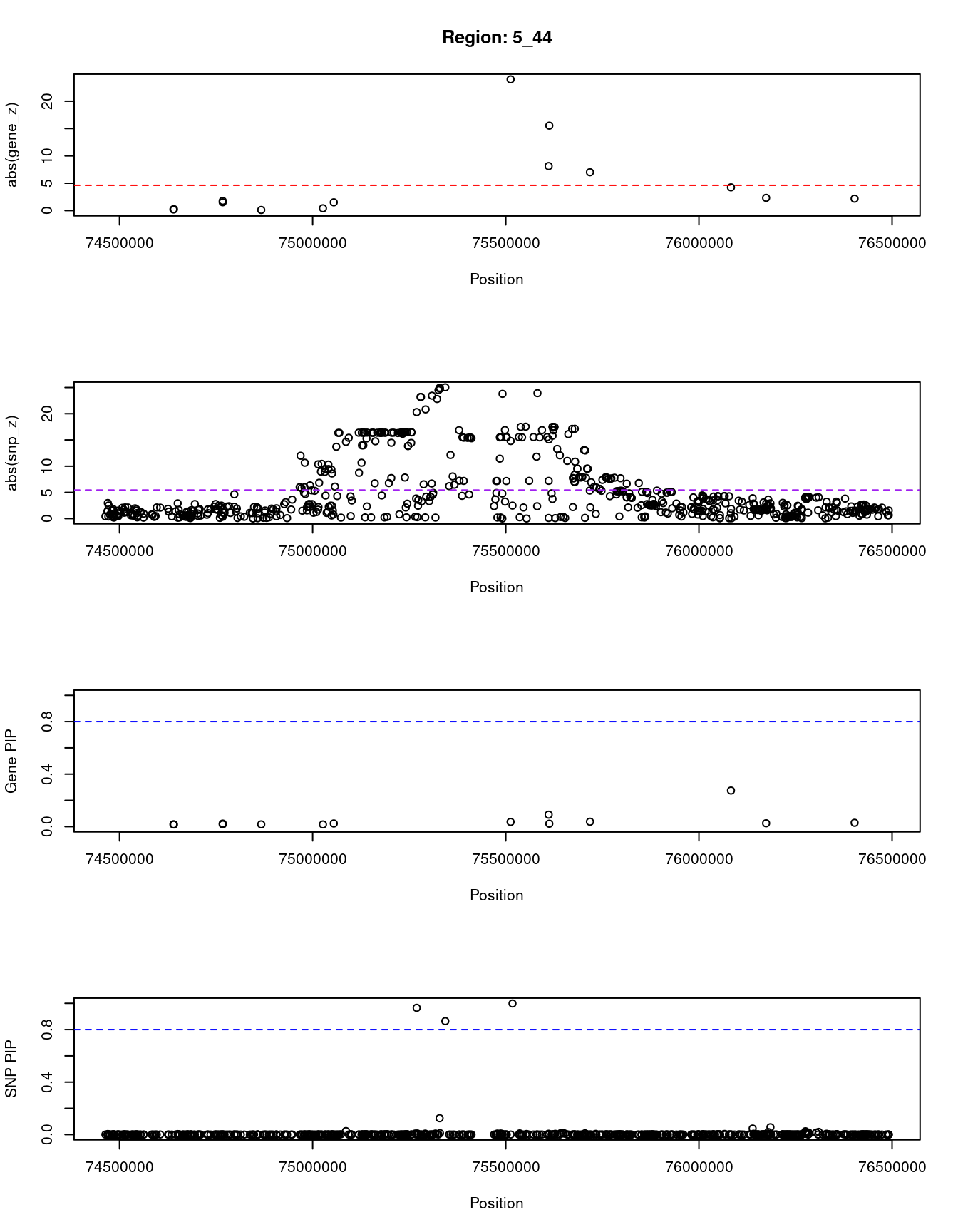
[1] "Region: 1_67"
genename region_tag susie_pip mu2 PVE
12677 RP11-356N1.2 1_67 0.008511622 4.878611 1.208450e-07
1230 SLC25A24 1_67 0.110623224 19.040173 6.129676e-06
7834 FAM102B 1_67 0.011788953 15.577365 5.344284e-07
7835 HENMT1 1_67 0.025890373 14.844196 1.118447e-06
3407 STXBP3 1_67 0.014885979 16.452942 7.127567e-07
3879 GPSM2 1_67 0.010770874 6.955047 2.180074e-07
7836 AKNAD1 1_67 0.010770874 6.955047 2.180074e-07
3878 CLCC1 1_67 0.008350193 5.020227 1.219945e-07
11729 TAF13 1_67 0.012348876 13.705360 4.925362e-07
12493 TMEM167B 1_67 0.011335261 15.309187 5.050146e-07
366 SARS 1_67 0.008842491 24.257145 6.242156e-07
4984 GSTM1 1_67 0.026030135 38.410146 2.909663e-06
6093 CELSR2 1_67 0.008522725 109.890397 2.725577e-06
4991 PSRC1 1_67 0.276724165 192.009108 1.546284e-04
12546 MYBPHL 1_67 0.008420809 21.076428 5.165010e-07
4994 SORT1 1_67 0.008445869 20.739040 5.097454e-07
6084 SYPL2 1_67 0.016453770 49.423212 2.366555e-06
7840 ATXN7L2 1_67 0.011482988 14.603293 4.880070e-07
9731 CYB561D1 1_67 0.008784524 51.319596 1.311964e-06
10499 AMIGO1 1_67 0.015914284 29.501073 1.366297e-06
7232 GPR61 1_67 0.008286089 9.151074 2.206693e-07
680 GNAI3 1_67 0.059461335 36.597463 6.332948e-06
8972 GSTM4 1_67 0.019958084 16.303653 9.469435e-07
3409 AMPD2 1_67 0.057412268 44.195010 7.384112e-06
12337 GSTM2 1_67 0.025354958 25.302853 1.867036e-06
4987 GSTM3 1_67 0.010048438 7.235686 2.115917e-07
4986 GSTM5 1_67 0.008934125 9.401227 2.444313e-07
z
12677 -0.2308337
1230 -0.6406888
7834 -4.6211180
7835 -1.8537405
3407 2.9914703
3879 1.0104135
7836 1.0104135
3878 -1.7197469
11729 -2.8413870
12493 1.5934686
366 -7.0857282
4984 6.5012768
6093 13.7257529
4991 -22.9681237
12546 -7.1894797
4994 -7.1373768
6084 9.7928029
7840 5.9371453
9731 9.9823505
10499 -3.1539161
7232 -4.2425343
680 -3.8408490
8972 -0.3531182
3409 8.4673955
12337 4.6633215
4987 -1.0142898
4986 2.0991563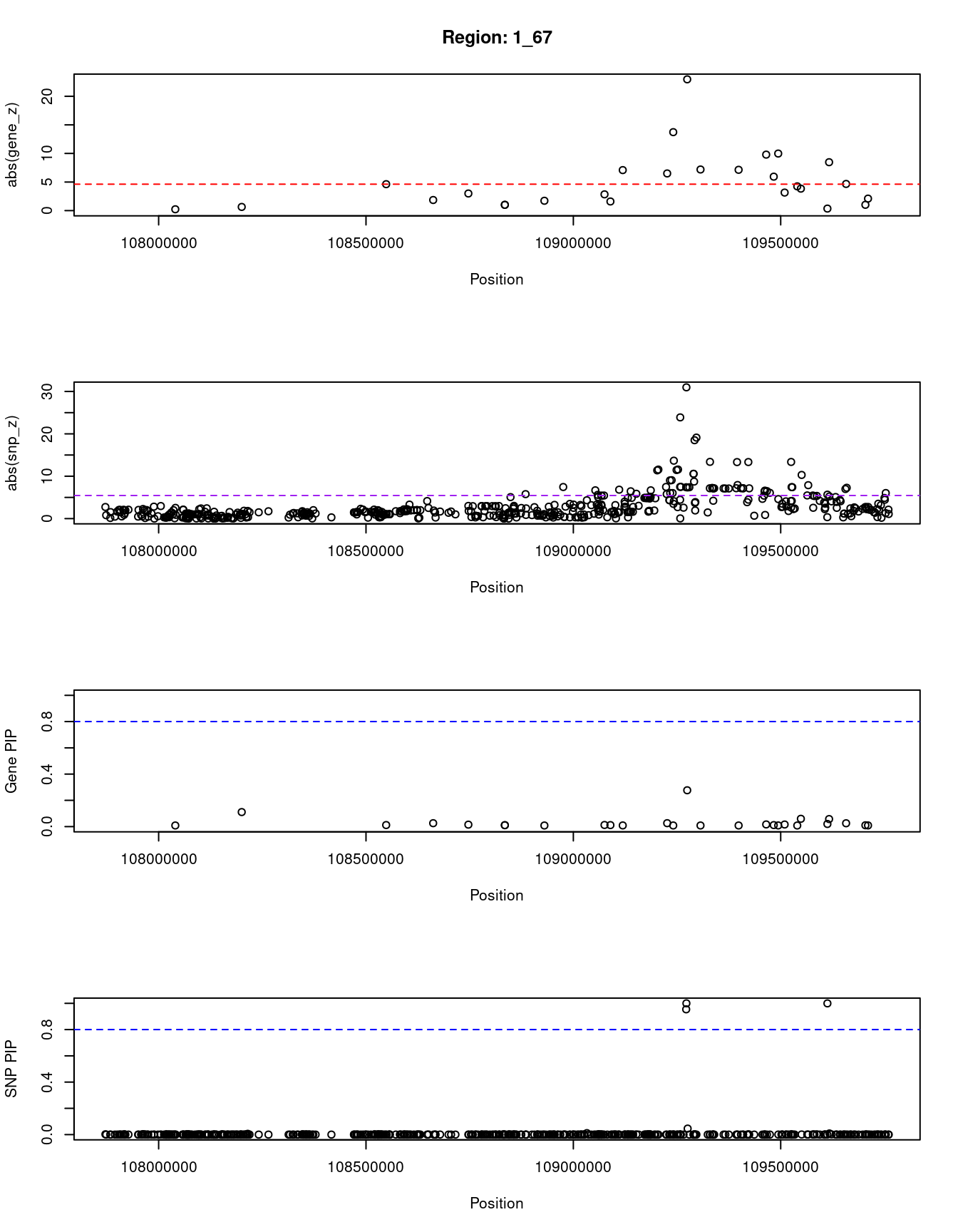
[1] "Region: 1_34"
genename region_tag susie_pip mu2 PVE
587 YIPF1 1_34 8.060219e-14 10.381893 2.435251e-18
1144 HSPB11 1_34 4.374279e-14 5.108528 6.503131e-19
3401 LRRC42 1_34 4.352074e-14 5.262271 6.664842e-19
3399 TCEANC2 1_34 4.196643e-14 4.741349 5.790610e-19
3400 TMEM59 1_34 4.152234e-14 4.644457 5.612251e-19
7325 CDCP2 1_34 9.736656e-14 12.363077 3.503134e-18
12499 CYB5RL 1_34 1.219025e-13 14.007672 4.969342e-18
3403 MRPL37 1_34 4.407585e-14 4.994136 6.405919e-19
7327 SSBP3 1_34 4.063416e-14 4.635790 5.481953e-19
7784 ACOT11 1_34 1.343370e-13 17.148420 6.704093e-18
10806 MROH7 1_34 1.868505e-12 45.488781 2.473540e-16
7785 FAM151A 1_34 4.327649e-13 27.822867 3.504082e-17
13202 TTC4 1_34 4.096723e-14 4.885584 5.824697e-19
7786 PARS2 1_34 8.781864e-14 10.323831 2.638444e-18
108 TTC22 1_34 1.326717e-13 15.011189 5.795802e-18
3387 DHCR24 1_34 6.250556e-14 9.053461 1.646848e-18
9036 PCSK9 1_34 8.982431e-01 105.002053 2.744808e-04
12945 RP11-90C4.1 1_34 5.484502e-14 7.932894 1.266162e-18
z
587 1.0226599
1144 -0.2303441
3401 0.2854315
3399 -0.1853540
3400 0.2100252
7325 -1.2055941
12499 1.1595815
3403 0.5344672
7327 -0.6008954
7784 -1.8518413
10806 3.9761380
7785 2.0257851
13202 -0.1056123
7786 -1.1551925
108 1.1662870
3387 -0.3806125
9036 17.2108693
12945 -0.1113431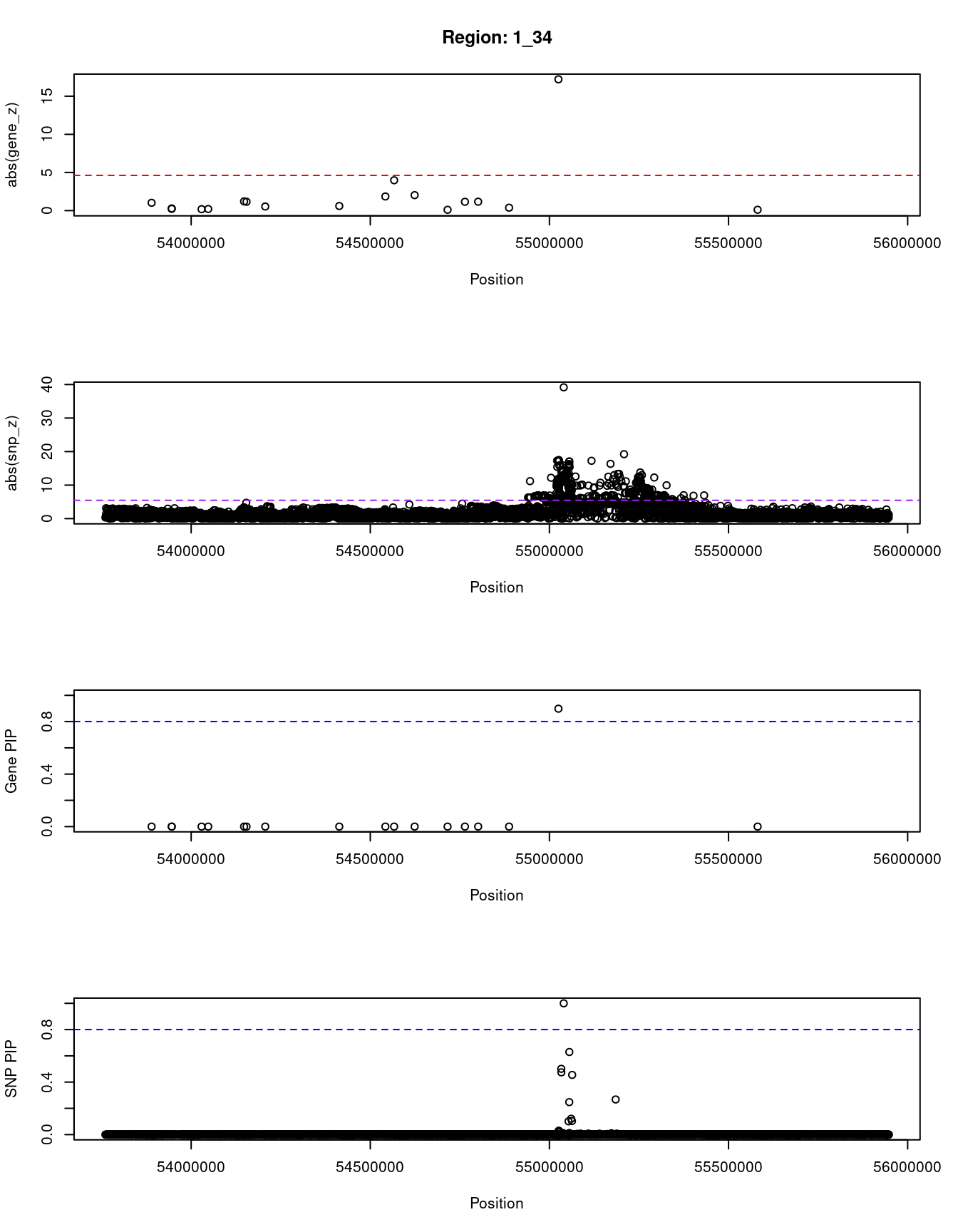
SNPs with highest PIPs
#snps with PIP>0.8 or 20 highest PIPs
head(ctwas_snp_res[order(-ctwas_snp_res$susie_pip),report_cols_snps],
max(sum(ctwas_snp_res$susie_pip>0.8), 20)) id region_tag susie_pip mu2 PVE
30777 rs611917 1_67 1.0000000 1138.55893 3.313415e-03
71715 rs1042034 2_13 1.0000000 268.95086 7.826962e-04
71721 rs934197 2_13 1.0000000 412.65819 1.200911e-03
322093 rs115740542 6_20 1.0000000 188.10302 5.474142e-04
367498 rs12208357 6_103 1.0000000 298.30479 8.681215e-04
404987 rs763798411 7_65 1.0000000 75043.88217 2.183914e-01
786687 rs73013176 19_9 1.0000000 242.45975 7.056022e-04
786725 rs137992968 19_9 1.0000000 244.44260 7.113727e-04
788882 rs3794991 19_15 1.0000000 512.84094 1.492461e-03
795899 rs62117204 19_31 1.0000000 829.08849 2.412799e-03
795917 rs111794050 19_31 1.0000000 826.73709 2.405956e-03
795950 rs814573 19_31 1.0000000 2427.15171 7.063456e-03
795952 rs113345881 19_31 1.0000000 845.59050 2.460823e-03
795955 rs12721109 19_31 1.0000000 1458.48490 4.244458e-03
853040 rs11591147 1_34 1.0000000 1354.64033 3.942251e-03
871345 rs528437193 2_54 1.0000000 5898.10409 1.716456e-02
976871 rs144230436 10_61 1.0000000 5154.29904 1.499995e-02
1100993 rs374141296 19_34 1.0000000 18407.46817 5.356910e-02
1106858 rs202143810 20_38 1.0000000 16806.02686 4.890861e-02
1061513 rs77542162 17_39 1.0000000 243.99385 7.100668e-04
806227 rs34507316 20_13 1.0000000 102.21251 2.974571e-04
322826 rs454182 6_22 1.0000000 167.03515 4.861029e-04
71666 rs11679386 2_12 1.0000000 174.05511 5.065322e-04
71724 rs548145 2_13 1.0000000 727.86017 2.118206e-03
494719 rs2437818 9_53 1.0000000 84.96770 2.472715e-04
502644 rs115478735 9_70 1.0000000 350.82136 1.020954e-03
71801 rs1848922 2_13 1.0000000 245.31857 7.139219e-04
729234 rs12149380 16_38 1.0000000 142.14901 4.136796e-04
796290 rs150262789 19_32 1.0000000 82.44781 2.399382e-04
59047 rs822928 1_121 1.0000000 121.96034 3.549269e-04
754280 rs1801689 17_38 1.0000000 89.97567 2.618457e-04
443015 rs4738679 8_45 1.0000000 120.62778 3.510489e-04
426829 rs7012814 8_12 1.0000000 102.22222 2.974854e-04
1061518 rs740516 17_39 1.0000000 118.59904 3.451449e-04
404998 rs4997569 7_65 1.0000000 74949.10279 2.181156e-01
1079090 rs148356565 19_10 1.0000000 105.20233 3.061580e-04
79162 rs72800939 2_28 1.0000000 63.04812 1.834816e-04
584116 rs4937122 11_77 0.9999997 80.73680 2.349588e-04
7646 rs79598313 1_18 0.9999996 52.85775 1.538257e-04
441620 rs140753685 8_42 0.9999995 62.77798 1.826953e-04
1100990 rs113176985 19_34 0.9999991 18388.02029 5.351246e-02
54649 rs2807848 1_112 0.9999989 63.98624 1.862115e-04
461125 rs6470359 8_83 0.9999956 362.17786 1.053999e-03
461128 rs13252684 8_83 0.9999849 296.19267 8.619618e-04
806226 rs6075251 20_13 0.9999844 74.50597 2.168226e-04
1078684 rs379309 19_10 0.9999831 76.42620 2.224105e-04
788913 rs113619686 19_15 0.9999828 78.84578 2.294517e-04
58997 rs6586405 1_121 0.9999652 56.24873 1.636884e-04
348935 rs9496567 6_67 0.9999646 43.39708 1.262890e-04
318364 rs11376017 6_13 0.9999343 74.89139 2.179333e-04
322072 rs72834643 6_20 0.9999228 47.56769 1.384200e-04
871350 rs2232748 2_54 0.9999040 5528.30463 1.608683e-02
698787 rs2070895 15_27 0.9998265 64.94659 1.889737e-04
367682 rs56393506 6_104 0.9997910 137.95054 4.013774e-04
367646 rs117733303 6_104 0.9997270 100.58291 2.926348e-04
729277 rs57186116 16_38 0.9997010 77.37089 2.250961e-04
30845 rs41313290 1_67 0.9996527 44.81969 1.303882e-04
538411 rs17875416 10_71 0.9996040 42.77777 1.244418e-04
494692 rs2297400 9_53 0.9992951 44.85553 1.304458e-04
79026 rs139029940 2_27 0.9991948 41.70698 1.212772e-04
1079069 rs36022659 19_10 0.9991446 163.28530 4.747836e-04
1091202 rs62115559 19_30 0.9990805 218.93562 6.365569e-04
280072 rs7701166 5_44 0.9989458 41.74451 1.213561e-04
788522 rs2302209 19_14 0.9988206 47.40383 1.377911e-04
404990 rs10274607 7_65 0.9985122 74854.32653 2.175157e-01
380879 rs56130071 7_19 0.9984407 107.73708 3.130457e-04
461116 rs2980875 8_83 0.9980352 633.92948 1.841226e-03
734729 rs2255451 16_48 0.9977545 42.31186 1.228587e-04
324048 rs28780090 6_26 0.9972192 64.61971 1.875322e-04
581455 rs75542613 11_71 0.9972110 39.46218 1.145219e-04
786708 rs147985405 19_9 0.9971834 2991.91019 8.682482e-03
431347 rs1495743 8_20 0.9968520 45.28373 1.313691e-04
581450 rs3135506 11_71 0.9968166 165.18459 4.791871e-04
811180 rs76981217 20_24 0.9957238 37.86348 1.097185e-04
621586 rs653178 12_67 0.9956545 108.48633 3.143431e-04
1090774 rs55840997 19_30 0.9950079 73.08471 2.116281e-04
442983 rs56386732 8_45 0.9924999 36.63704 1.058208e-04
138888 rs709149 3_9 0.9908560 39.07459 1.126744e-04
811131 rs6029132 20_24 0.9900871 43.93722 1.265978e-04
605002 rs7397189 12_36 0.9894455 36.53653 1.052058e-04
324071 rs62407548 6_26 0.9889379 84.75000 2.439097e-04
30111 rs1730862 1_66 0.9877001 31.82720 9.148373e-05
145898 rs9834932 3_24 0.9869064 73.91419 2.122873e-04
590025 rs11048034 12_9 0.9840744 41.02285 1.174827e-04
661133 rs2332328 14_3 0.9833522 49.88471 1.427568e-04
729010 rs4396539 16_37 0.9821794 31.88560 9.113931e-05
564249 rs174553 11_34 0.9767736 160.89318 4.573534e-04
323234 rs28986304 6_23 0.9767213 49.61396 1.410246e-04
79039 rs13430143 2_27 0.9759468 106.74594 3.031781e-04
796273 rs34942359 19_32 0.9717124 63.05336 1.783061e-04
624540 rs1169300 12_74 0.9711999 76.44526 2.160625e-04
477044 rs1556516 9_16 0.9700584 82.27290 2.322603e-04
79042 rs4076834 2_27 0.9687378 497.58791 1.402802e-03
221301 rs1458038 4_54 0.9674394 57.84775 1.628660e-04
757507 rs4969183 17_44 0.9672484 53.57873 1.508171e-04
321911 rs75080831 6_19 0.9662625 64.69946 1.819349e-04
280013 rs10062361 5_44 0.9658709 235.78515 6.627593e-04
403917 rs3197597 7_61 0.9644754 30.84290 8.656986e-05
1091285 rs185920692 19_30 0.9624772 62.06563 1.738449e-04
30776 rs614174 1_67 0.9543018 997.00335 2.768871e-03
244030 rs114756490 4_100 0.9495856 28.70009 7.931177e-05
871140 rs116298919 2_54 0.9479192 195.51625 5.393547e-04
633761 rs1012130 13_11 0.9451492 52.27240 1.437782e-04
469180 rs7024888 9_3 0.9373299 28.96077 7.899924e-05
799682 rs34003091 19_39 0.9369114 112.80320 3.075674e-04
729275 rs9652628 16_38 0.9368747 141.57287 3.859951e-04
624874 rs11057830 12_76 0.9362409 28.65852 7.808394e-05
1078955 rs146898772 19_10 0.9335190 84.70661 2.301234e-04
786713 rs34008246 19_9 0.9307697 430.07037 1.164936e-03
819537 rs62219001 21_2 0.9289824 28.88066 7.807911e-05
388248 rs141379002 7_33 0.9239433 28.57089 7.682266e-05
367492 rs9456502 6_103 0.9214539 35.21585 9.443481e-05
811184 rs73124945 20_24 0.9201328 34.63702 9.274945e-05
796190 rs377297589 19_32 0.9148043 55.56087 1.479168e-04
323263 rs3130253 6_23 0.9116315 30.12442 7.992053e-05
507594 rs10905277 10_8 0.9110444 30.82776 8.173381e-05
565079 rs6591179 11_36 0.9084156 29.38535 7.768475e-05
744739 rs117859452 17_17 0.9055281 28.07439 7.398310e-05
600089 rs2638250 12_25 0.9044876 29.71427 7.821463e-05
170070 rs189174 3_74 0.9040516 48.84111 1.284988e-04
494712 rs2777788 9_53 0.8964806 70.33165 1.834898e-04
71718 rs78610189 2_13 0.8947460 65.27825 1.699764e-04
195069 rs36205397 4_4 0.8941437 43.54356 1.133056e-04
577719 rs201912654 11_59 0.8853021 43.61893 1.123794e-04
815319 rs10641149 20_32 0.8769782 30.21548 7.711496e-05
619679 rs1196760 12_63 0.8759164 28.99816 7.391855e-05
485030 rs11144506 9_35 0.8649418 29.86618 7.517734e-05
280036 rs3843482 5_44 0.8645555 453.70001 1.141516e-03
806207 rs78348000 20_13 0.8593281 33.23065 8.310328e-05
1052156 rs2908806 17_7 0.8535522 41.23789 1.024346e-04
351671 rs12199109 6_73 0.8455182 28.95792 7.125422e-05
584119 rs74612335 11_77 0.8435624 78.48155 1.926660e-04
633753 rs1799955 13_11 0.8396620 85.78265 2.096159e-04
729215 rs12708919 16_38 0.8379278 165.05612 4.024932e-04
71518 rs6531234 2_12 0.8373807 44.76669 1.090933e-04
357874 rs9321207 6_87 0.8346463 33.48227 8.132756e-05
809925 rs11167269 20_21 0.8289012 62.92394 1.517885e-04
827915 rs149577713 21_19 0.8265083 35.40947 8.517005e-05
39376 rs1795240 1_84 0.8253529 28.82322 6.923130e-05
534888 rs10882161 10_59 0.8247620 32.49835 7.800281e-05
811149 rs6102034 20_24 0.8240734 107.53455 2.578898e-04
811325 rs11086801 20_25 0.8166312 120.68906 2.868231e-04
826778 rs2835302 21_16 0.8144176 28.99333 6.871722e-05
426840 rs13265179 8_12 0.8121150 39.53294 9.343230e-05
976844 rs915506 10_61 0.8046116 5153.93515 1.206828e-02
z
30777 30.975273
71715 -16.573036
71721 -33.060888
322093 12.532321
367498 -12.282337
404987 -3.272149
786687 16.232742
786725 10.752566
788882 21.492060
795899 44.672230
795917 33.599649
795950 -55.537887
795952 34.318568
795955 46.325818
853040 39.164934
871345 -1.194340
976871 -4.153975
1100993 2.412211
1106858 5.163023
1061513 -16.625724
806227 6.814661
322826 -4.779053
71666 -11.909428
71724 -33.086010
494719 -6.333973
502644 -19.011790
71801 -25.412292
729234 4.164582
796290 10.898464
59047 -12.369141
754280 -9.396430
443015 11.699924
426829 -10.906064
1061518 11.279140
404998 2.984117
1079090 10.875418
79162 7.845728
584116 -12.147947
7646 -7.024638
441620 -7.799241
1100990 -2.692042
54649 7.882775
461125 -9.646876
461128 -11.964411
806226 2.329832
1078684 8.426981
788913 -0.593903
58997 -8.960936
348935 6.340216
318364 8.507919
322072 6.048695
871350 4.165781
698787 -7.734663
367682 -14.088321
367646 -10.097959
729277 -7.714555
30845 5.646803
538411 6.266313
494692 -6.605676
79026 -6.814991
1079069 9.873513
1091202 14.946045
280072 2.484790
788522 -6.636049
404990 2.866958
380879 -10.978916
461116 22.102229
734729 6.362828
324048 -6.871357
581455 6.534400
786708 48.935175
431347 6.515969
581450 -12.372986
811180 -7.692477
621586 -11.050062
1090774 8.331826
442983 7.012272
138888 6.781974
811131 6.762459
605002 5.770964
324071 -8.257335
30111 5.284644
145898 8.481579
590025 -6.133690
661133 -7.035042
729010 5.232860
564249 12.656944
323234 -7.382502
79039 3.344504
796273 7.009565
624540 -8.685477
477044 8.992146
79042 20.108567
221301 7.417851
757507 -7.169275
321911 7.906709
280013 -20.320600
403917 5.045242
1091285 7.096889
30776 -7.395089
244030 -4.988910
871140 -1.134879
633761 2.781022
469180 5.055827
799682 10.423688
729275 -11.950504
624874 -4.929635
1078955 -3.660667
786713 -12.967795
819537 4.948445
388248 -4.896981
367492 -5.963991
811184 7.775426
796190 6.786505
323263 -5.641451
507594 -5.125802
565079 -4.893333
744739 3.851670
600089 5.037754
170070 -6.767794
494712 5.737015
71718 8.385467
195069 -6.159378
577719 6.305597
815319 -5.075761
619679 4.866700
485030 -5.042667
280036 -25.034352
806207 -5.220624
1052156 6.026359
351671 -4.857045
584119 -11.904831
633753 6.693636
729215 -11.302762
71518 7.170830
357874 -5.401634
809925 7.795037
827915 -3.316824
39376 4.846186
534888 5.475649
811149 11.189979
811325 -10.975177
826778 4.653743
426840 7.414877
976844 4.639318SNPs with largest effect sizes
#plot PIP vs effect size
#plot(ctwas_snp_res$susie_pip, ctwas_snp_res$mu2, xlab="PIP", ylab="mu^2", main="SNP PIPs vs Effect Size")
#SNPs with 50 largest effect sizes
head(ctwas_snp_res[order(-ctwas_snp_res$mu2),report_cols_snps],50) id region_tag susie_pip mu2 PVE
404987 rs763798411 7_65 1.000000e+00 75043.88 2.183914e-01
404998 rs4997569 7_65 1.000000e+00 74949.10 2.181156e-01
404990 rs10274607 7_65 9.985122e-01 74854.33 2.175157e-01
405005 rs6952534 7_65 3.330669e-16 74720.13 7.242516e-17
404993 rs13230660 7_65 3.876101e-03 74695.51 8.425776e-04
405004 rs4730069 7_65 0.000000e+00 74656.20 0.000000e+00
404997 rs10242713 7_65 0.000000e+00 74394.16 0.000000e+00
405000 rs10249965 7_65 0.000000e+00 73784.47 0.000000e+00
405012 rs1013016 7_65 0.000000e+00 70758.63 0.000000e+00
405037 rs8180737 7_65 0.000000e+00 67146.84 0.000000e+00
405030 rs17778396 7_65 0.000000e+00 67130.43 0.000000e+00
405031 rs2237621 7_65 0.000000e+00 67100.37 0.000000e+00
405002 rs71562637 7_65 0.000000e+00 67067.58 0.000000e+00
405064 rs10224564 7_65 0.000000e+00 66978.19 0.000000e+00
405049 rs10255779 7_65 0.000000e+00 66943.24 0.000000e+00
405066 rs78132606 7_65 0.000000e+00 66623.96 0.000000e+00
405069 rs4610671 7_65 0.000000e+00 66540.27 0.000000e+00
405071 rs12669532 7_65 0.000000e+00 63778.61 0.000000e+00
405028 rs2237618 7_65 0.000000e+00 62736.31 0.000000e+00
405073 rs118089279 7_65 0.000000e+00 62114.71 0.000000e+00
405060 rs73188303 7_65 0.000000e+00 62067.31 0.000000e+00
405070 rs560364150 7_65 0.000000e+00 49230.37 0.000000e+00
405056 rs10261738 7_65 0.000000e+00 40201.47 0.000000e+00
405011 rs368909701 7_65 0.000000e+00 30863.84 0.000000e+00
405010 rs2299297 7_65 0.000000e+00 24319.08 0.000000e+00
404996 rs6961668 7_65 0.000000e+00 22263.45 0.000000e+00
405054 rs56384866 7_65 0.000000e+00 20005.36 0.000000e+00
1100993 rs374141296 19_34 1.000000e+00 18407.47 5.356910e-02
1100990 rs113176985 19_34 9.999991e-01 18388.02 5.351246e-02
1100983 rs35295508 19_34 1.073992e-05 18300.22 5.719756e-07
1100997 rs2946865 19_34 8.376866e-06 18264.01 4.452439e-07
1100988 rs73056069 19_34 1.448471e-05 18257.96 7.696307e-07
1100981 rs61371437 19_34 3.338087e-04 18235.00 1.771429e-05
1100985 rs2878354 19_34 1.465398e-05 18203.70 7.763107e-07
1100971 rs739349 19_34 3.329284e-04 18160.68 1.759557e-05
1100972 rs756628 19_34 3.055162e-04 18160.50 1.614665e-05
1100968 rs739347 19_34 1.263878e-04 18127.40 6.667472e-06
1100969 rs2073614 19_34 7.032501e-05 18111.89 3.706756e-06
1100974 rs2077300 19_34 8.011674e-04 18067.36 4.212483e-05
1100978 rs73056059 19_34 6.301036e-04 18032.90 3.306723e-05
1100964 rs4802613 19_34 9.588347e-05 18032.59 5.031787e-06
1100998 rs60815603 19_34 3.754717e-06 18004.99 1.967389e-07
1101001 rs1316885 19_34 5.002071e-06 17915.21 2.607907e-07
1101003 rs60746284 19_34 7.229350e-06 17898.70 3.765660e-07
1101006 rs2946863 19_34 5.368889e-06 17885.94 2.794580e-07
1100999 rs35443645 19_34 3.817582e-06 17865.82 1.984868e-07
1100962 rs10403394 19_34 1.262844e-04 17776.26 6.532967e-06
1100963 rs17555056 19_34 4.863870e-05 17774.77 2.515975e-06
1100979 rs73056062 19_34 8.292969e-06 17561.20 4.238231e-07
1101009 rs553431297 19_34 2.623183e-05 17361.21 1.325344e-06
z
404987 -3.2721491
404998 2.9841166
404990 2.8669582
405005 2.8884240
404993 2.9479628
405004 2.8658735
404997 2.8123983
405000 2.8497381
405012 -2.3988524
405037 2.8328454
405030 2.7980012
405031 2.8029605
405002 2.6635936
405064 2.7911904
405049 2.8135791
405066 2.7728082
405069 2.7249742
405071 2.7702573
405028 2.4663255
405073 2.6667208
405060 2.4217031
405070 1.8694582
405056 2.6665109
405011 0.7778883
405010 -0.7963506
404996 3.2318586
405054 1.8825782
1100993 2.4122106
1100990 -2.6920415
1100983 -2.6673841
1100997 -2.7042362
1100988 -2.8620364
1100981 -2.4817929
1100985 -2.8021669
1100971 -2.4874835
1100972 -2.4791524
1100968 -2.4351089
1100969 -2.4259236
1100974 -2.6423569
1100978 -2.6287434
1100964 -2.4736415
1100998 -2.5756724
1101001 -2.6081557
1101003 -2.7767892
1101006 -2.6328340
1100999 -2.4998457
1100962 -2.4990920
1100963 -2.4726709
1100979 -2.1759694
1101009 -2.4664028SNPs with highest PVE
#SNPs with 50 highest pve
head(ctwas_snp_res[order(-ctwas_snp_res$PVE),report_cols_snps],50) id region_tag susie_pip mu2 PVE
404987 rs763798411 7_65 1.000000000 75043.8822 0.2183914318
404998 rs4997569 7_65 0.999999999 74949.1028 0.2181156062
404990 rs10274607 7_65 0.998512209 74854.3265 0.2175156901
1100993 rs374141296 19_34 1.000000000 18407.4682 0.0535691013
1100990 rs113176985 19_34 0.999999118 18388.0203 0.0535124572
1106858 rs202143810 20_38 1.000000000 16806.0269 0.0489086140
1106855 rs6089961 20_38 0.659268971 16617.4341 0.0318820989
1106857 rs2738758 20_38 0.659268971 16617.4341 0.0318820989
1106836 rs35201382 20_38 0.390730204 16612.9410 0.0188905155
871345 rs528437193 2_54 1.000000000 5898.1041 0.0171645624
871350 rs2232748 2_54 0.999903973 5528.3046 0.0160868334
976871 rs144230436 10_61 1.000000000 5154.2990 0.0149999535
1106838 rs2750483 20_38 0.273949328 16612.0846 0.0132438629
976844 rs915506 10_61 0.804611585 5153.9351 0.0120682843
1106837 rs67468102 20_38 0.224353422 16610.0367 0.0108448511
976875 rs1047370 10_61 0.657939484 5153.0692 0.0098667069
786708 rs147985405 19_9 0.997183368 2991.9102 0.0086824818
1106833 rs2315009 20_38 0.171477459 16607.2822 0.0082875452
795950 rs814573 19_31 1.000000000 2427.1517 0.0070634557
795955 rs12721109 19_31 1.000000000 1458.4849 0.0042444580
871334 rs14976 2_54 0.343874039 4237.4593 0.0042405798
871333 rs2289976 2_54 0.327595532 4232.7082 0.0040353072
853040 rs11591147 1_34 1.000000000 1354.6403 0.0039422513
30777 rs611917 1_67 1.000000000 1138.5589 0.0033134149
30776 rs614174 1_67 0.954301843 997.0033 0.0027688707
795952 rs113345881 19_31 1.000000000 845.5905 0.0024608231
795899 rs62117204 19_31 1.000000000 829.0885 0.0024127992
795917 rs111794050 19_31 1.000000000 826.7371 0.0024059562
871335 rs72843872 2_54 0.189897665 4244.9608 0.0023459222
71724 rs548145 2_13 1.000000000 727.8602 0.0021182063
461116 rs2980875 8_83 0.998035201 633.9295 0.0018412261
871331 rs1374370 2_54 0.138632765 4224.5803 0.0017043930
788882 rs3794991 19_15 1.000000000 512.8409 0.0014924610
79042 rs4076834 2_27 0.968737828 497.5879 0.0014028020
871461 rs7316 2_54 0.506706141 895.8882 0.0013210836
71721 rs934197 2_13 1.000000000 412.6582 0.0012009108
786713 rs34008246 19_9 0.930769714 430.0704 0.0011649360
280036 rs3843482 5_44 0.864555544 453.7000 0.0011415160
461125 rs6470359 8_83 0.999995563 362.1779 0.0010539992
786698 rs8102273 19_9 0.590586660 600.1720 0.0010315248
502644 rs115478735 9_70 1.000000000 350.8214 0.0010209544
871467 rs3024821 2_54 0.387897543 896.0980 0.0010115628
367498 rs12208357 6_103 1.000000000 298.3048 0.0008681215
461128 rs13252684 8_83 0.999984856 296.1927 0.0008619618
404993 rs13230660 7_65 0.003876101 74695.5132 0.0008425776
976820 rs10786262 10_61 0.052942128 5146.0216 0.0007928541
71715 rs1042034 2_13 1.000000000 268.9509 0.0007826962
71801 rs1848922 2_13 1.000000000 245.3186 0.0007139219
786725 rs137992968 19_9 1.000000000 244.4426 0.0007113727
1061513 rs77542162 17_39 1.000000000 243.9938 0.0007100668
z
404987 -3.2721491
404998 2.9841166
404990 2.8669582
1100993 2.4122106
1100990 -2.6920415
1106858 5.1630228
1106855 -5.4768594
1106857 -5.4768594
1106836 -5.4829409
871345 -1.1943396
871350 4.1657806
976871 -4.1539753
1106838 -5.4803179
976844 4.6393182
1106837 -5.4817963
976875 4.6387213
786708 48.9351750
1106833 -5.4827739
795950 -55.5378874
795955 46.3258178
871334 2.7468945
871333 2.7557689
853040 39.1649340
30777 30.9752731
30776 -7.3950888
795952 34.3185677
795899 44.6722304
795917 33.5996491
871335 2.7104140
71724 -33.0860100
461116 22.1022294
871331 2.7663193
788882 21.4920605
79042 20.1085673
871461 0.9857653
71721 -33.0608878
786713 -12.9677952
280036 -25.0343522
461125 -9.6468762
786698 14.1676792
502644 -19.0117902
871467 0.9590447
367498 -12.2823367
461128 -11.9644114
404993 2.9479628
976820 4.5865580
71715 -16.5730356
71801 -25.4122923
786725 10.7525659
1061513 -16.6257239SNPs with largest z scores
#histogram of (abs) SNP z scores
hist(abs(ctwas_snp_res$z))
#SNPs with 50 largest z scores
head(ctwas_snp_res[order(-abs(ctwas_snp_res$z)),report_cols_snps],50) id region_tag susie_pip mu2 PVE
795950 rs814573 19_31 1.000000e+00 2427.1517 7.063456e-03
786708 rs147985405 19_9 9.971834e-01 2991.9102 8.682482e-03
786703 rs73015020 19_9 2.395051e-03 2979.8260 2.076949e-05
786701 rs138175288 19_9 2.645903e-04 2975.6937 2.291302e-06
786702 rs138294113 19_9 5.610593e-05 2972.8789 4.854073e-07
786704 rs77140532 19_9 7.644535e-05 2973.0842 6.614219e-07
786705 rs112552009 19_9 1.004570e-05 2969.7824 8.682104e-08
786706 rs10412048 19_9 1.439373e-05 2969.8131 1.244007e-07
786700 rs55997232 19_9 2.788378e-10 2949.5890 2.393501e-12
795955 rs12721109 19_31 1.000000e+00 1458.4849 4.244458e-03
795899 rs62117204 19_31 1.000000e+00 829.0885 2.412799e-03
795886 rs1551891 19_31 0.000000e+00 476.0861 0.000000e+00
786709 rs17248769 19_9 0.000000e+00 2121.0722 0.000000e+00
786710 rs2228671 19_9 0.000000e+00 2109.8025 0.000000e+00
853040 rs11591147 1_34 1.000000e+00 1354.6403 3.942251e-03
786699 rs9305020 19_9 0.000000e+00 1898.6023 0.000000e+00
795946 rs405509 19_31 0.000000e+00 963.4870 0.000000e+00
795952 rs113345881 19_31 1.000000e+00 845.5905 2.460823e-03
795870 rs62120566 19_31 0.000000e+00 1415.6597 0.000000e+00
795917 rs111794050 19_31 1.000000e+00 826.7371 2.405956e-03
71724 rs548145 2_13 1.000000e+00 727.8602 2.118206e-03
795923 rs4802238 19_31 0.000000e+00 981.1816 0.000000e+00
71721 rs934197 2_13 1.000000e+00 412.6582 1.200911e-03
795864 rs188099946 19_31 0.000000e+00 1356.4681 0.000000e+00
795934 rs2972559 19_31 0.000000e+00 1356.8465 0.000000e+00
795858 rs201314191 19_31 0.000000e+00 1256.0146 0.000000e+00
795925 rs56394238 19_31 0.000000e+00 986.2288 0.000000e+00
795902 rs2965169 19_31 0.000000e+00 321.6927 0.000000e+00
795926 rs3021439 19_31 0.000000e+00 867.1722 0.000000e+00
30777 rs611917 1_67 1.000000e+00 1138.5589 3.313415e-03
71751 rs12997242 2_13 1.834533e-12 373.0173 1.991474e-15
795933 rs12162222 19_31 0.000000e+00 1157.0572 0.000000e+00
71725 rs478588 2_13 2.058931e-11 673.6905 4.036663e-14
795863 rs62119327 19_31 0.000000e+00 1103.0302 0.000000e+00
71726 rs56350433 2_13 4.322098e-13 349.6183 4.397533e-16
71731 rs56079819 2_13 4.312106e-13 348.8427 4.377633e-16
71735 rs2337383 2_13 4.420908e-13 341.8865 4.398592e-16
71736 rs56090741 2_13 4.413137e-13 341.3520 4.383996e-16
71740 rs7568899 2_13 4.609646e-13 332.3664 4.458666e-16
71741 rs62135036 2_13 4.624079e-13 332.1316 4.469467e-16
71747 rs11687710 2_13 4.570788e-13 331.3359 4.407374e-16
71752 rs532300 2_13 7.592815e-13 619.1780 1.368166e-15
71753 rs558130 2_13 7.592815e-13 619.1778 1.368165e-15
71754 rs533211 2_13 7.592815e-13 619.1778 1.368165e-15
71775 rs574461 2_13 7.817080e-13 618.7263 1.407549e-15
71777 rs494465 2_13 7.772671e-13 618.5566 1.399169e-15
71755 rs528113 2_13 7.551737e-13 618.8891 1.360129e-15
71760 rs1652418 2_13 7.539525e-13 618.5494 1.357184e-15
71762 rs563696 2_13 7.510659e-13 618.4287 1.351724e-15
71750 rs312979 2_13 7.348566e-13 618.4019 1.322494e-15
z
795950 -55.53789
786708 48.93517
786703 48.79563
786701 48.78069
786702 48.75193
786704 48.73799
786705 48.70516
786706 48.70123
786700 48.52431
795955 46.32582
795899 44.67223
795886 42.26680
786709 40.84249
786710 40.70262
853040 39.16493
786699 34.84073
795946 34.63979
795952 34.31857
795870 33.73539
795917 33.59965
71724 -33.08601
795923 -33.07569
71721 -33.06089
795864 33.04407
795934 -32.28660
795858 32.06858
795925 -31.55187
795902 31.38057
795926 -31.04506
30777 30.97527
71751 -30.81528
795933 -30.49671
71725 -30.48811
795863 30.41868
71726 -30.23229
71731 -30.19307
71735 -29.88780
71736 -29.86248
71740 -29.70268
71741 -29.69428
71747 -29.63286
71752 -29.56783
71753 -29.56783
71754 -29.56783
71775 -29.56585
71777 -29.56317
71755 -29.56210
71760 -29.55946
71762 -29.55712
71750 -29.55544Gene set enrichment for genes with PIP>0.8
#GO enrichment analysis
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
#number of genes for gene set enrichment
length(genes)[1] 36if (length(genes)>0){
GO_enrichment <- enrichr(genes, dbs)
for (db in dbs){
print(db)
df <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
df <- df[df$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(df)
}
#DisGeNET enrichment
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <-disease_enrichment(entities=genes, vocabulary = "HGNC",
database = "CURATED" )
df <- res_enrich@qresult[1:10, c("Description", "FDR", "Ratio", "BgRatio")]
print(df)
#WebGestalt enrichment
library(WebGestaltR)
background <- ctwas_gene_res$genename
#listGeneSet()
databases <- c("pathway_KEGG", "disease_GLAD4U", "disease_OMIM")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
print(enrichResult[,c("description", "size", "overlap", "FDR", "database", "userId")])
}Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
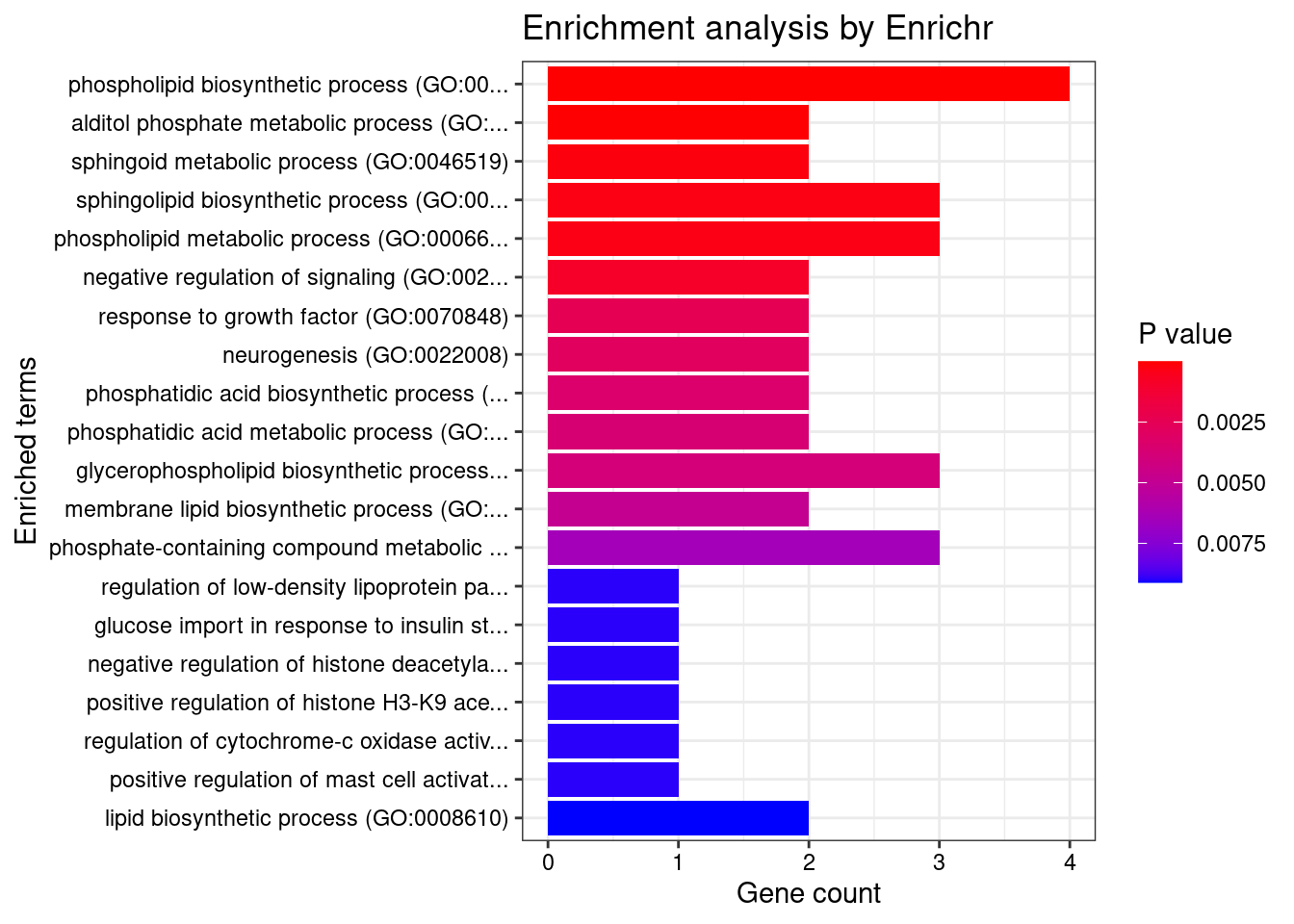
[1] "GO_Biological_Process_2021"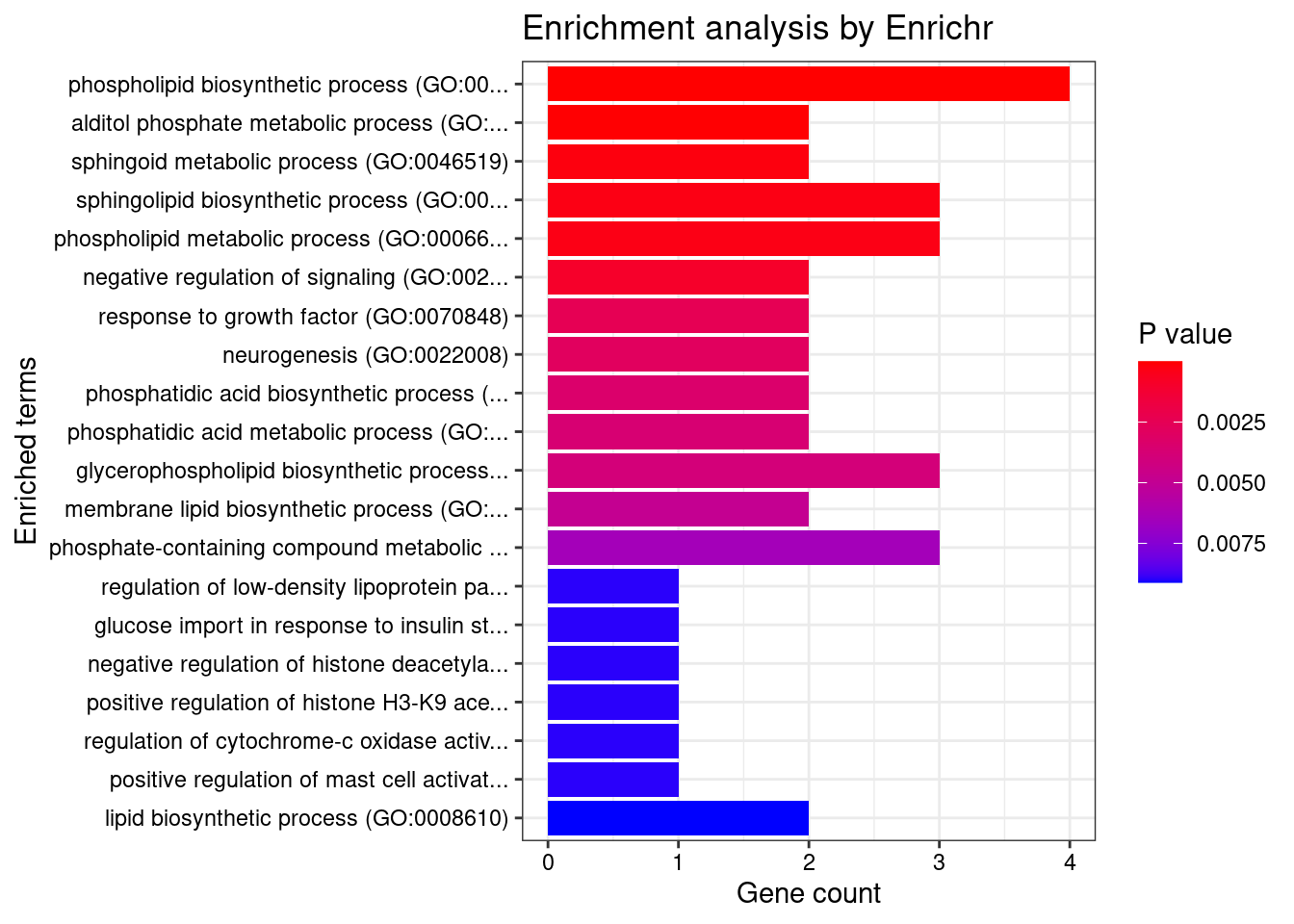
Term Overlap
1 phospholipid biosynthetic process (GO:0008654) 4/37
2 alditol phosphate metabolic process (GO:0052646) 2/5
3 sphingoid metabolic process (GO:0046519) 2/12
4 sphingolipid biosynthetic process (GO:0030148) 3/74
5 phospholipid metabolic process (GO:0006644) 3/76
Adjusted.P.value Genes
1 0.0002131762 GPAM;SPHK2;CPNE3;ABCA8
2 0.0059803621 GPAM;ACP6
3 0.0261060926 SPHK2;KDSR
4 0.0262131634 SPHK2;KDSR;ABCA8
5 0.0262131634 PLPPR2;GPAM;ACP6
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)SPTY2D1 gene(s) from the input list not found in DisGeNET CURATEDMPND gene(s) from the input list not found in DisGeNET CURATEDFCGRT gene(s) from the input list not found in DisGeNET CURATEDNTN5 gene(s) from the input list not found in DisGeNET CURATEDTMED4 gene(s) from the input list not found in DisGeNET CURATEDFAM117B gene(s) from the input list not found in DisGeNET CURATEDPELO gene(s) from the input list not found in DisGeNET CURATEDACP6 gene(s) from the input list not found in DisGeNET CURATEDLINC01184 gene(s) from the input list not found in DisGeNET CURATEDAC007950.2 gene(s) from the input list not found in DisGeNET CURATEDPARP9 gene(s) from the input list not found in DisGeNET CURATEDUBASH3B gene(s) from the input list not found in DisGeNET CURATEDC10orf88 gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
48 Learning Disorders 0.02645503 2/22 29/9703
73 Relapsing Fever 0.02645503 1/22 1/9703
75 Sclerosis 0.02645503 1/22 1/9703
132 Schnitzler Syndrome 0.02645503 1/22 1/9703
145 Adult Learning Disorders 0.02645503 2/22 29/9703
146 Learning Disturbance 0.02645503 2/22 29/9703
147 Learning Disabilities 0.02645503 2/22 29/9703
188 Developmental Academic Disorder 0.02645503 2/22 29/9703
199 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2I 0.02645503 1/22 1/9703
200 MUSCULAR DYSTROPHY, CONGENITAL, 1C 0.02645503 1/22 1/9703******************************************
* *
* Welcome to WebGestaltR ! *
* *
******************************************Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet,
minNum = minNum, : No significant gene set is identified based on FDR 0.05!
NULLSensitivity, specificity and precision for silver standard genes
library("readxl")
known_annotations <- read_xlsx("data/summary_known_genes_annotations.xlsx", sheet="LDL")New names:
* `` -> ...4
* `` -> ...5known_annotations <- unique(known_annotations$`Gene Symbol`)
unrelated_genes <- ctwas_gene_res$genename[!(ctwas_gene_res$genename %in% known_annotations)]
#number of genes in known annotations
print(length(known_annotations))[1] 69#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 40#assign ctwas, TWAS, and bystander genes
ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z)>sig_thresh]
novel_genes <- ctwas_genes[!(ctwas_genes %in% twas_genes)]
#significance threshold for TWAS
print(sig_thresh)[1] 4.618904#number of ctwas genes
length(ctwas_genes)[1] 36#number of TWAS genes
length(twas_genes)[1] 227#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z
10255 KLHDC7A 1_13 0.9544827 18.66743 5.185288e-05 4.124187
7879 ACP6 1_73 0.9932775 21.77810 6.295220e-05 4.613976
5327 IL1RN 2_67 0.9410797 20.79413 5.694919e-05 4.455379
4022 ACVR1C 2_94 0.9057641 19.38221 5.109033e-05 -4.185879
8904 PDHB 3_40 0.9140947 24.02176 6.390228e-05 3.361078
5607 PARP9 3_76 0.8452748 22.78261 5.604304e-05 3.744644
13258 LINC01184 5_78 0.8593862 18.23008 4.559291e-05 -3.918127
1003 TPD52 8_57 0.9370510 18.68606 5.095669e-05 -4.121885
1237 CPNE3 8_62 0.8147843 18.04139 4.277922e-05 3.753687
1559 SCD 10_64 0.8966611 18.64373 4.864984e-05 -4.541468
3724 GPAM 10_70 0.8129956 19.45034 4.601885e-05 4.000718
3633 CCND2 12_4 0.9318759 19.26549 5.224666e-05 -4.128258
11865 C2CD4A 15_28 0.8713009 21.59986 5.476958e-05 4.535165
3674 KDSR 18_35 0.9199769 18.92509 5.066818e-05 -4.119957
161 MPND 19_5 0.9197113 19.83813 5.309731e-05 -4.322018
2313 PLPPR2 19_10 0.9790947 28.36151 8.081172e-05 3.965665
10447 FKRP 19_33 0.9482623 20.61788 5.689744e-05 4.425174
2199 FCGRT 19_34 0.9967913 10378.34712 3.010598e-02 -4.165895
num_eqtl
10255 1
7879 4
5327 1
4022 1
8904 2
5607 1
13258 2
1003 2
1237 3
1559 1
3724 1
3633 2
11865 1
3674 2
161 2
2313 1
10447 5
2199 2#sensitivity / recall
sensitivity <- rep(NA,2)
names(sensitivity) <- c("ctwas", "TWAS")
sensitivity["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
sensitivity["TWAS"] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
sensitivity ctwas TWAS
0.01449275 0.18840580 #specificity
specificity <- rep(NA,2)
names(specificity) <- c("ctwas", "TWAS")
specificity["ctwas"] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
specificity["TWAS"] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
specificity ctwas TWAS
0.9972912 0.9834378 #precision / PPV
precision <- rep(NA,2)
names(precision) <- c("ctwas", "TWAS")
precision["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
precision["TWAS"] <- sum(twas_genes %in% known_annotations)/length(twas_genes)
precision ctwas TWAS
0.02777778 0.05726872 #ROC curves
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1))
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=2)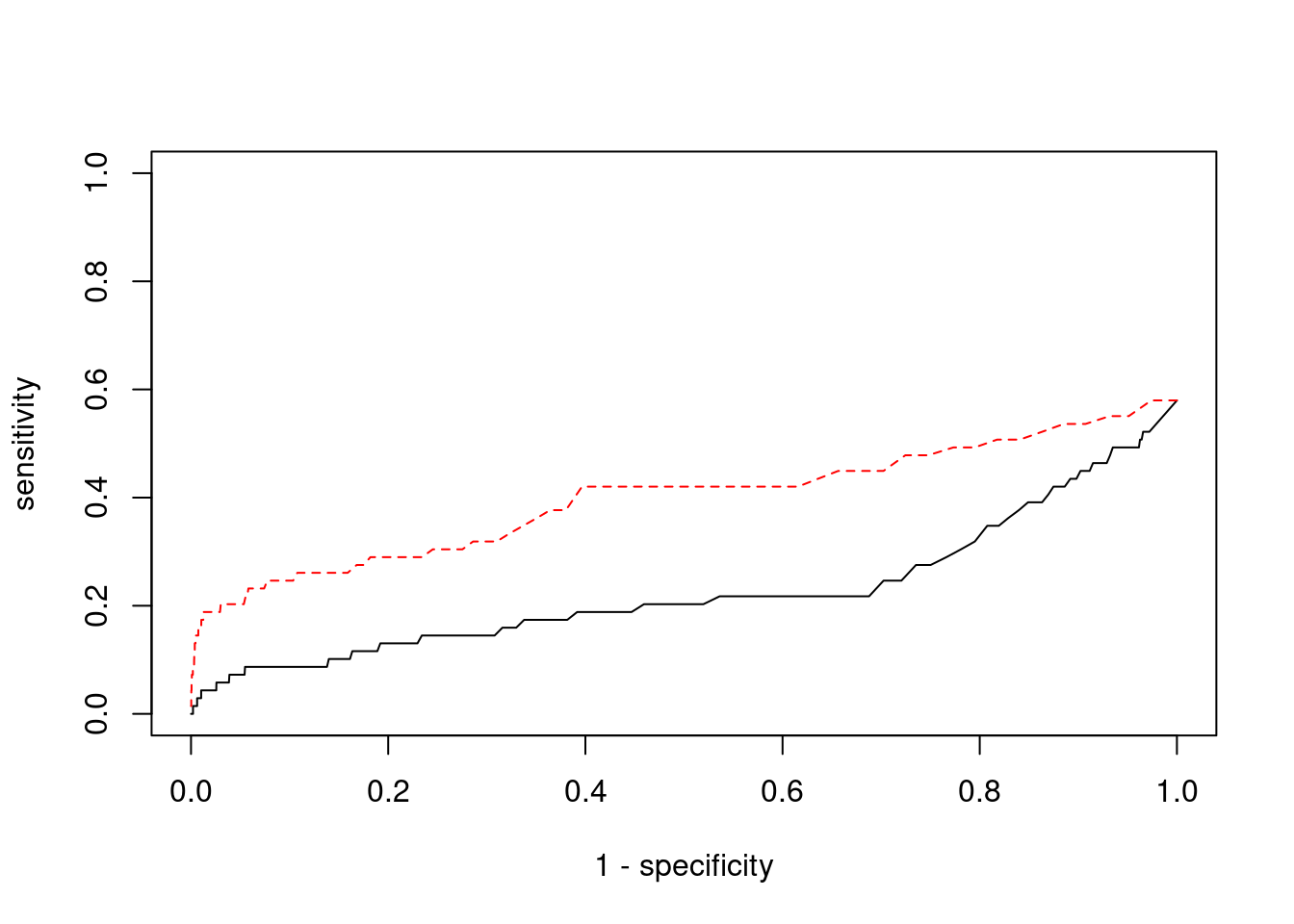
Sensitivity, specificity and precision for silver standard genes - bystanders only
This section first uses imputed silver standard genes to identify bystander genes within 1Mb. The bystander gene list is then subset to only genes with imputed expression in this analysis. Then, the ctwas and TWAS gene lists from this analysis are subset to only genes that are in the (subset) silver standard and bystander genes. These gene lists are then used to compute sensitivity, specificity and precision for ctwas and TWAS.
library(biomaRt)
library(GenomicRanges)Loading required package: stats4Loading required package: BiocGenericsLoading required package: parallel
Attaching package: 'BiocGenerics'The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLBThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, basename, cbind,
colnames, dirname, do.call, duplicated, eval, evalq, Filter,
Find, get, grep, grepl, intersect, is.unsorted, lapply, Map,
mapply, match, mget, order, paste, pmax, pmax.int, pmin,
pmin.int, Position, rank, rbind, Reduce, rownames, sapply,
setdiff, sort, table, tapply, union, unique, unsplit, which,
which.max, which.minLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following object is masked from 'package:base':
expand.gridLoading required package: IRangesLoading required package: GenomeInfoDbensembl <- useEnsembl(biomart="ENSEMBL_MART_ENSEMBL", dataset="hsapiens_gene_ensembl")
G_list <- getBM(filters= "chromosome_name", attributes= c("hgnc_symbol","chromosome_name","start_position","end_position","gene_biotype"), values=1:22, mart=ensembl)
G_list <- G_list[G_list$hgnc_symbol!="",]
G_list <- G_list[G_list$gene_biotype %in% c("protein_coding","lncRNA"),]
G_list$start <- G_list$start_position
G_list$end <- G_list$end_position
G_list_granges <- makeGRangesFromDataFrame(G_list, keep.extra.columns=T)
#remove genes without imputed expression from gene lists
known_annotations <- known_annotations[known_annotations %in% ctwas_gene_res$genename]
known_annotations_positions <- G_list[G_list$hgnc_symbol %in% known_annotations,]
half_window <- 1000000
known_annotations_positions$start <- known_annotations_positions$start_position - half_window
known_annotations_positions$end <- known_annotations_positions$end_position + half_window
known_annotations_positions$start[known_annotations_positions$start<1] <- 1
known_annotations_granges <- makeGRangesFromDataFrame(known_annotations_positions, keep.extra.columns=T)
bystanders <- findOverlaps(known_annotations_granges,G_list_granges)
bystanders <- unique(subjectHits(bystanders))
bystanders <- G_list$hgnc_symbol[bystanders]
bystanders <- unique(bystanders[!(bystanders %in% known_annotations)])
unrelated_genes <- bystanders
#save gene lists
save(known_annotations, file=paste0(results_dir, "/known_annotations.Rd"))
save(unrelated_genes, file=paste0(results_dir, "/bystanders.Rd"))
load(paste0(results_dir, "/known_annotations.Rd"))
load(paste0(results_dir, "/bystanders.Rd"))
#remove genes without imputed expression from bystander list
unrelated_genes <- unrelated_genes[unrelated_genes %in% ctwas_gene_res$genename]
#number of genes in known annotations (with imputed expression)
print(length(known_annotations))[1] 40#number of bystander genes (with imputed expression)
print(length(unrelated_genes))[1] 676#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.618904#number of ctwas genes (in known annotations or bystanders)
length(ctwas_genes)[1] 3#number of TWAS genes (in known annotations or bystanders)
length(twas_genes)[1] 60#sensitivity / recall
sensitivity <- rep(NA,2)
names(sensitivity) <- c("ctwas", "TWAS")
sensitivity["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
sensitivity["TWAS"] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
sensitivityctwas TWAS
0.025 0.325 #specificity / (1 - False Positive Rate)
specificity <- rep(NA,2)
names(specificity) <- c("ctwas", "TWAS")
specificity["ctwas"] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
specificity["TWAS"] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
specificity ctwas TWAS
0.9970414 0.9304734 #precision / PPV / (1 - False Discovery Rate)
precision <- rep(NA,2)
names(precision) <- c("ctwas", "TWAS")
precision["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
precision["TWAS"] <- sum(twas_genes %in% known_annotations)/length(twas_genes)
precision ctwas TWAS
0.3333333 0.2166667 #store sensitivity and specificity calculations for plots
sensitivity_plot <- sensitivity
specificity_plot <- specificity
#precision / PPV by PIP bin
pip_range <- c(0.2, 0.4, 0.6, 0.8, 1)
precision_range <- rep(NA, length(pip_range))
for (i in 1:length(pip_range)){
pip_upper <- pip_range[i]
if (i==1){
pip_lower <- 0
} else {
pip_lower <- pip_range[i-1]
}
#assign ctwas genes in PIP bin
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip_lower & ctwas_gene_res_subset$susie_pip<pip_upper]
precision_range[i] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
}
names(precision_range) <- paste(c(0, pip_range[-length(pip_range)]), pip_range,sep=" - ")
barplot(precision_range, ylim=c(0,1), main="Precision by PIP Range", xlab="PIP Range", ylab="Precision")
abline(h=0.2, lty=2)
abline(h=0.4, lty=2)
abline(h=0.6, lty=2)
abline(h=0.8, lty=2)
barplot(precision_range, add=T, col="darkgrey")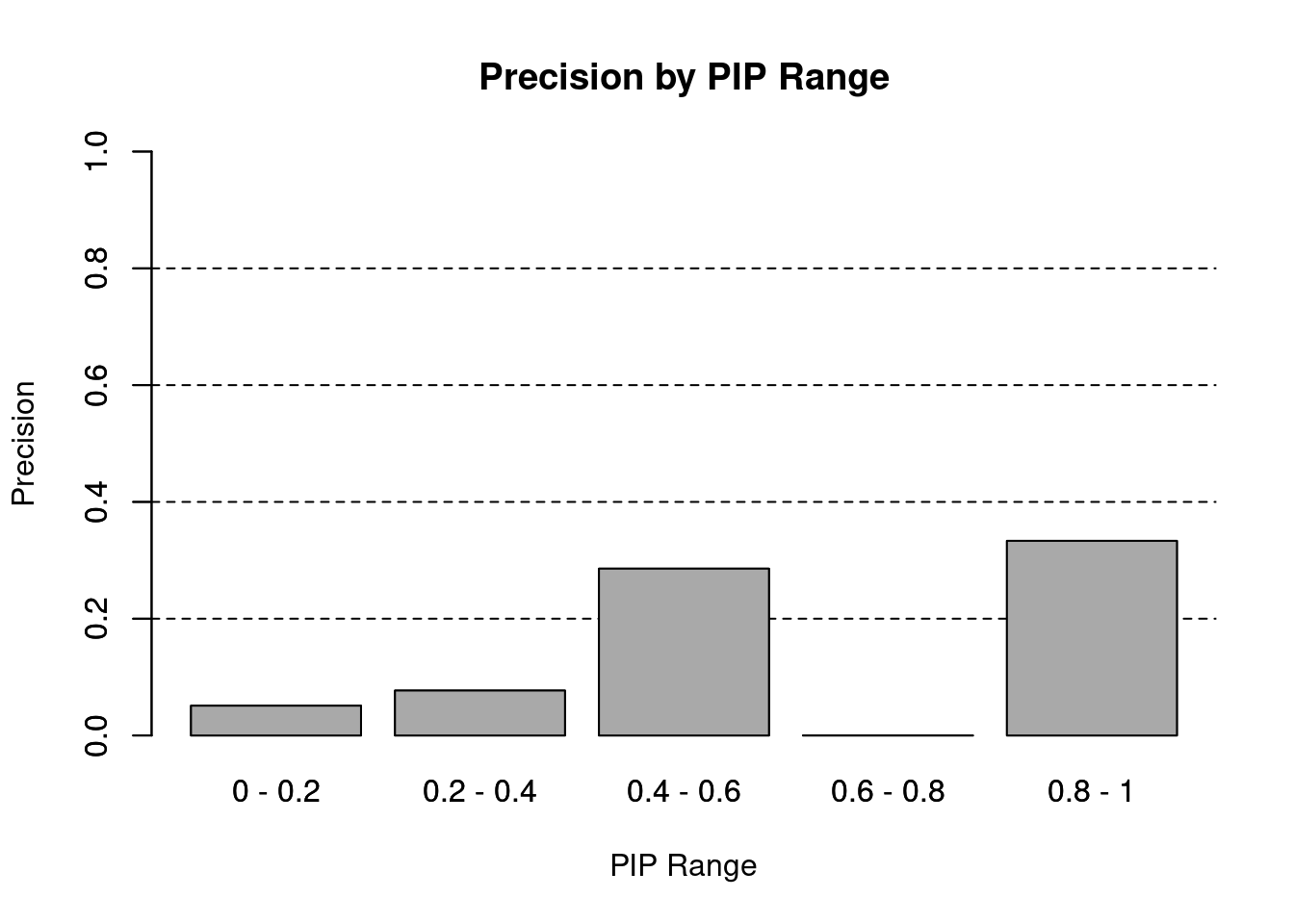
#precision / PPV by PIP threshold
#pip_range <- c(0.2, 0.4, 0.6, 0.8, 1)
pip_range <- c(0.5, 0.8, 1)
precision_range <- rep(NA, length(pip_range))
number_detected <- rep(NA, length(pip_range))
for (i in 1:length(pip_range)){
pip_upper <- pip_range[i]
if (i==1){
pip_lower <- 0
} else {
pip_lower <- pip_range[i-1]
}
#assign ctwas genes using PIP threshold
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip_lower]
number_detected[i] <- length(ctwas_genes)
precision_range[i] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
}
names(precision_range) <- paste0(">= ", c(0, pip_range[-length(pip_range)]))
precision_range <- precision_range*100
precision_range <- c(precision_range, precision["TWAS"]*100)
names(precision_range)[4] <- "TWAS Bonferroni"
number_detected <- c(number_detected, length(twas_genes))
barplot(precision_range, ylim=c(0,100), main="Precision for Distinguishing Silver Standard and Bystander Genes", xlab="PIP Threshold for Detection", ylab="% of Detected Genes in Silver Standard")
abline(h=20, lty=2)
abline(h=40, lty=2)
abline(h=60, lty=2)
abline(h=80, lty=2)
xx <- barplot(precision_range, add=T, col=c(rep("darkgrey",3), "white"))
text(x = xx, y = rep(0, length(number_detected)), label = paste0(number_detected, " detected"), pos = 3, cex=0.8)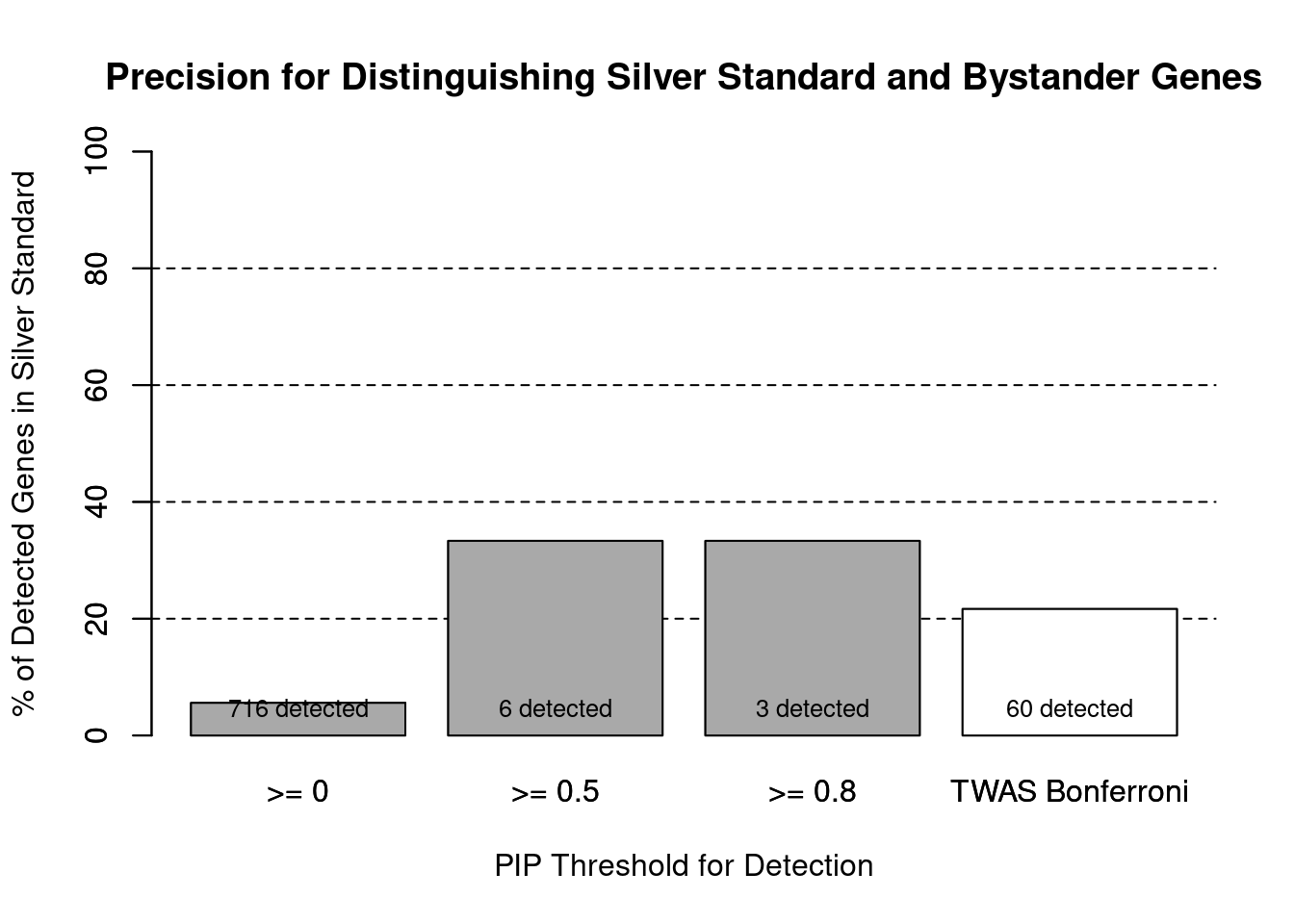
#text(x = xx, y = precision_range, label = paste0(round(precision_range,1), "%"), pos = 3, cex=0.8, offset = 1.5)
#false discovery rate by PIP threshold
barplot(100-precision_range, ylim=c(0,100), main="False Discovery Rate for Distinguishing Silver Standard and Bystander Genes", xlab="PIP Threshold for Detection", ylab="% Bystanders in Detected Genes")
abline(h=20, lty=2)
abline(h=40, lty=2)
abline(h=60, lty=2)
abline(h=80, lty=2)
xx <- barplot(100-precision_range, add=T, col=c(rep("darkgrey",3), "white"))
text(x = xx, y = rep(0, length(number_detected)), label = paste0(number_detected, " detected"), pos = 3, cex=0.8)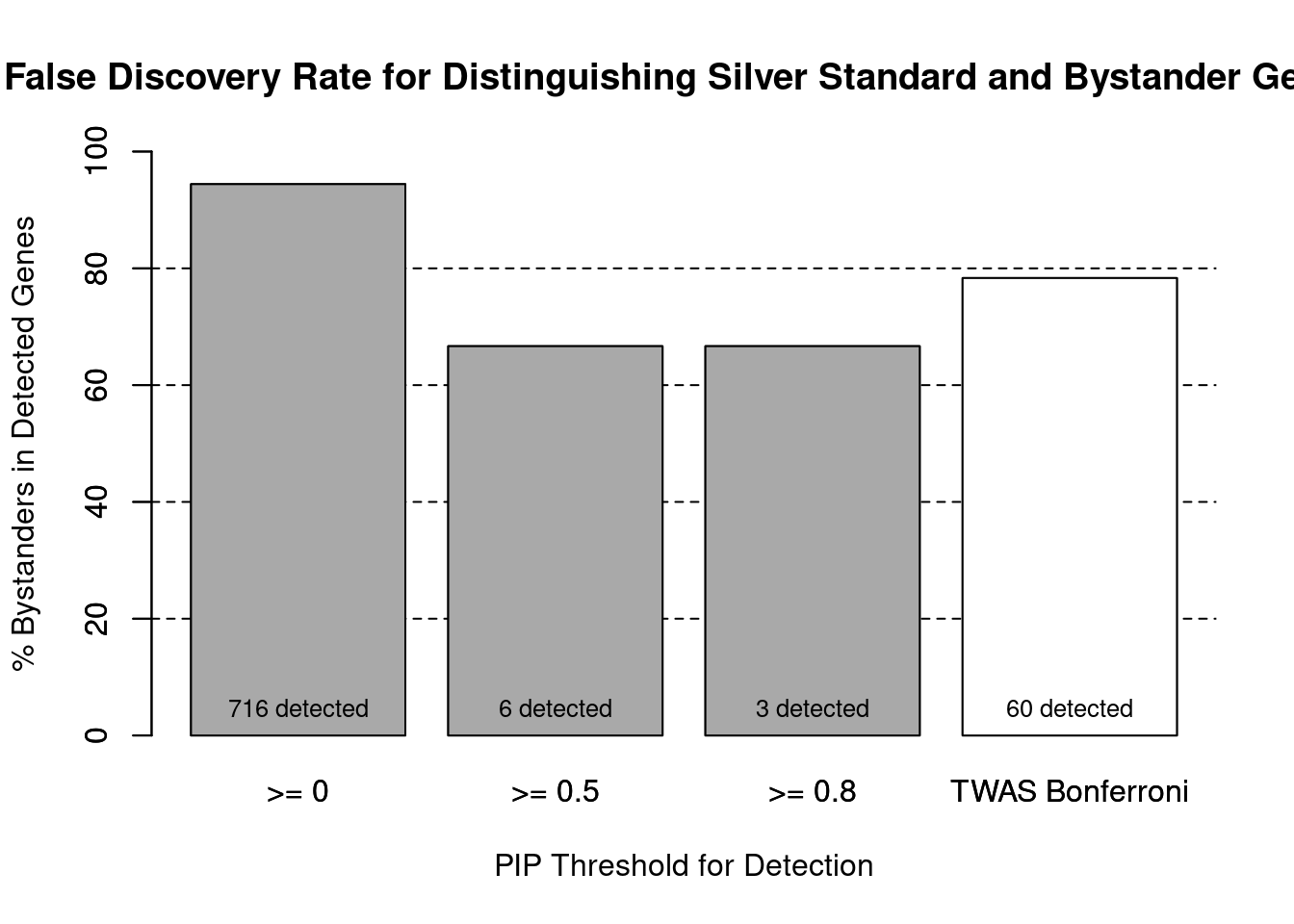
#text(x = xx, y = precision_range, label = paste0(round(precision_range,1), "%"), pos = 3, cex=0.8, offset = 1.5)
#ROC curves
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1), main="", xlab="1 - Specificity", ylab="Sensitivity")
title(expression("ROC Curve for cTWAS (black) and TWAS (" * phantom("red") * ")"))
title(expression(phantom("ROC Curve for cTWAS (black) and TWAS (") * "red" * phantom(")")), col.main="red")
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res_subset$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=1)
abline(a=0,b=1,lty=3)
#add previously computed points from the analysis
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
points(1-specificity_plot["ctwas"], sensitivity_plot["ctwas"], pch=21, bg="black")
points(1-specificity_plot["TWAS"], sensitivity_plot["TWAS"], pch=21, bg="red")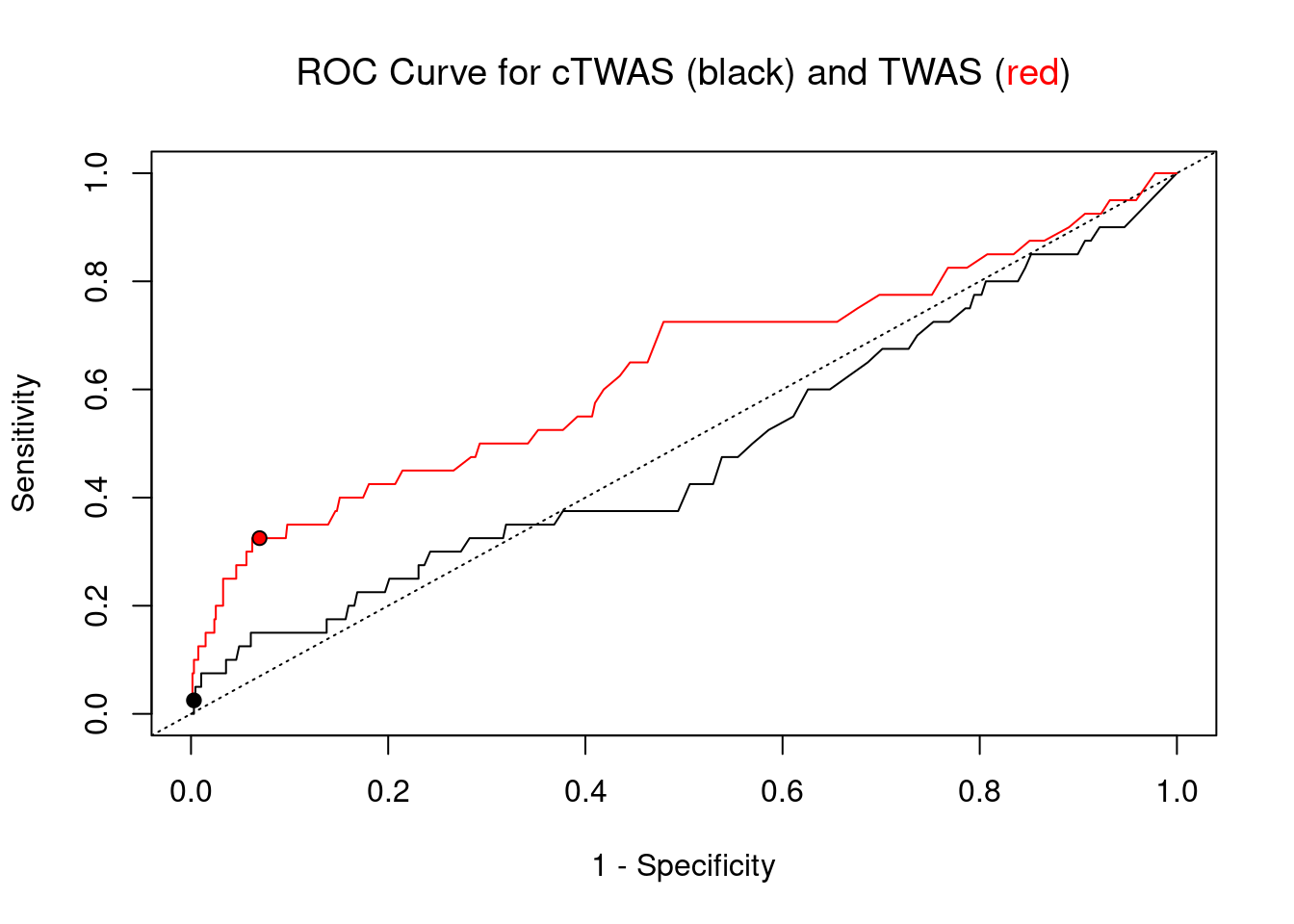
PIP Manhattan Plot
library(tibble)
library(tidyverse)── Attaching packages ────────────────────────────────── tidyverse 1.3.0 ──✔ tidyr 1.1.0 ✔ dplyr 1.0.7
✔ readr 1.4.0 ✔ stringr 1.4.0
✔ purrr 0.3.4 ✔ forcats 0.4.0── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::collapse() masks IRanges::collapse()
✖ dplyr::combine() masks BiocGenerics::combine()
✖ dplyr::desc() masks IRanges::desc()
✖ tidyr::expand() masks S4Vectors::expand()
✖ tidyr::extract() masks disgenet2r::extract()
✖ dplyr::filter() masks stats::filter()
✖ dplyr::first() masks S4Vectors::first()
✖ dplyr::lag() masks stats::lag()
✖ BiocGenerics::Position() masks ggplot2::Position(), base::Position()
✖ purrr::reduce() masks GenomicRanges::reduce(), IRanges::reduce()
✖ dplyr::rename() masks S4Vectors::rename()
✖ dplyr::select() masks biomaRt::select()
✖ dplyr::slice() masks IRanges::slice()full.gene.pip.summary <- data.frame(gene_name = ctwas_gene_res$genename,
gene_pip = ctwas_gene_res$susie_pip,
gene_id = ctwas_gene_res$id,
chr = as.integer(ctwas_gene_res$chrom),
start = ctwas_gene_res$pos / 1e3,
is_highlight = F, stringsAsFactors = F) %>% as_tibble()
full.gene.pip.summary$is_highlight <- full.gene.pip.summary$gene_pip > 0.80
don <- full.gene.pip.summary %>%
# Compute chromosome size
group_by(chr) %>%
summarise(chr_len=max(start)) %>%
# Calculate cumulative position of each chromosome
mutate(tot=cumsum(chr_len)-chr_len) %>%
dplyr::select(-chr_len) %>%
# Add this info to the initial dataset
left_join(full.gene.pip.summary, ., by=c("chr"="chr")) %>%
# Add a cumulative position of each SNP
arrange(chr, start) %>%
mutate( BPcum=start+tot)
axisdf <- don %>% group_by(chr) %>% summarize(center=( max(BPcum) + min(BPcum) ) / 2 )
x_axis_labels <- axisdf$chr
x_axis_labels[seq(1,21,2)] <- ""
ggplot(don, aes(x=BPcum, y=gene_pip)) +
# Show all points
ggrastr::geom_point_rast(aes(color=as.factor(chr)), size=2) +
scale_color_manual(values = rep(c("grey", "skyblue"), 22 )) +
# custom X axis:
# scale_x_continuous(label = axisdf$chr,
# breaks= axisdf$center,
# guide = guide_axis(n.dodge = 2)) +
scale_x_continuous(label = x_axis_labels,
breaks = axisdf$center) +
scale_y_continuous(expand = c(0, 0), limits = c(0,1.25), breaks=(1:5)*0.2, minor_breaks=(1:10)*0.1) + # remove space between plot area and x axis
# Add highlighted points
ggrastr::geom_point_rast(data=subset(don, is_highlight==T), color="orange", size=2) +
# Add label using ggrepel to avoid overlapping
ggrepel::geom_label_repel(data=subset(don, is_highlight==T),
aes(label=gene_name),
size=4,
min.segment.length = 0,
label.size = NA,
fill = alpha(c("white"),0)) +
# Custom the theme:
theme_bw() +
theme(
text = element_text(size = 14),
legend.position="none",
panel.border = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
) +
xlab("Chromosome") +
ylab("cTWAS PIP")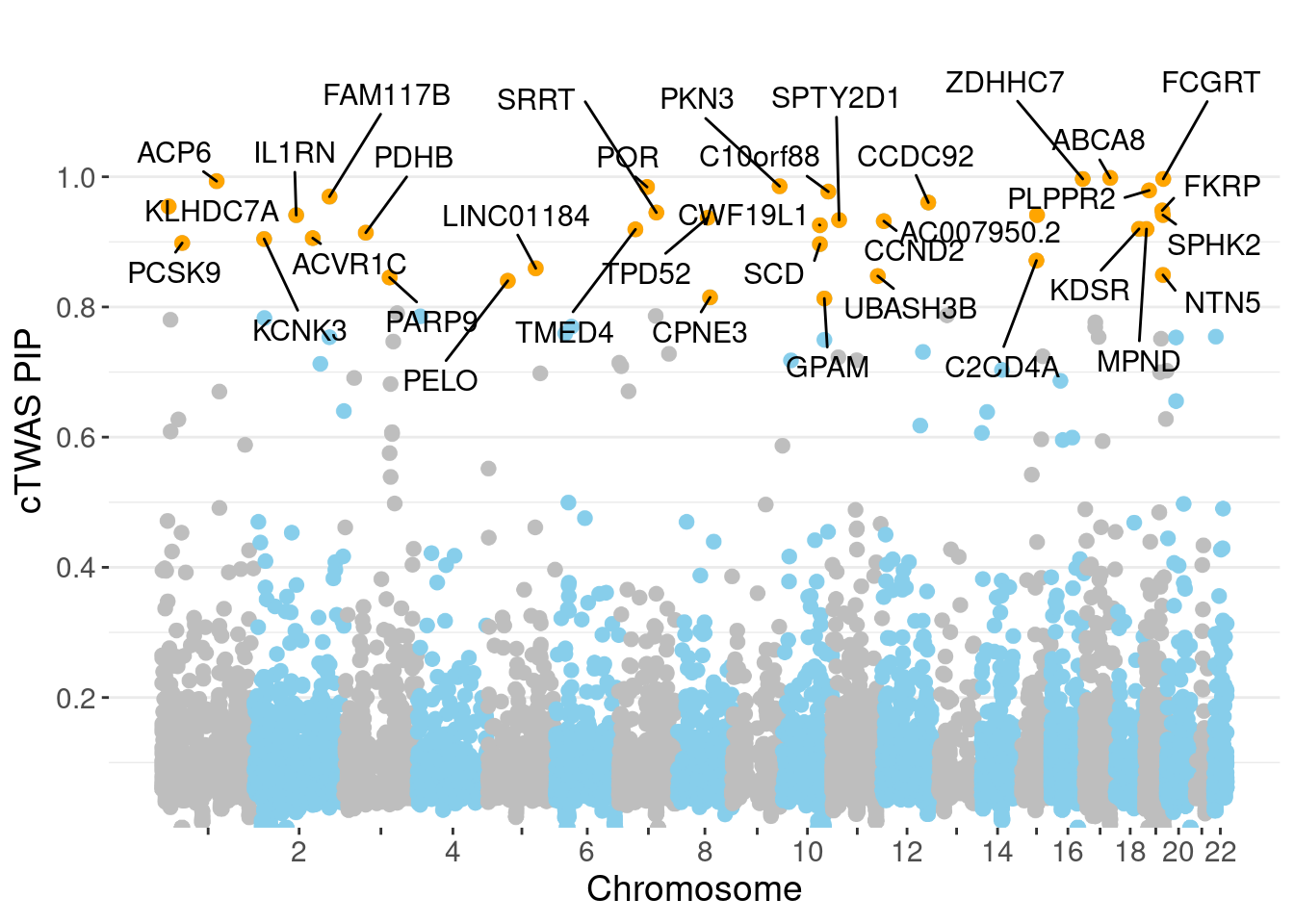
Locus Plots - 1_67
library(ctwas)
Attaching package: 'ctwas'The following object is masked _by_ '.GlobalEnv':
z_snplocus_plot <- function(region_tag, rerun_ctwas = F, plot_eqtl = T, label="cTWAS"){
region_tag1 <- unlist(strsplit(region_tag, "_"))[1]
region_tag2 <- unlist(strsplit(region_tag, "_"))[2]
a <- ctwas_res[ctwas_res$region_tag==region_tag,]
regionlist <- readRDS(paste0(results_dir, "/", analysis_id, "_ctwas.regionlist.RDS"))
region <- regionlist[[as.numeric(region_tag1)]][[region_tag2]]
R_snp_info <- do.call(rbind, lapply(region$regRDS, function(x){data.table::fread(paste0(tools::file_path_sans_ext(x), ".Rvar"))}))
if (isTRUE(rerun_ctwas)){
ld_exprfs <- paste0(results_dir, "/", analysis_id, "_expr_chr", 1:22, ".expr.gz")
temp_reg <- data.frame("chr" = paste0("chr",region_tag1), "start" = region$start, "stop" = region$stop)
write.table(temp_reg,
#file= paste0(results_dir, "/", analysis_id, "_ctwas.temp.reg.txt") ,
file= "temp_reg.txt",
row.names=F, col.names=T, sep="\t", quote = F)
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
z_gene_temp <- z_gene[z_gene$id %in% a$id[a$type=="gene"],]
z_snp_temp <- z_snp[z_snp$id %in% R_snp_info$id,]
ctwas_rss(z_gene_temp, z_snp_temp, ld_exprfs, ld_pgenfs = NULL,
ld_R_dir = dirname(region$regRDS)[1],
ld_regions_custom = "temp_reg.txt", thin = 1,
outputdir = ".", outname = "temp", ncore = 1, ncore.rerun = 1, prob_single = 0,
group_prior = estimated_group_prior, group_prior_var = estimated_group_prior_var,
estimate_group_prior = F, estimate_group_prior_var = F)
a <- data.table::fread("temp.susieIrss.txt", header = T)
rownames(z_snp_temp) <- z_snp_temp$id
z_snp_temp <- z_snp_temp[a$id[a$type=="SNP"],]
z_gene_temp <- z_gene_temp[a$id[a$type=="gene"],]
a$z <- NA
a$z[a$type=="SNP"] <- z_snp_temp$z
a$z[a$type=="gene"] <- z_gene_temp$z
}
a$ifcausal <- 0
focus <- a$id[a$type=="gene"][which.max(abs(a$z[a$type=="gene"]))]
a$ifcausal <- as.numeric(a$id==focus)
a$PVALUE <- (-log(2) - pnorm(abs(a$z), lower.tail=F, log.p=T))/log(10)
R_gene <- readRDS(region$R_g_file)
R_snp_gene <- readRDS(region$R_sg_file)
R_snp <- as.matrix(Matrix::bdiag(lapply(region$regRDS, readRDS)))
rownames(R_gene) <- region$gid
colnames(R_gene) <- region$gid
rownames(R_snp_gene) <- R_snp_info$id
colnames(R_snp_gene) <- region$gid
rownames(R_snp) <- R_snp_info$id
colnames(R_snp) <- R_snp_info$id
a$r2max <- NA
a$r2max[a$type=="gene"] <- R_gene[focus,a$id[a$type=="gene"]]
a$r2max[a$type=="SNP"] <- R_snp_gene[a$id[a$type=="SNP"],focus]
r2cut <- 0.4
colorsall <- c("#7fc97f", "#beaed4", "#fdc086")
layout(matrix(1:2, ncol = 1), widths = 1, heights = c(1.5,1.5), respect = FALSE)
par(mar = c(0, 4.1, 4.1, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 19, xlab=paste0("Chromosome ", region_tag1, " Position"),frame.plot=FALSE, col = "white", ylim= c(-0.1,1.1), ylab = "cTWAS PIP", xaxt = 'n')
grid()
points(a$pos[a$type=="SNP"], a$susie_pip[a$type == "SNP"], pch = 21, xlab="Genomic position", bg = colorsall[1])
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$susie_pip[a$type == "SNP" & a$r2max >r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$susie_pip[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$susie_pip[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$susie_pip[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$susie_pip[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
if (isTRUE(plot_eqtl)){
for (cgene in a[a$type=="gene" & a$ifcausal == 1, ]$id){
load(paste0(results_dir, "/",analysis_id, "_expr_chr", region_tag1, ".exprqc.Rd"))
eqtls <- rownames(wgtlist[[cgene]])
points(a[a$id %in% eqtls,]$pos, rep( -0.15, nrow(a[a$id %in% eqtls,])), pch = "|", col = "salmon", cex = 1.5)
}
}
legend(min(a$pos), y= 1.1 ,c("Gene", "SNP"), pch = c(22,21), title="Shape Legend", bty ='n', cex=0.6, title.adj = 0)
legend(min(a$pos), y= 0.7 ,c("Lead TWAS Gene", "R2 > 0.4", "R2 <= 0.4"), pch = 19, col = c("salmon", "purple", colorsall[1]), title="Color Legend", bty ='n', cex=0.6, title.adj = 0)
if (label=="cTWAS"){
text(a$pos[a$id==focus], a$susie_pip[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
par(mar = c(4.1, 4.1, 0.5, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 21, xlab=paste0("Chromosome ", region_tag1, " Position"), frame.plot=FALSE, bg = colorsall[1], ylab = "TWAS -log10(p value)", panel.first = grid(), ylim =c(0, max(a$PVALUE)*1.2))
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$PVALUE[a$type == "SNP" & a$r2max > r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$PVALUE[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$PVALUE[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$PVALUE[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$PVALUE[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
abline(h=-log10(alpha/nrow(ctwas_gene_res)), col ="red", lty = 2)
if (label=="TWAS"){
text(a$pos[a$id==focus], a$PVALUE[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
}
locus_plot("1_67", label="TWAS")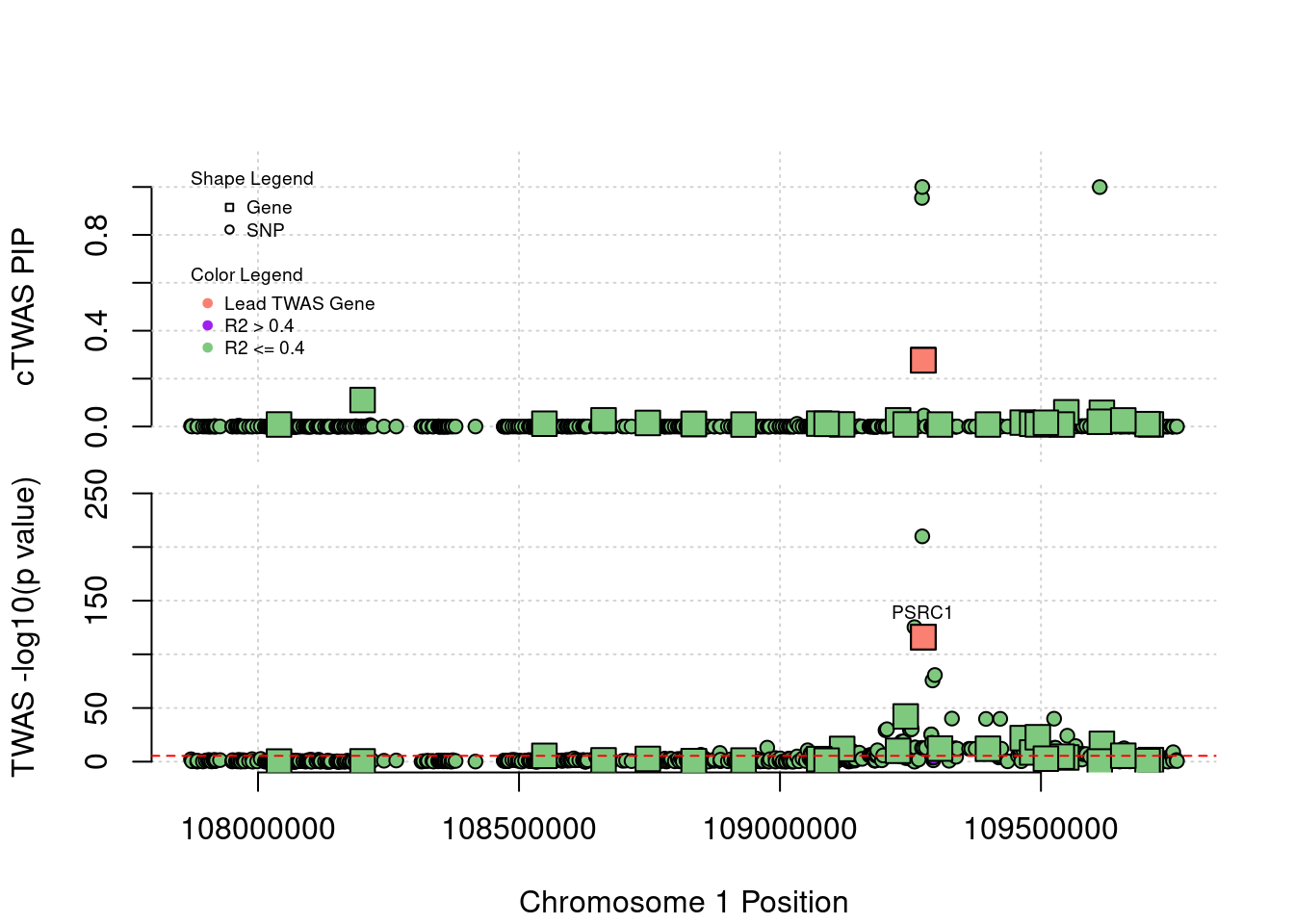
Locus Plots - 5_45 - Thin
locus_plot("5_45", label="TWAS")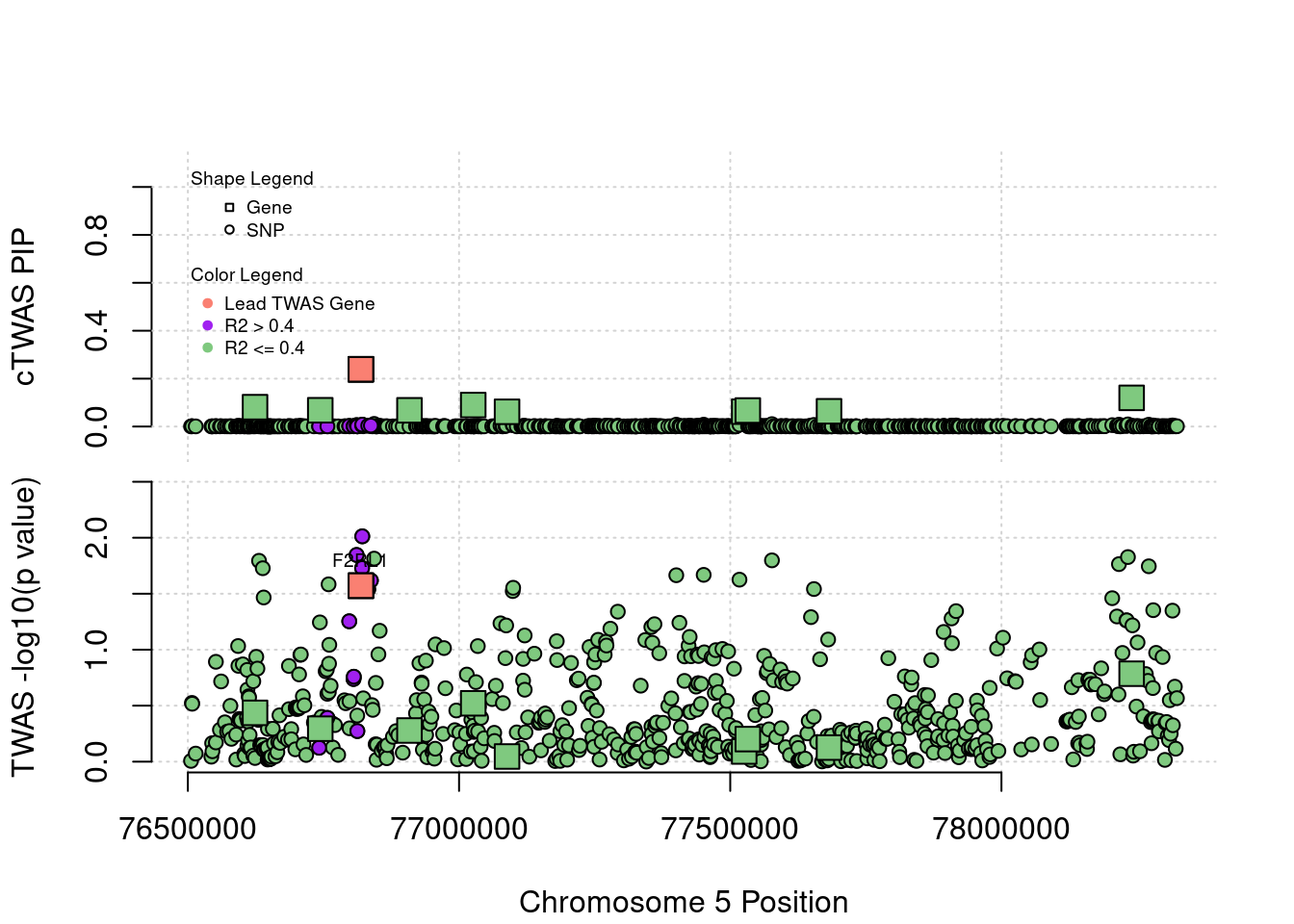
Locus Plots - 5_45 - Re-run
#locus_plot("5_45", label="TWAS", rerun_ctwas = T)Locus Plots - 8_12
locus_plot4 <- function(region_tag, rerun_ctwas = F, plot_eqtl = T, label="cTWAS"){
region_tag1 <- unlist(strsplit(region_tag, "_"))[1]
region_tag2 <- unlist(strsplit(region_tag, "_"))[2]
a <- ctwas_res[ctwas_res$region_tag==region_tag,]
regionlist <- readRDS(paste0(results_dir, "/", analysis_id, "_ctwas.regionlist.RDS"))
region <- regionlist[[as.numeric(region_tag1)]][[region_tag2]]
R_snp_info <- do.call(rbind, lapply(region$regRDS, function(x){data.table::fread(paste0(tools::file_path_sans_ext(x), ".Rvar"))}))
if (isTRUE(rerun_ctwas)){
ld_exprfs <- paste0(results_dir, "/", analysis_id, "_expr_chr", 1:22, ".expr.gz")
temp_reg <- data.frame("chr" = paste0("chr",region_tag1), "start" = region$start, "stop" = region$stop)
write.table(temp_reg,
#file= paste0(results_dir, "/", analysis_id, "_ctwas.temp.reg.txt") ,
file= "temp_reg.txt",
row.names=F, col.names=T, sep="\t", quote = F)
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
z_gene_temp <- z_gene[z_gene$id %in% a$id[a$type=="gene"],]
z_snp_temp <- z_snp[z_snp$id %in% R_snp_info$id,]
ctwas_rss(z_gene_temp, z_snp_temp, ld_exprfs, ld_pgenfs = NULL,
ld_R_dir = dirname(region$regRDS)[1],
ld_regions_custom = "temp_reg.txt", thin = 1,
outputdir = ".", outname = "temp", ncore = 1, ncore.rerun = 1, prob_single = 0,
group_prior = estimated_group_prior, group_prior_var = estimated_group_prior_var,
estimate_group_prior = F, estimate_group_prior_var = F)
a <- data.table::fread("temp.susieIrss.txt", header = T)
rownames(z_snp_temp) <- z_snp_temp$id
z_snp_temp <- z_snp_temp[a$id[a$type=="SNP"],]
z_gene_temp <- z_gene_temp[a$id[a$type=="gene"],]
a$z <- NA
a$z[a$type=="SNP"] <- z_snp_temp$z
a$z[a$type=="gene"] <- z_gene_temp$z
}
a$ifcausal <- 0
focus <- a$id[a$type=="gene"][which.max(abs(a$z[a$type=="gene"]))]
a$ifcausal <- as.numeric(a$id==focus)
a$PVALUE <- (-log(2) - pnorm(abs(a$z), lower.tail=F, log.p=T))/log(10)
R_gene <- readRDS(region$R_g_file)
R_snp_gene <- readRDS(region$R_sg_file)
R_snp <- as.matrix(Matrix::bdiag(lapply(region$regRDS, readRDS)))
rownames(R_gene) <- region$gid
colnames(R_gene) <- region$gid
rownames(R_snp_gene) <- R_snp_info$id
colnames(R_snp_gene) <- region$gid
rownames(R_snp) <- R_snp_info$id
colnames(R_snp) <- R_snp_info$id
a$r2max <- NA
a$r2max[a$type=="gene"] <- R_gene[focus,a$id[a$type=="gene"]]
a$r2max[a$type=="SNP"] <- R_snp_gene[a$id[a$type=="SNP"],focus]
r2cut <- 0.4
colorsall <- c("#7fc97f", "#beaed4", "#fdc086")
layout(matrix(1:2, ncol = 1), widths = 1, heights = c(1.5,1.5), respect = FALSE)
par(mar = c(0, 4.1, 4.1, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 19, xlab=paste0("Chromosome ", region_tag1, " Position"),frame.plot=FALSE, col = "white", ylim= c(-0.1,1.1), ylab = "cTWAS PIP", xaxt = 'n')
grid()
points(a$pos[a$type=="SNP"], a$susie_pip[a$type == "SNP"], pch = 21, xlab="Genomic position", bg = colorsall[1])
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$susie_pip[a$type == "SNP" & a$r2max >r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$susie_pip[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$susie_pip[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$susie_pip[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$susie_pip[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
if (isTRUE(plot_eqtl)){
for (cgene in a[a$type=="gene" & a$ifcausal == 1, ]$id){
load(paste0(results_dir, "/",analysis_id, "_expr_chr", region_tag1, ".exprqc.Rd"))
eqtls <- rownames(wgtlist[[cgene]])
points(a[a$id %in% eqtls,]$pos, rep( -0.15, nrow(a[a$id %in% eqtls,])), pch = "|", col = "salmon", cex = 1.5)
}
}
#legend(min(a$pos), y= 1.1 ,c("Gene", "SNP"), pch = c(22,21), title="Shape Legend", bty ='n', cex=0.6, title.adj = 0)
#legend(min(a$pos), y= 0.7 ,c("Lead TWAS Gene", "R2 > 0.4", "R2 <= 0.4"), pch = 19, col = c("salmon", "purple", colorsall[1]), title="Color Legend", bty ='n', cex=0.6, title.adj = 0)
legend(max(a$pos)-0.2*(max(a$pos)-min(a$pos)), y= 1.1 ,c("Gene", "SNP"), pch = c(22,21), title="Shape Legend", bty ='n', cex=0.6, title.adj = 0)
legend(max(a$pos)-0.2*(max(a$pos)-min(a$pos)), y= 0.7 ,c("Lead TWAS Gene", "R2 > 0.4", "R2 <= 0.4"), pch = 19, col = c("salmon", "purple", colorsall[1]), title="Color Legend", bty ='n', cex=0.6, title.adj = 0)
if (label=="cTWAS"){
text(a$pos[a$id==focus], a$susie_pip[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
par(mar = c(4.1, 4.1, 0.5, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 21, xlab=paste0("Chromosome ", region_tag1, " Position"), frame.plot=FALSE, bg = colorsall[1], ylab = "TWAS -log10(p value)", panel.first = grid(), ylim =c(0, max(a$PVALUE)*1.2))
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$PVALUE[a$type == "SNP" & a$r2max > r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$PVALUE[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$PVALUE[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$PVALUE[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$PVALUE[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
abline(h=-log10(alpha/nrow(ctwas_gene_res)), col ="red", lty = 2)
if (label=="TWAS"){
text(a$pos[a$id==focus], a$PVALUE[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
}
#locus_plot4("8_12", label="cTWAS")locus_plot5 <- function(region_tag, rerun_ctwas = F, plot_eqtl = T, label="cTWAS", focus){
region_tag1 <- unlist(strsplit(region_tag, "_"))[1]
region_tag2 <- unlist(strsplit(region_tag, "_"))[2]
a <- ctwas_res[ctwas_res$region_tag==region_tag,]
regionlist <- readRDS(paste0(results_dir, "/", analysis_id, "_ctwas.regionlist.RDS"))
region <- regionlist[[as.numeric(region_tag1)]][[region_tag2]]
R_snp_info <- do.call(rbind, lapply(region$regRDS, function(x){data.table::fread(paste0(tools::file_path_sans_ext(x), ".Rvar"))}))
if (isTRUE(rerun_ctwas)){
ld_exprfs <- paste0(results_dir, "/", analysis_id, "_expr_chr", 1:22, ".expr.gz")
temp_reg <- data.frame("chr" = paste0("chr",region_tag1), "start" = region$start, "stop" = region$stop)
write.table(temp_reg,
#file= paste0(results_dir, "/", analysis_id, "_ctwas.temp.reg.txt") ,
file= "temp_reg.txt",
row.names=F, col.names=T, sep="\t", quote = F)
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
z_gene_temp <- z_gene[z_gene$id %in% a$id[a$type=="gene"],]
z_snp_temp <- z_snp[z_snp$id %in% R_snp_info$id,]
ctwas_rss(z_gene_temp, z_snp_temp, ld_exprfs, ld_pgenfs = NULL,
ld_R_dir = dirname(region$regRDS)[1],
ld_regions_custom = "temp_reg.txt", thin = 1,
outputdir = ".", outname = "temp", ncore = 1, ncore.rerun = 1, prob_single = 0,
group_prior = estimated_group_prior, group_prior_var = estimated_group_prior_var,
estimate_group_prior = F, estimate_group_prior_var = F)
a <- data.table::fread("temp.susieIrss.txt", header = T)
rownames(z_snp_temp) <- z_snp_temp$id
z_snp_temp <- z_snp_temp[a$id[a$type=="SNP"],]
z_gene_temp <- z_gene_temp[a$id[a$type=="gene"],]
a$z <- NA
a$z[a$type=="SNP"] <- z_snp_temp$z
a$z[a$type=="gene"] <- z_gene_temp$z
}
a$ifcausal <- 0
focus <- a$id[which(a$genename==focus)]
a$ifcausal <- as.numeric(a$id==focus)
a$PVALUE <- (-log(2) - pnorm(abs(a$z), lower.tail=F, log.p=T))/log(10)
R_gene <- readRDS(region$R_g_file)
R_snp_gene <- readRDS(region$R_sg_file)
R_snp <- as.matrix(Matrix::bdiag(lapply(region$regRDS, readRDS)))
rownames(R_gene) <- region$gid
colnames(R_gene) <- region$gid
rownames(R_snp_gene) <- R_snp_info$id
colnames(R_snp_gene) <- region$gid
rownames(R_snp) <- R_snp_info$id
colnames(R_snp) <- R_snp_info$id
a$r2max <- NA
a$r2max[a$type=="gene"] <- R_gene[focus,a$id[a$type=="gene"]]
a$r2max[a$type=="SNP"] <- R_snp_gene[a$id[a$type=="SNP"],focus]
r2cut <- 0.4
colorsall <- c("#7fc97f", "#beaed4", "#fdc086")
layout(matrix(1:2, ncol = 1), widths = 1, heights = c(1.5,1.5), respect = FALSE)
par(mar = c(0, 4.1, 4.1, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 19, xlab=paste0("Chromosome ", region_tag1, " Position"),frame.plot=FALSE, col = "white", ylim= c(-0.1,1.1), ylab = "cTWAS PIP", xaxt = 'n')
grid()
points(a$pos[a$type=="SNP"], a$susie_pip[a$type == "SNP"], pch = 21, xlab="Genomic position", bg = colorsall[1])
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$susie_pip[a$type == "SNP" & a$r2max >r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$susie_pip[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$susie_pip[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$susie_pip[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$susie_pip[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
if (isTRUE(plot_eqtl)){
for (cgene in a[a$type=="gene" & a$ifcausal == 1, ]$id){
load(paste0(results_dir, "/",analysis_id, "_expr_chr", region_tag1, ".exprqc.Rd"))
eqtls <- rownames(wgtlist[[cgene]])
points(a[a$id %in% eqtls,]$pos, rep( -0.15, nrow(a[a$id %in% eqtls,])), pch = "|", col = "salmon", cex = 1.5)
}
}
legend(max(a$pos)-0.2*(max(a$pos)-min(a$pos)), y= 1.1 ,c("Gene", "SNP"), pch = c(22,21), title="Shape Legend", bty ='n', cex=0.6, title.adj = 0)
legend(max(a$pos)-0.2*(max(a$pos)-min(a$pos)), y= 0.7 ,c("Focal Gene", "R2 > 0.4", "R2 <= 0.4"), pch = 19, col = c("salmon", "purple", colorsall[1]), title="Color Legend", bty ='n', cex=0.6, title.adj = 0)
if (label=="cTWAS"){
text(a$pos[a$id==focus], a$susie_pip[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
par(mar = c(4.1, 4.1, 0.5, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 21, xlab=paste0("Chromosome ", region_tag1, " Position"), frame.plot=FALSE, bg = colorsall[1], ylab = "TWAS -log10(p value)", panel.first = grid(), ylim =c(0, max(a$PVALUE)*1.2))
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$PVALUE[a$type == "SNP" & a$r2max > r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$PVALUE[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$PVALUE[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$PVALUE[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$PVALUE[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
abline(h=-log10(alpha/nrow(ctwas_gene_res)), col ="red", lty = 2)
if (label=="TWAS"){
text(a$pos[a$id==focus], a$PVALUE[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
}
locus_plot5("19_33", focus="PRKD2")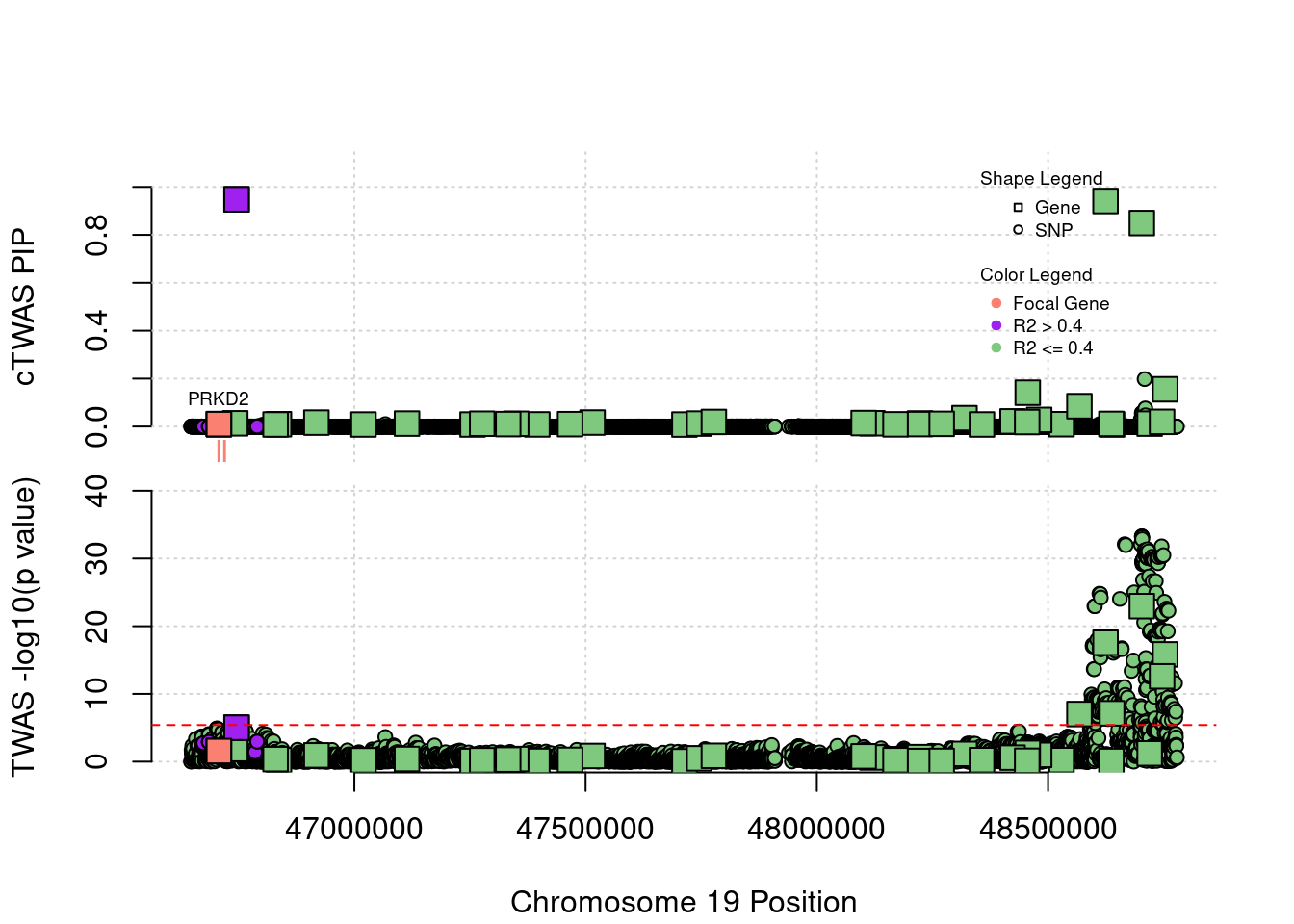
Locus Plots - Exploring known annotations
This section produces locus plots for all silver standard genes with known annotations. The highlighted gene at each region is the silver standard gene. Note that if no genes in a region have PIP>0.8, then only the result using thinned SNPs is displayed.
locus_plot3 <- function(region_tag, rerun_ctwas = F, plot_eqtl = T, label="cTWAS", focus){
region_tag1 <- unlist(strsplit(region_tag, "_"))[1]
region_tag2 <- unlist(strsplit(region_tag, "_"))[2]
a <- ctwas_res[ctwas_res$region_tag==region_tag,]
regionlist <- readRDS(paste0(results_dir, "/", analysis_id, "_ctwas.regionlist.RDS"))
region <- regionlist[[as.numeric(region_tag1)]][[region_tag2]]
R_snp_info <- do.call(rbind, lapply(region$regRDS, function(x){data.table::fread(paste0(tools::file_path_sans_ext(x), ".Rvar"))}))
if (isTRUE(rerun_ctwas)){
ld_exprfs <- paste0(results_dir, "/", analysis_id, "_expr_chr", 1:22, ".expr.gz")
temp_reg <- data.frame("chr" = paste0("chr",region_tag1), "start" = region$start, "stop" = region$stop)
write.table(temp_reg,
#file= paste0(results_dir, "/", analysis_id, "_ctwas.temp.reg.txt") ,
file= "temp_reg.txt",
row.names=F, col.names=T, sep="\t", quote = F)
load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
z_gene_temp <- z_gene[z_gene$id %in% a$id[a$type=="gene"],]
z_snp_temp <- z_snp[z_snp$id %in% R_snp_info$id,]
ctwas_rss(z_gene_temp, z_snp_temp, ld_exprfs, ld_pgenfs = NULL,
ld_R_dir = dirname(region$regRDS)[1],
ld_regions_custom = "temp_reg.txt", thin = 1,
outputdir = ".", outname = "temp", ncore = 1, ncore.rerun = 1, prob_single = 0,
group_prior = estimated_group_prior, group_prior_var = estimated_group_prior_var,
estimate_group_prior = F, estimate_group_prior_var = F)
a <- data.table::fread("temp.susieIrss.txt", header = T)
rownames(z_snp_temp) <- z_snp_temp$id
z_snp_temp <- z_snp_temp[a$id[a$type=="SNP"],]
z_gene_temp <- z_gene_temp[a$id[a$type=="gene"],]
a$z <- NA
a$z[a$type=="SNP"] <- z_snp_temp$z
a$z[a$type=="gene"] <- z_gene_temp$z
}
a$ifcausal <- 0
focus <- a$id[which(a$genename==focus)]
a$ifcausal <- as.numeric(a$id==focus)
a$PVALUE <- (-log(2) - pnorm(abs(a$z), lower.tail=F, log.p=T))/log(10)
R_gene <- readRDS(region$R_g_file)
R_snp_gene <- readRDS(region$R_sg_file)
R_snp <- as.matrix(Matrix::bdiag(lapply(region$regRDS, readRDS)))
rownames(R_gene) <- region$gid
colnames(R_gene) <- region$gid
rownames(R_snp_gene) <- R_snp_info$id
colnames(R_snp_gene) <- region$gid
rownames(R_snp) <- R_snp_info$id
colnames(R_snp) <- R_snp_info$id
a$r2max <- NA
a$r2max[a$type=="gene"] <- R_gene[focus,a$id[a$type=="gene"]]
a$r2max[a$type=="SNP"] <- R_snp_gene[a$id[a$type=="SNP"],focus]
r2cut <- 0.4
colorsall <- c("#7fc97f", "#beaed4", "#fdc086")
layout(matrix(1:2, ncol = 1), widths = 1, heights = c(1.5,1.5), respect = FALSE)
par(mar = c(0, 4.1, 4.1, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 19, xlab=paste0("Chromosome ", region_tag1, " Position"),frame.plot=FALSE, col = "white", ylim= c(-0.1,1.1), ylab = "cTWAS PIP", xaxt = 'n')
grid()
points(a$pos[a$type=="SNP"], a$susie_pip[a$type == "SNP"], pch = 21, xlab="Genomic position", bg = colorsall[1])
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$susie_pip[a$type == "SNP" & a$r2max >r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$susie_pip[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$susie_pip[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$susie_pip[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$susie_pip[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
if (isTRUE(plot_eqtl)){
for (cgene in a[a$type=="gene" & a$ifcausal == 1, ]$id){
load(paste0(results_dir, "/",analysis_id, "_expr_chr", region_tag1, ".exprqc.Rd"))
eqtls <- rownames(wgtlist[[cgene]])
points(a[a$id %in% eqtls,]$pos, rep( -0.15, nrow(a[a$id %in% eqtls,])), pch = "|", col = "salmon", cex = 1.5)
}
}
legend(min(a$pos), y= 1.1 ,c("Gene", "SNP"), pch = c(22,21), title="Shape Legend", bty ='n', cex=0.6, title.adj = 0)
legend(min(a$pos), y= 0.7 ,c("Lead TWAS Gene", "R2 > 0.4", "R2 <= 0.4"), pch = 19, col = c("salmon", "purple", colorsall[1]), title="Color Legend", bty ='n', cex=0.6, title.adj = 0)
if (label=="cTWAS"){
text(a$pos[a$id==focus], a$susie_pip[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
par(mar = c(4.1, 4.1, 0.5, 2.1))
plot(a$pos[a$type=="SNP"], a$PVALUE[a$type == "SNP"], pch = 21, xlab=paste0("Chromosome ", region_tag1, " Position"), frame.plot=FALSE, bg = colorsall[1], ylab = "TWAS -log10(p value)", panel.first = grid(), ylim =c(0, max(a$PVALUE)*1.2))
points(a$pos[a$type=="SNP" & a$r2max > r2cut], a$PVALUE[a$type == "SNP" & a$r2max > r2cut], pch = 21, bg = "purple")
points(a$pos[a$type=="SNP" & a$ifcausal == 1], a$PVALUE[a$type == "SNP" & a$ifcausal == 1], pch = 21, bg = "salmon")
points(a$pos[a$type=="gene"], a$PVALUE[a$type == "gene"], pch = 22, bg = colorsall[1], cex = 2)
points(a$pos[a$type=="gene" & a$r2max > r2cut], a$PVALUE[a$type == "gene" & a$r2max > r2cut], pch = 22, bg = "purple", cex = 2)
points(a$pos[a$type=="gene" & a$ifcausal == 1], a$PVALUE[a$type == "gene" & a$ifcausal == 1], pch = 22, bg = "salmon", cex = 2)
abline(h=-log10(alpha/nrow(ctwas_gene_res)), col ="red", lty = 2)
if (label=="TWAS"){
text(a$pos[a$id==focus], a$PVALUE[a$id==focus], labels=ctwas_gene_res$genename[ctwas_gene_res$id==focus], pos=3, cex=0.6)
}
}load(paste0(results_dir, "/known_annotations.Rd"))
load(paste0(results_dir, "/bystanders.Rd"))
for (i in 1:length(known_annotations)){
focus <- known_annotations[i]
region_tag <- ctwas_res$region_tag[which(ctwas_res$genename==focus)]
locus_plot3(region_tag, focus=focus)
mtext(text=region_tag)
print(focus)
print(region_tag)
print(ctwas_gene_res[ctwas_gene_res$region_tag==region_tag,report_cols,])
}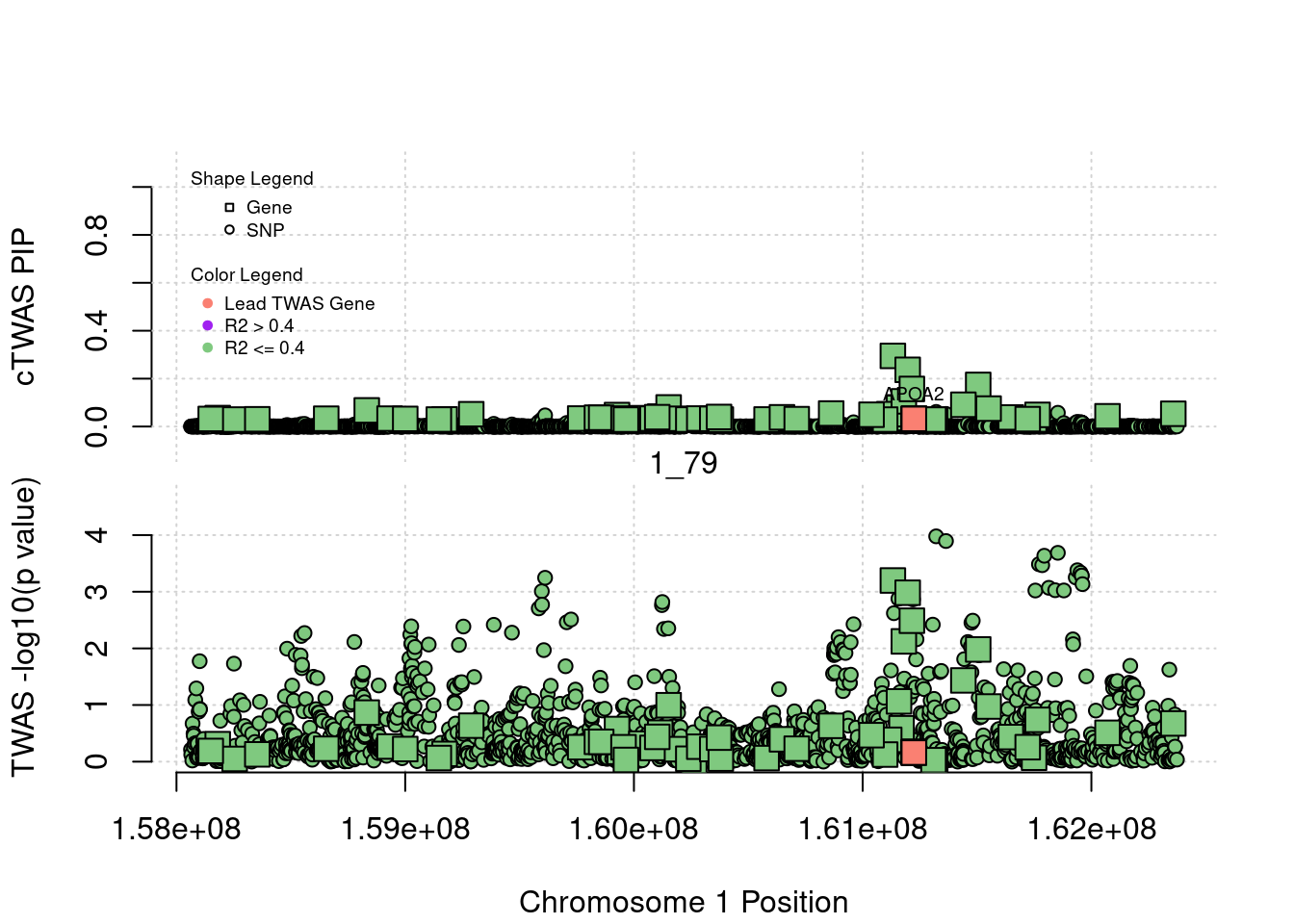
[1] "APOA2"
[1] "1_79"
genename region_tag susie_pip mu2 PVE
1233 IGSF9 1_79 0.03110337 5.084947 4.602715e-07
7429 CD1D 1_79 0.03537426 6.240779 6.424606e-07
7430 CD1A 1_79 0.02919040 4.515464 3.835859e-07
7433 CD1E 1_79 0.03033675 4.860996 4.291554e-07
7446 TAGLN2 1_79 0.04802009 8.995736 1.257129e-06
7449 DUSP23 1_79 0.03347673 5.745257 5.597225e-07
7850 CADM3 1_79 0.03000307 4.761767 4.157710e-07
8003 SPTA1 1_79 0.03266344 5.524340 5.251249e-07
8005 MNDA 1_79 0.06744246 12.078001 2.370548e-06
8006 PYHIN1 1_79 0.03309172 5.641341 5.432778e-07
8007 IFI16 1_79 0.03188884 5.308824 4.926713e-07
8008 AIM2 1_79 0.02931334 4.553157 3.884171e-07
10318 FCER1A 1_79 0.05156339 9.639737 1.446528e-06
11240 C1orf204 1_79 0.03692451 6.626546 7.120694e-07
12830 LINC01704 1_79 0.03157013 5.218651 4.794629e-07
13180 VSIG8 1_79 0.03676695 6.588073 7.049143e-07
281 ATP1A2 1_79 0.04141732 7.660566 9.233432e-07
716 CD84 1_79 0.02941505 4.584217 3.924235e-07
885 FCGR2B 1_79 0.03789131 6.859073 7.563544e-07
1137 DUSP12 1_79 0.02955346 4.626317 3.978910e-07
3484 SLAMF1 1_79 0.03862498 7.031769 7.904114e-07
3485 CD48 1_79 0.03079331 4.995056 4.476279e-07
3575 ATF6 1_79 0.04877379 9.136466 1.296836e-06
3897 COPA 1_79 0.03017585 4.813279 4.226888e-07
3898 CD244 1_79 0.05443100 10.129825 1.604606e-06
4840 ATP1A4 1_79 0.07971043 13.605335 3.156056e-06
4843 DCAF8 1_79 0.02893496 4.436645 3.735922e-07
6111 UFC1 1_79 0.08002033 13.640961 3.176622e-06
6112 PPOX 1_79 0.03542025 6.252557 6.445098e-07
6113 FCGR2A 1_79 0.17382070 20.886380 1.056538e-05
6115 SDHC 1_79 0.02895066 4.441512 3.742051e-07
6121 PIGM 1_79 0.03487332 6.112551 6.203490e-07
6122 CASQ1 1_79 0.03253114 5.487889 5.195471e-07
7454 NIT1 1_79 0.05159135 9.644154 1.447976e-06
7455 DEDD 1_79 0.29482827 26.094504 2.238920e-05
7461 B4GALT3 1_79 0.10518720 16.159216 4.946562e-06
7464 NDUFS2 1_79 0.23672774 23.887945 1.645691e-05
7466 FCER1G 1_79 0.15695991 19.910967 9.094973e-06
7467 APOA2 1_79 0.03194668 5.324987 4.950677e-07
7468 TOMM40L 1_79 0.03241361 5.455268 5.145929e-07
7469 MPZ 1_79 0.02904782 4.471559 3.780009e-07
7854 SLAMF9 1_79 0.03315572 5.658699 5.460034e-07
7855 KCNJ9 1_79 0.03305103 5.630291 5.415470e-07
7856 IGSF8 1_79 0.03917127 7.158225 8.160058e-07
7858 PEA15 1_79 0.02913932 4.499756 3.815826e-07
7859 PEX19 1_79 0.03339954 5.724511 5.564155e-07
7860 NCSTN 1_79 0.03218695 5.392365 5.051023e-07
7861 VANGL2 1_79 0.02924946 4.533590 3.859049e-07
7863 FCRLB 1_79 0.03090852 5.028508 4.523115e-07
7865 KLHDC9 1_79 0.02905842 4.474840 3.784163e-07
9413 NHLH1 1_79 0.04000960 7.348963 8.556784e-07
11982 NOS1AP 1_79 0.04286845 7.971076 9.944318e-07
12013 FCGR3A 1_79 0.07489710 13.035035 2.841172e-06
12497 TSTD1 1_79 0.04933698 9.240222 1.326708e-06
12576 LINC01133 1_79 0.02900654 4.458800 3.763866e-07
13085 C1orf226 1_79 0.05328643 9.937417 1.541028e-06
14716 RP11-122G18.11 1_79 0.09095682 14.817781 3.922281e-06
z num_eqtl
1233 0.42528829 2
7429 -0.68914550 1
7430 0.13177842 1
7433 -0.32627268 3
7446 -1.11380907 1
7449 0.58886854 2
7850 0.34581896 2
8003 0.53584957 2
8005 -1.49070119 1
8006 0.61251073 2
8007 -0.52613904 1
8008 0.17115180 2
10318 1.19652845 1
11240 0.76234673 4
12830 -0.45705216 1
13180 0.76032649 2
281 1.01961433 2
716 -0.17012652 1
885 0.90105676 1
1137 0.18491305 3
3484 0.83666570 1
3485 -0.53813609 1
3575 1.33216623 3
3897 -0.33313590 1
3898 -1.19306430 1
4840 1.64106571 2
4843 0.02937989 1
6111 -1.71610924 1
6112 -1.13273833 2
6113 -2.56082676 2
6115 -0.02197264 1
6121 0.70491904 2
6122 0.52195982 2
7454 0.82125790 2
7455 -3.41702724 1
7461 -2.67250298 3
7464 -3.27895520 2
7466 -2.94328519 3
7467 -0.39581982 1
7468 0.43065543 1
7469 0.07283357 1
7854 -0.58691461 2
7855 0.56272965 1
7856 -0.90635405 1
7858 0.14094214 1
7859 -0.60553082 1
7860 -0.52575638 2
7861 0.14603238 2
7863 -0.58867628 3
7865 0.31833172 1
9413 0.88415968 1
11982 -1.01549743 1
12013 1.61487487 1
12497 -0.95257428 3
12576 0.08332223 1
13085 -1.24890307 1
14716 -2.08810018 1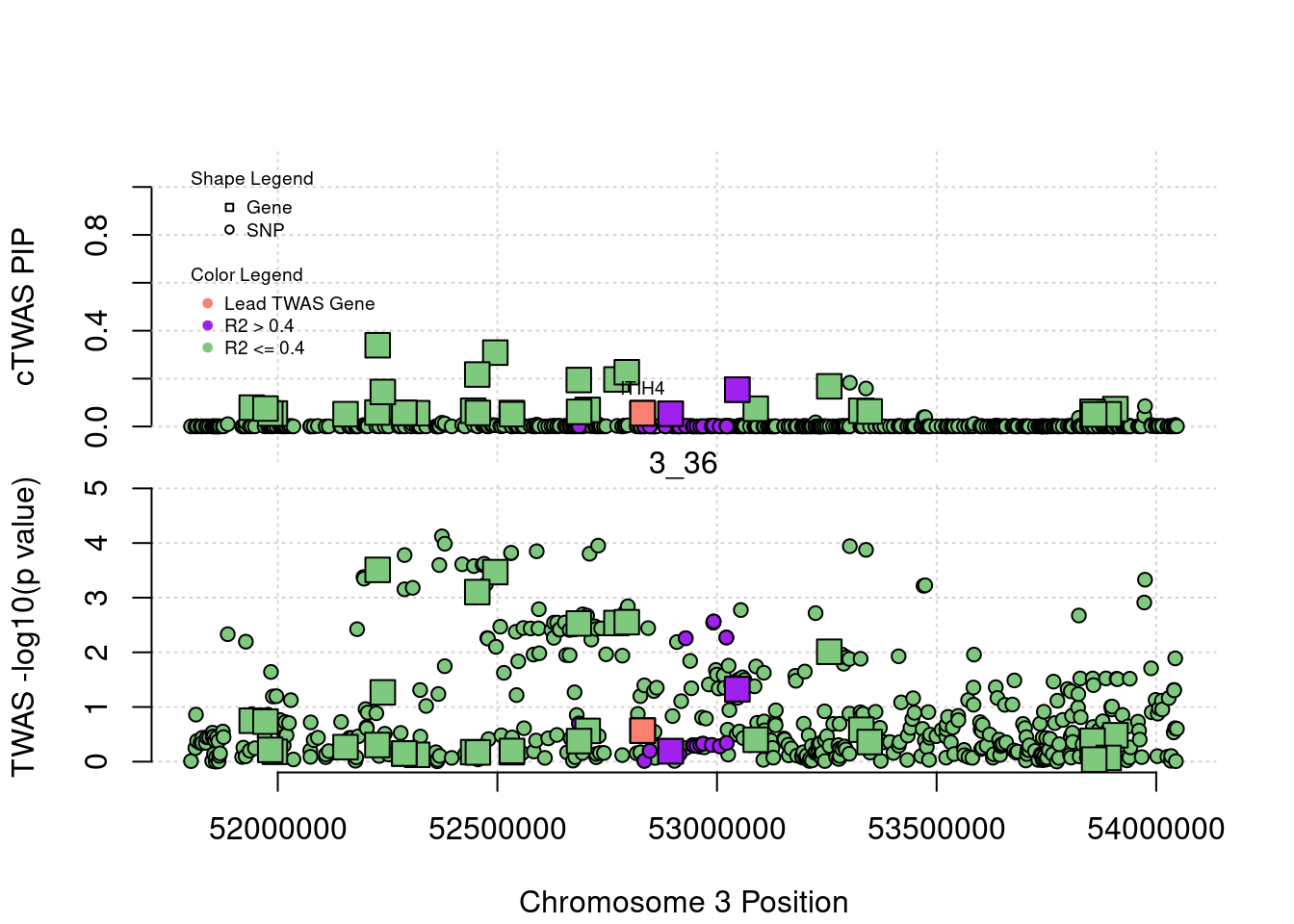
[1] "ITIH4"
[1] "3_36"
genename region_tag susie_pip mu2 PVE z
195 SEMA3G 3_36 0.06621250 6.650062 1.281404e-06 0.4278002
196 NISCH 3_36 0.05674459 5.378396 8.881728e-07 -0.4130717
197 STAB1 3_36 0.30850946 16.677819 1.497366e-05 3.5822738
271 CHDH 3_36 0.07287796 7.671453 1.627025e-06 -0.9771110
274 GLT8D1 3_36 0.06969832 7.208039 1.462042e-06 1.0942021
436 PARP3 3_36 0.07896936 8.633668 1.984149e-06 -1.3456620
561 ITIH4 3_36 0.05544470 5.244164 8.461680e-07 1.1005563
566 IL17RB 3_36 0.06168370 6.275767 1.126568e-06 -0.8151013
3156 SELENOK 3_36 0.05108440 4.524134 6.725801e-07 -0.1830178
3157 ACTR8 3_36 0.05057829 4.450069 6.550150e-07 -0.1041810
3228 DNAH1 3_36 0.05579829 5.249189 8.523803e-07 0.3177441
3231 TNNC1 3_36 0.21711961 14.758573 9.325320e-06 -3.3561809
3236 NEK4 3_36 0.19658902 16.662554 9.532814e-06 -2.9773988
7341 CACNA1D 3_36 0.06595840 6.984083 1.340602e-06 1.1251378
7769 RPL29 3_36 0.05443710 5.106519 8.089846e-07 -0.4392119
7770 ITIH3 3_36 0.22674584 17.946333 1.184228e-05 -2.9881334
8102 TKT 3_36 0.16791039 14.104279 6.892055e-06 2.5841158
8103 RFT1 3_36 0.07443114 7.760287 1.680942e-06 0.8461193
8104 SFMBT1 3_36 0.15396484 15.003409 6.722515e-06 -1.9840557
8105 GNL3 3_36 0.19420560 16.546270 9.351519e-06 -2.9712715
8106 PBRM1 3_36 0.06140594 5.817619 1.039623e-06 -0.8048656
8150 POC1A 3_36 0.05171592 4.804022 7.230188e-07 0.6100943
8151 PPM1M 3_36 0.05863926 5.627273 9.602996e-07 -0.6814726
8153 WDR82 3_36 0.05708876 5.441720 9.040804e-07 0.3708077
8891 GLYCTK 3_36 0.05708876 5.441720 9.040804e-07 0.3708077
8897 NT5DC2 3_36 0.05651802 5.704254 9.382231e-07 0.4249165
8898 SMIM4 3_36 0.05170980 4.634961 6.974920e-07 -0.4707184
12351 TMEM110 3_36 0.05340778 5.178894 8.049370e-07 -0.4603354
13080 TLR9 3_36 0.33929847 17.172140 1.695613e-05 3.6067545
13212 ACY1 3_36 0.05452035 5.141837 8.158254e-07 -0.5115172
13284 TWF2 3_36 0.14436032 13.891841 5.836170e-06 -1.9263835
13300 ABHD14A 3_36 0.07535098 8.206826 1.799635e-06 1.3240881
14324 DCP1A 3_36 0.06331768 6.463491 1.191002e-06 0.7749500
num_eqtl
195 1
196 2
197 1
271 1
274 1
436 2
561 1
566 1
3156 2
3157 4
3228 1
3231 2
3236 1
7341 1
7769 1
7770 1
8102 1
8103 2
8104 2
8105 1
8106 1
8150 1
8151 3
8153 1
8891 1
8897 2
8898 2
12351 1
13080 1
13212 1
13284 1
13300 1
14324 1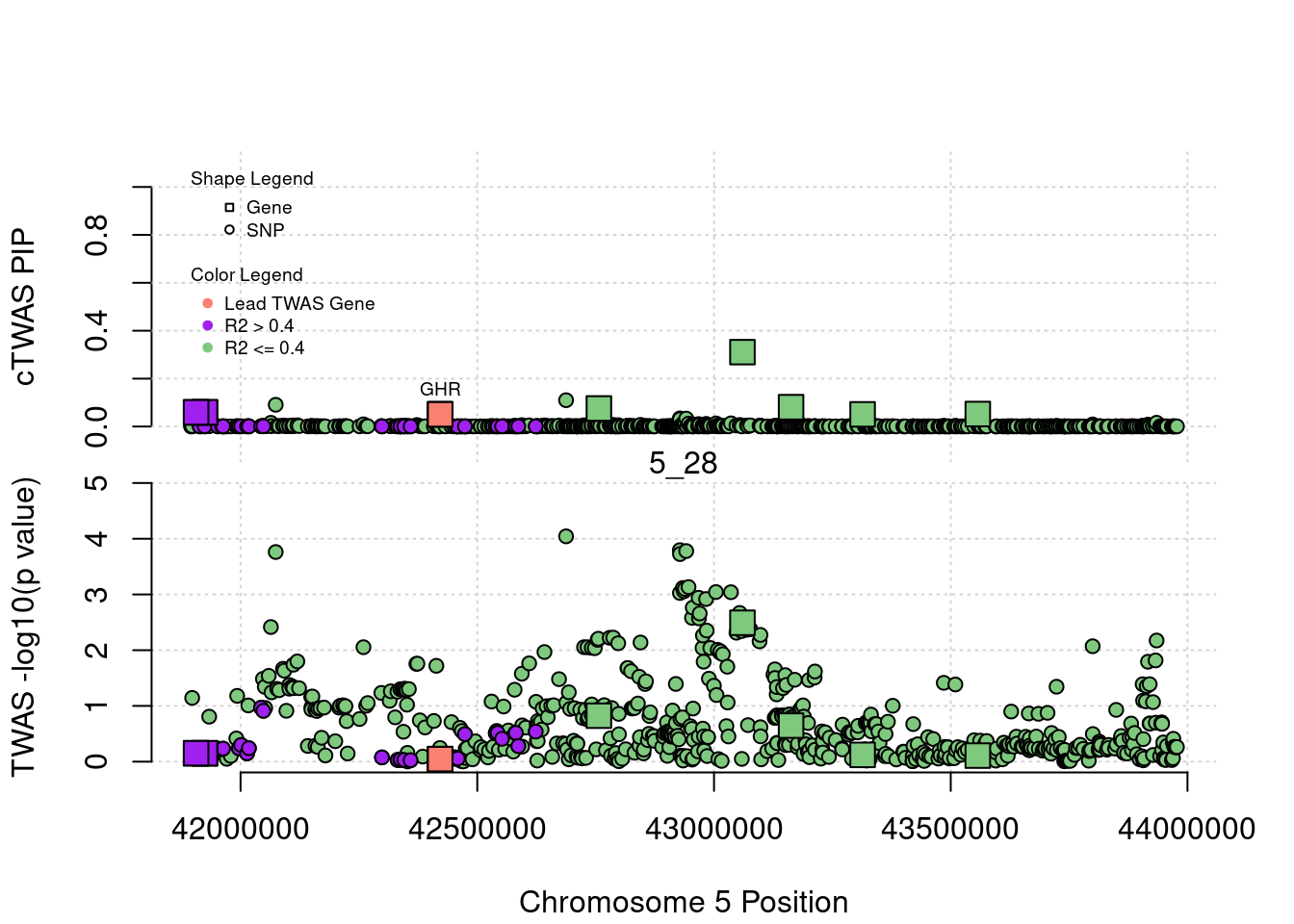
[1] "GHR"
[1] "5_28"
genename region_tag susie_pip mu2 PVE
3077 GHR 5_28 0.05087052 4.479922 6.632190e-07
3078 HMGCS1 5_28 0.05093065 4.490625 6.655892e-07
6895 FBXO4 5_28 0.05918183 5.851818 1.007858e-06
9483 PAIP1 5_28 0.05214027 4.703107 7.136387e-07
10076 NIM1K 5_28 0.08092327 8.706397 2.050370e-06
11960 CCDC152 5_28 0.07552907 8.074666 1.774839e-06
12254 C5orf51 5_28 0.05918738 5.852669 1.008099e-06
14230 CTD-2035E11.5 5_28 0.31057739 21.597060 1.952022e-05
z num_eqtl
3077 -0.1262247 1
3078 -0.3102555 2
6895 0.3758312 1
9483 0.2818347 1
10076 1.2121911 3
11960 -1.4400434 1
12254 0.3759901 1
14230 2.9460695 1
[1] "EPHX2"
[1] "8_27"
genename region_tag susie_pip mu2 PVE z
2123 TRIM35 8_27 0.08266517 8.014845 1.928137e-06 -1.03320305
3806 CLU 8_27 0.08108999 7.838367 1.849750e-06 -0.90762086
3808 PTK2B 8_27 0.07331590 6.915841 1.475582e-06 -0.80784214
3812 EPHX2 8_27 0.06538932 5.872090 1.117429e-06 -0.62557250
4964 ELP3 8_27 0.16265255 14.327535 6.781920e-06 2.12504185
6538 CCDC25 8_27 0.12619481 11.932307 4.382140e-06 -1.53830095
8872 SCARA3 8_27 0.06332663 5.580318 1.028408e-06 -0.51876612
8873 PBK 8_27 0.06094660 5.231949 9.279687e-07 0.48335074
8874 SCARA5 8_27 0.05581829 4.433925 7.202531e-07 -0.01387977
9354 ESCO2 8_27 0.05632461 4.515829 7.402117e-07 -0.18809073
11401 NUGGC 8_27 0.26113373 18.945058 1.439724e-05 -2.49551570
num_eqtl
2123 2
3806 1
3808 1
3812 1
4964 1
6538 1
8872 3
8873 1
8874 1
9354 1
11401 1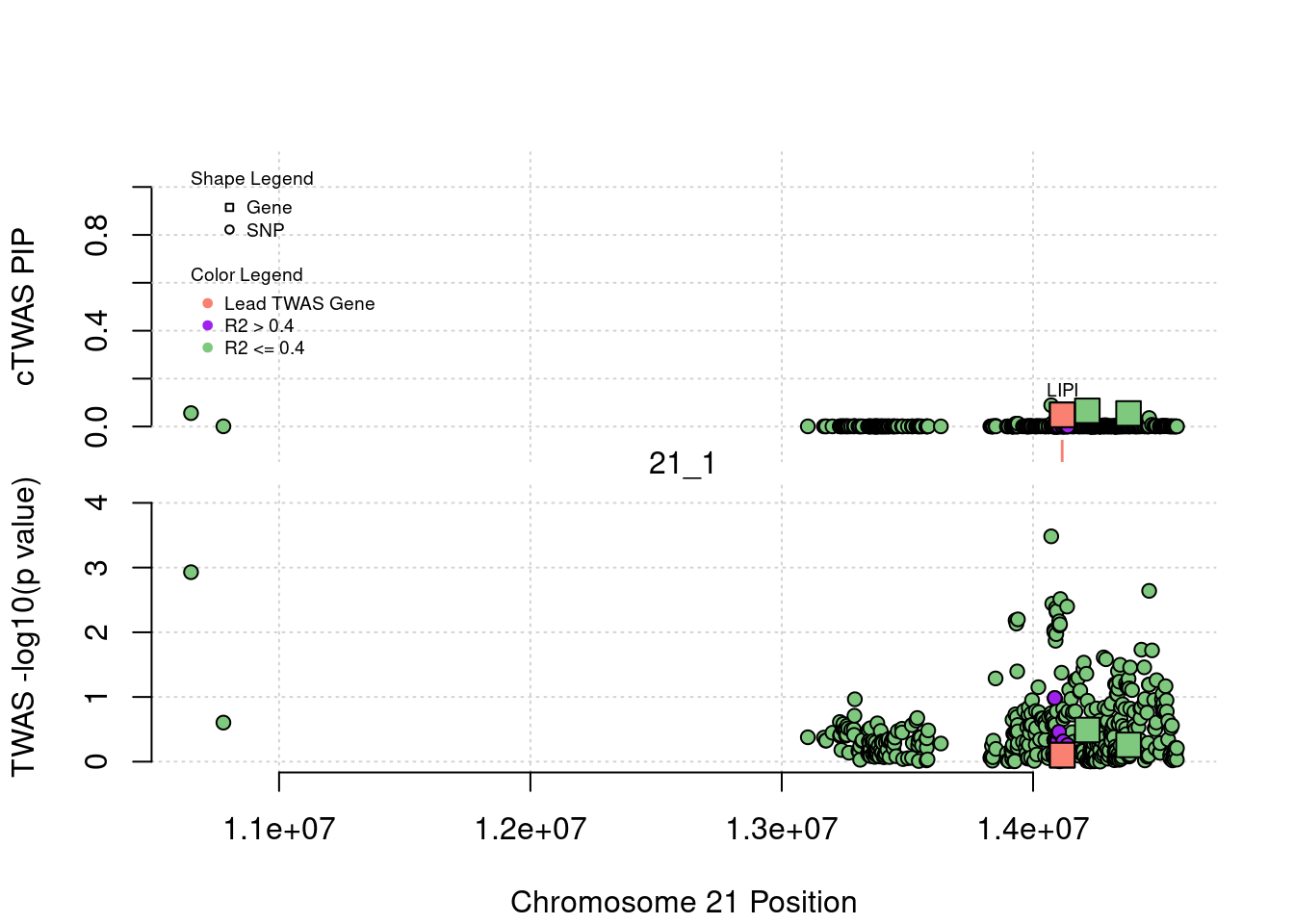
[1] "LIPI"
[1] "21_1"
genename region_tag susie_pip mu2 PVE z
7176 HSPA13 21_1 0.05468812 5.715365 9.096143e-07 -0.5976853
10910 RBM11 21_1 0.06575440 7.389707 1.414075e-06 -0.9860076
11369 LIPI 21_1 0.04864069 4.654479 6.588569e-07 -0.2618739
num_eqtl
7176 2
10910 2
11369 1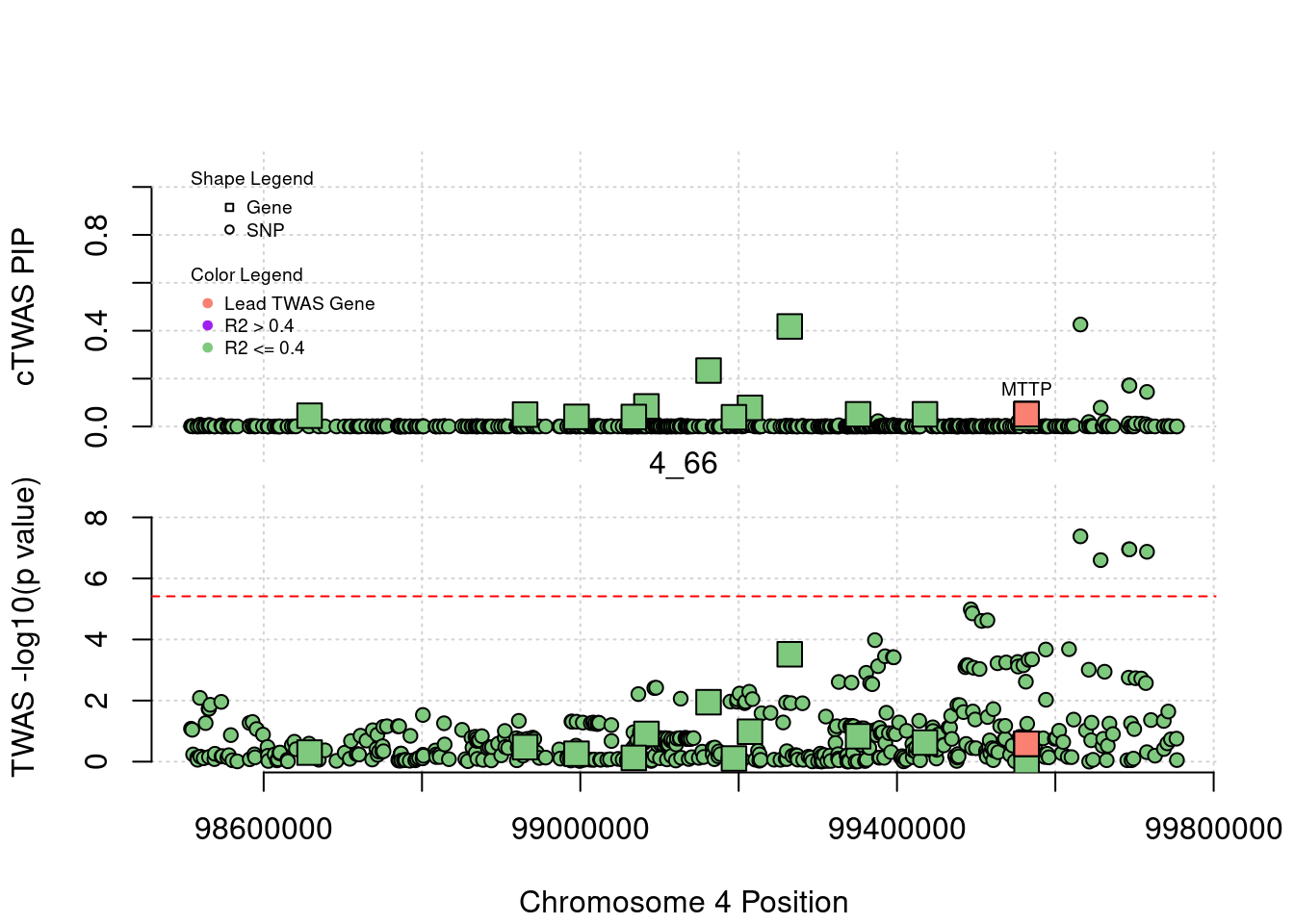
[1] "MTTP"
[1] "4_66"
genename region_tag susie_pip mu2 PVE
5664 MTTP 4_66 0.05177806 7.559951 1.139161e-06
6378 TRMT10A 4_66 0.03740272 4.436930 4.829543e-07
6825 EIF4E 4_66 0.04995378 7.038158 1.023170e-06
8125 METAP1 4_66 0.04047155 5.165848 6.084315e-07
8978 TSPAN5 4_66 0.04530893 6.211309 8.190063e-07
9588 ADH6 4_66 0.07808769 11.029611 2.506473e-06
11210 ADH1A 4_66 0.03871596 4.778017 5.383417e-07
11474 ADH7 4_66 0.05109442 7.576550 1.126588e-06
11535 ADH1B 4_66 0.41779633 26.754222 3.252949e-05
11752 ADH5 4_66 0.08411582 11.850860 2.901001e-06
11796 ADH4 4_66 0.23368026 20.516922 1.395258e-05
13290 ADH1C 4_66 0.05128665 7.205360 1.075425e-06
14305 RP11-571L19.8 4_66 0.03997627 4.979432 5.792985e-07
z num_eqtl
5664 1.12400758 1
6378 -0.06765527 2
6825 0.96532383 1
8125 0.59927970 2
8978 -0.66850706 2
9588 1.62276202 3
11210 -0.27018216 1
11474 1.18093280 1
11535 -3.61112568 2
11752 1.54003066 3
11796 -2.53274599 1
13290 -1.44685831 2
14305 -0.31136184 4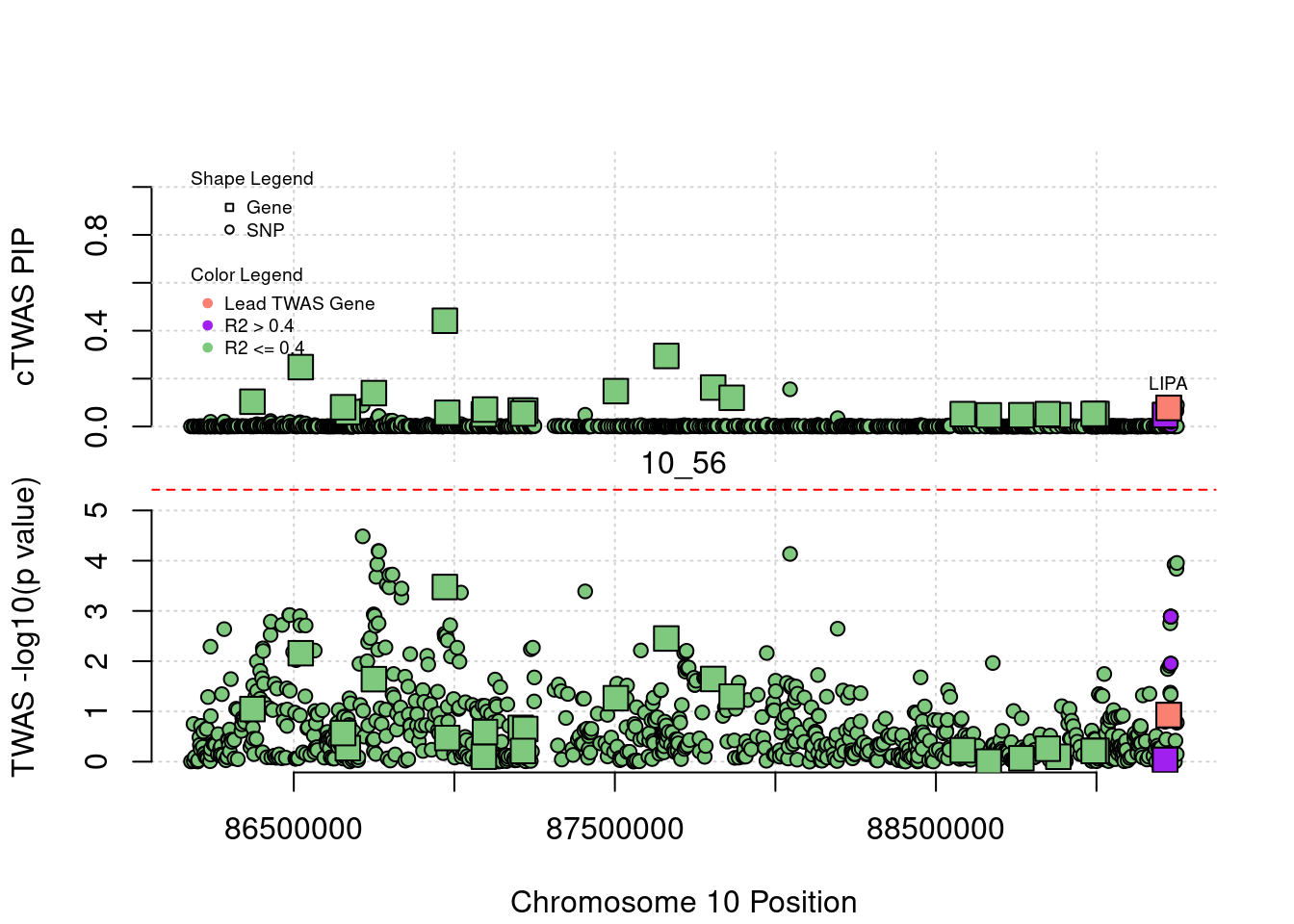
[1] "LIPA"
[1] "10_56"
genename region_tag susie_pip mu2 PVE z
628 WAPL 10_56 0.24805434 20.026732 1.445697e-05 -2.6996472034
2552 BMPR1A 10_56 0.13958471 14.474163 5.879652e-06 2.2764891466
3904 LDB3 10_56 0.06035660 6.738376 1.183587e-06 -0.6140380900
3905 OPN4 10_56 0.08057165 9.374366 2.198085e-06 -1.1028000389
3906 FAM35A 10_56 0.05091595 5.196063 7.699252e-07 -0.2590730319
6647 GLUD1 10_56 0.07106199 8.225758 1.701115e-06 -1.1325627492
9631 MMRN2 10_56 0.44161458 26.094511 3.353612e-05 -3.5874382677
10614 GRID1 10_56 0.10374476 11.703627 3.533515e-06 -1.6960715699
10862 NUTM2A 10_56 0.06704642 7.695141 1.501456e-06 1.2462484873
11256 FAM25A 10_56 0.05738894 6.280771 1.048966e-06 0.9566667456
12445 NUTM2D 10_56 0.06651215 7.622273 1.475386e-06 1.2154178990
12596 LINC00863 10_56 0.05429394 5.777828 9.129275e-07 -0.5054439421
336 FAS 10_56 0.05244707 5.464347 8.340264e-07 0.5111463639
2553 MINPP1 10_56 0.14783611 15.016142 6.460397e-06 1.9218952071
2554 ACTA2 10_56 0.05273099 5.513248 8.460456e-07 0.5063182504
2555 LIPA 10_56 0.07773327 9.045550 2.046267e-06 -1.5612453556
5560 STAMBPL1 10_56 0.04749276 4.566679 6.311726e-07 -0.2547553334
5561 CH25H 10_56 0.04756885 4.581162 6.341889e-07 0.0947268029
5562 ATAD1 10_56 0.16282481 15.932401 7.549568e-06 2.2801464509
6973 ANKRD22 10_56 0.05190164 5.369664 8.110516e-07 -0.5927424299
10568 LIPF 10_56 0.04679213 4.432392 6.035751e-07 0.0002399494
10841 RNLS 10_56 0.05192014 5.372906 8.118306e-07 0.5342430208
11898 PAPSS2 10_56 0.29463805 21.762928 1.866064e-05 -2.9168127698
12037 LIPN 10_56 0.04751161 4.570269 6.319196e-07 0.1807762682
12694 KLLN 10_56 0.12005752 13.061030 4.563385e-06 1.9541126075
num_eqtl
628 1
2552 2
3904 2
3905 4
3906 2
6647 2
9631 1
10614 1
10862 4
11256 2
12445 4
12596 3
336 1
2553 1
2554 1
2555 2
5560 1
5561 1
5562 1
6973 2
10568 1
10841 2
11898 4
12037 1
12694 1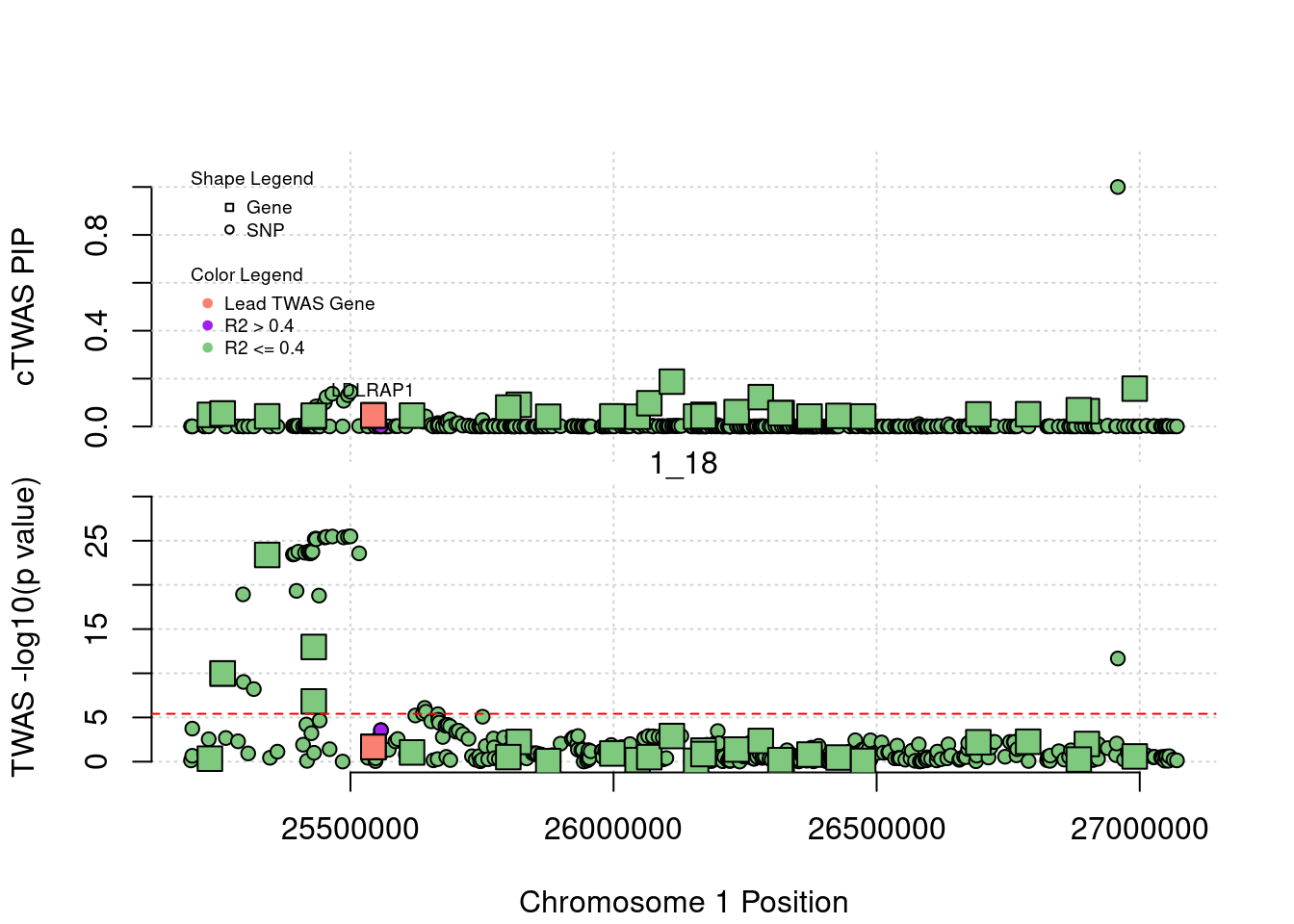
[1] "LDLRAP1"
[1] "1_18"
genename region_tag susie_pip mu2 PVE z
605 PIGV 1_18 0.05136519 12.572290 1.879332e-06 -2.7730250
1402 NUDC 1_18 0.06429513 13.388589 2.505147e-06 2.5764625
3544 SYF2 1_18 0.04879230 6.162460 8.750355e-07 0.7426202
3546 MTFR1L 1_18 0.09111064 13.683658 3.628203e-06 2.7576118
3547 MAN1C1 1_18 0.04573464 7.203217 9.587205e-07 1.6902246
3548 DHDDS 1_18 0.04439161 5.463763 7.058511e-07 0.9497144
3551 ARID1A 1_18 0.05090687 12.362214 1.831441e-06 -2.7281921
4633 CEP85 1_18 0.06025510 9.598386 1.683109e-06 2.0281607
6057 SH3BGRL3 1_18 0.12270489 15.770691 5.631614e-06 -2.8355236
6058 CNKSR1 1_18 0.04898008 7.118078 1.014618e-06 -1.9113034
6067 GPN2 1_18 0.06825325 8.119909 1.612853e-06 -0.5565182
7384 LDLRAP1 1_18 0.04661922 8.917102 1.209787e-06 2.2890064
7387 PAFAH2 1_18 0.04246450 5.849825 7.229182e-07 -1.5558136
7388 SLC30A2 1_18 0.04093828 4.581994 5.458890e-07 0.4276042
7390 TRIM63 1_18 0.09737485 10.995539 3.115901e-06 1.0142171
7395 UBXN11 1_18 0.05638504 6.292779 1.032587e-06 0.4067319
7793 SELENON 1_18 0.07928259 9.836167 2.269468e-06 1.0241546
9078 CD52 1_18 0.05561002 6.203147 1.003888e-06 -0.4221420
9837 PDIK1L 1_18 0.18734630 20.052390 1.093280e-05 3.2263042
9940 ZNF683 1_18 0.04171448 5.452621 6.619306e-07 -1.4082793
10610 PAQR7 1_18 0.04109575 4.534041 5.422538e-07 0.1738933
10735 TMEM50A 1_18 0.04258632 83.462767 1.034387e-05 10.1300072
11134 RHD 1_18 0.05441571 39.553117 6.263618e-06 -6.4603360
11321 RHCE 1_18 0.04107844 46.794836 5.594126e-06 7.4344194
11636 FAM110D 1_18 0.04178882 4.585999 5.577176e-07 -0.2619079
11942 HMGN2 1_18 0.04201176 4.591698 5.613897e-07 0.2239647
12053 TMEM57 1_18 0.04545133 26.749940 3.538260e-06 5.2485323
13008 RP11-96L14.7 1_18 0.04331332 5.808910 7.322113e-07 -1.4819009
13432 TRNP1 1_18 0.15821144 14.738355 6.785896e-06 -1.1357927
14265 RP3-465N24.6 1_18 0.05441571 39.553117 6.263618e-06 6.4603360
num_eqtl
605 1
1402 1
3544 1
3546 1
3547 1
3548 2
3551 1
4633 3
6057 1
6058 2
6067 2
7384 2
7387 1
7388 1
7390 1
7395 1
7793 2
9078 1
9837 1
9940 1
10610 1
10735 1
11134 1
11321 2
11636 3
11942 2
12053 2
13008 1
13432 1
14265 1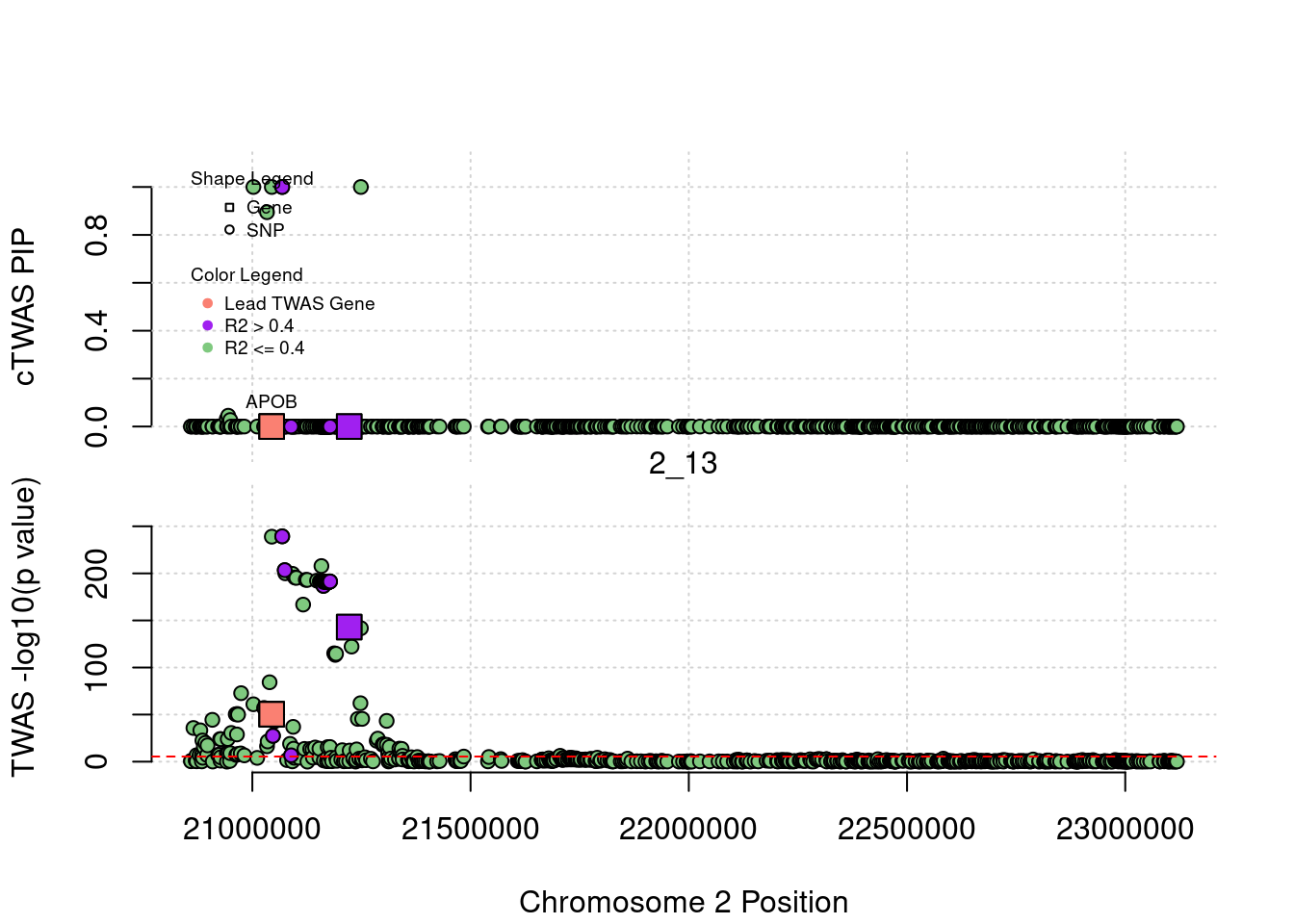
[1] "APOB"
[1] "2_13"
genename region_tag susie_pip mu2 PVE z
1211 APOB 2_13 2.837784e-10 181.8676 1.501948e-13 -15.03583
12880 AC067959.1 2_13 4.072320e-11 330.7175 3.919398e-14 -25.52145
num_eqtl
1211 2
12880 1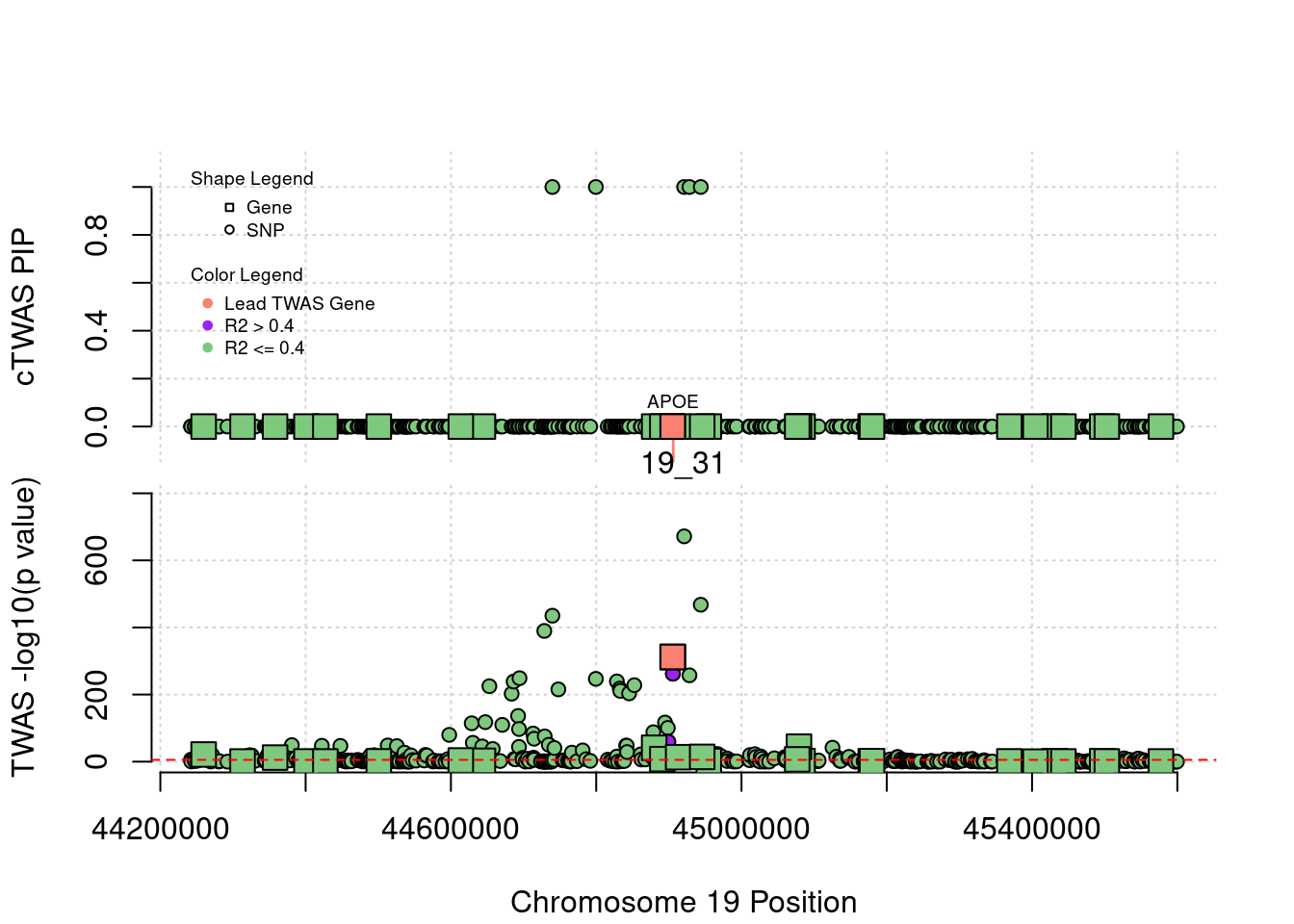
[1] "APOE"
[1] "19_31"
genename region_tag susie_pip mu2 PVE z num_eqtl
123 MARK4 19_31 0 20.312700 0 -2.24637681 1
131 TRAPPC6A 19_31 0 16.600235 0 1.92140959 1
229 ERCC1 19_31 0 13.134793 0 -2.19401016 2
624 ZNF112 19_31 0 72.068605 0 7.18938487 3
898 PVR 19_31 0 30.112592 0 -3.01973306 1
2196 CLPTM1 19_31 0 48.802068 0 -2.57517256 1
2198 PPP1R37 19_31 0 35.405859 0 -2.37518201 2
2203 ERCC2 19_31 0 13.615522 0 1.53401174 1
3560 CD3EAP 19_31 0 16.730266 0 0.05565582 4
4222 OPA3 19_31 0 11.581430 0 1.32335471 2
4224 RTN2 19_31 0 6.803450 0 2.07007103 1
4226 VASP 19_31 0 23.808738 0 1.26035740 1
4570 NECTIN2 19_31 0 55.577675 0 -13.59762224 1
4571 APOE 19_31 0 999.967785 0 37.80754977 2
4572 TOMM40 19_31 0 56.000201 0 5.53230466 2
4573 APOC1 19_31 0 296.620171 0 7.53014709 1
6024 GEMIN7 19_31 0 157.439551 0 14.13454209 2
7552 ZNF233 19_31 0 98.228951 0 -9.63193736 1
7553 ZNF235 19_31 0 8.185909 0 -0.97877558 1
8729 ZNF180 19_31 0 25.642421 0 1.41443321 2
9263 ZNF296 19_31 0 65.538646 0 5.25423092 2
11089 CEACAM19 19_31 0 13.759203 0 3.64086985 3
11388 BLOC1S3 19_31 0 8.975124 0 2.30141189 1
12385 PPM1N 19_31 0 20.227762 0 -1.22009641 2
12950 APOC2 19_31 0 99.467017 0 7.82468739 2
14021 ZNF285 19_31 0 10.013342 0 -1.26709542 2
14628 ZNF229 19_31 0 8.630054 0 1.23011935 2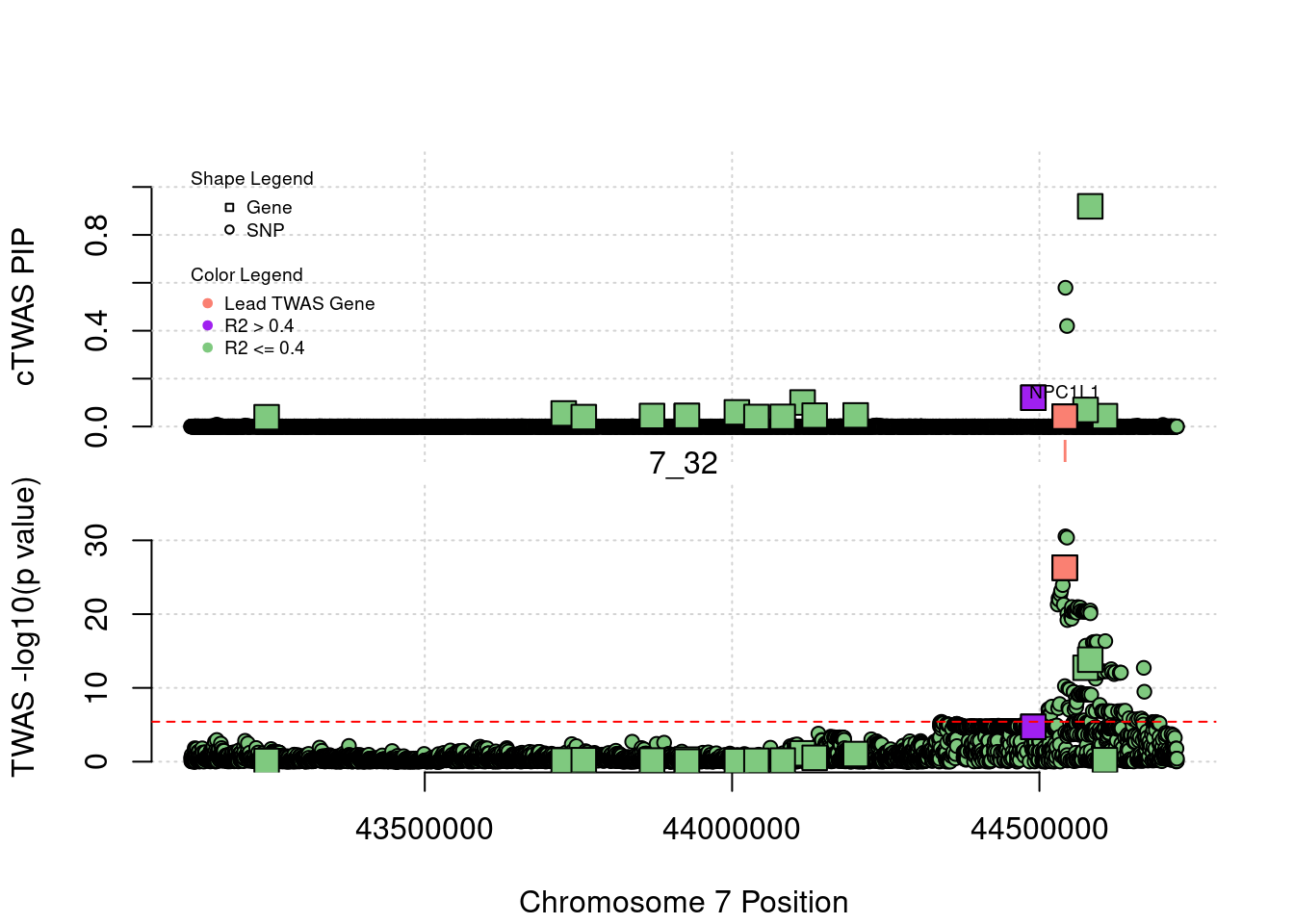
[1] "NPC1L1"
[1] "7_32"
genename region_tag susie_pip mu2 PVE z
17 HECW1 7_32 0.03879622 4.502064 5.083015e-07 -0.23376042
268 NPC1L1 7_32 0.04099715 82.556624 9.849767e-06 10.76193109
270 NUDCD3 7_32 0.12016967 17.520565 6.127217e-06 4.27589199
627 MRPS24 7_32 0.04400621 5.502620 7.046992e-07 0.38278180
1072 UBE2D4 7_32 0.04316907 5.512225 6.925003e-07 0.62909522
2384 OGDH 7_32 0.04345606 22.901957 2.896298e-06 0.47417198
2465 COA1 7_32 0.05470517 6.827556 1.086961e-06 -0.37405767
2466 BLVRA 7_32 0.03930484 4.699292 5.375251e-07 0.46600524
2467 URGCP 7_32 0.04456617 5.287322 6.857430e-07 0.03508092
2472 POLD2 7_32 0.10048790 13.317824 3.894640e-06 1.78074584
2474 GCK 7_32 0.04566065 6.311854 8.387246e-07 1.02935564
2476 YKT6 7_32 0.04642044 7.365875 9.950706e-07 1.67948623
3938 POLM 7_32 0.04055106 4.857444 5.732319e-07 -0.36899829
5285 DDX56 7_32 0.06932067 30.497390 6.152417e-06 7.35708820
7441 TMED4 7_32 0.91919811 36.878293 9.865071e-05 7.68827409
12733 AC004951.6 7_32 0.05933237 7.272867 1.255792e-06 0.22091509
12967 LINC00957 7_32 0.03984318 4.700790 5.450609e-07 0.29011976
num_eqtl
17 3
268 1
270 1
627 1
1072 1
2384 2
2465 2
2466 1
2467 2
2472 4
2474 2
2476 1
3938 3
5285 2
7441 2
12733 1
12967 1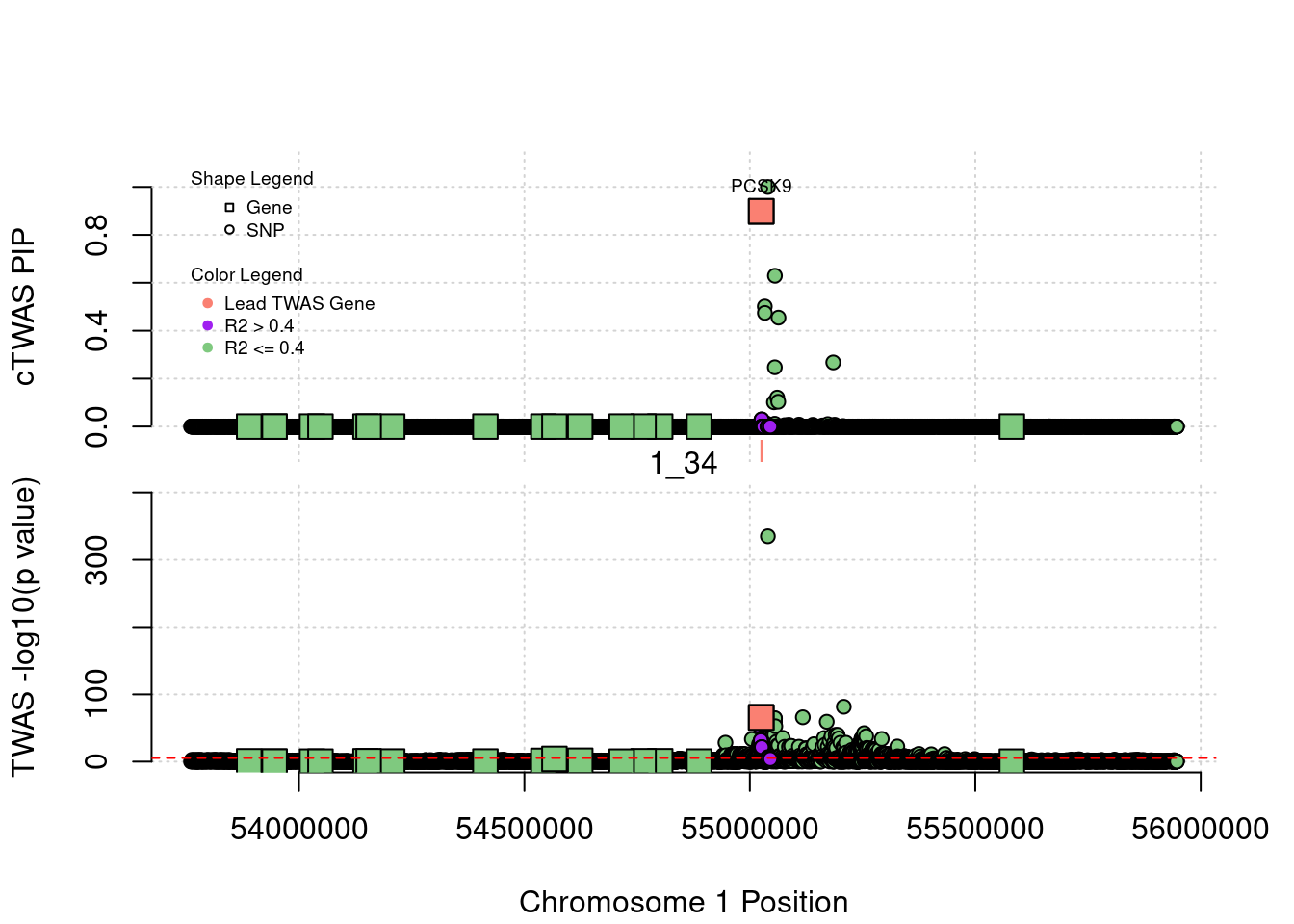
[1] "PCSK9"
[1] "1_34"
genename region_tag susie_pip mu2 PVE
108 TTC22 1_34 1.326717e-13 15.011189 5.795802e-18
587 YIPF1 1_34 8.060219e-14 10.381893 2.435251e-18
1144 HSPB11 1_34 4.374279e-14 5.108528 6.503131e-19
3387 DHCR24 1_34 6.250556e-14 9.053461 1.646848e-18
3399 TCEANC2 1_34 4.196643e-14 4.741349 5.790610e-19
3400 TMEM59 1_34 4.152234e-14 4.644457 5.612251e-19
3401 LRRC42 1_34 4.352074e-14 5.262271 6.664842e-19
3403 MRPL37 1_34 4.407585e-14 4.994136 6.405919e-19
7325 CDCP2 1_34 9.736656e-14 12.363077 3.503134e-18
7327 SSBP3 1_34 4.063416e-14 4.635790 5.481953e-19
7784 ACOT11 1_34 1.343370e-13 17.148420 6.704093e-18
7785 FAM151A 1_34 4.327649e-13 27.822867 3.504082e-17
7786 PARS2 1_34 8.781864e-14 10.323831 2.638444e-18
9036 PCSK9 1_34 8.982431e-01 105.002053 2.744808e-04
10806 MROH7 1_34 1.868505e-12 45.488781 2.473540e-16
12499 CYB5RL 1_34 1.219025e-13 14.007672 4.969342e-18
12945 RP11-90C4.1 1_34 5.484502e-14 7.932894 1.266162e-18
13202 TTC4 1_34 4.096723e-14 4.885584 5.824697e-19
z num_eqtl
108 1.1662870 2
587 1.0226599 3
1144 -0.2303441 1
3387 -0.3806125 1
3399 -0.1853540 1
3400 0.2100252 2
3401 0.2854315 2
3403 0.5344672 2
7325 -1.2055941 1
7327 -0.6008954 1
7784 -1.8518413 3
7785 2.0257851 1
7786 -1.1551925 2
9036 17.2108693 1
10806 3.9761380 4
12499 1.1595815 2
12945 -0.1113431 1
13202 -0.1056123 1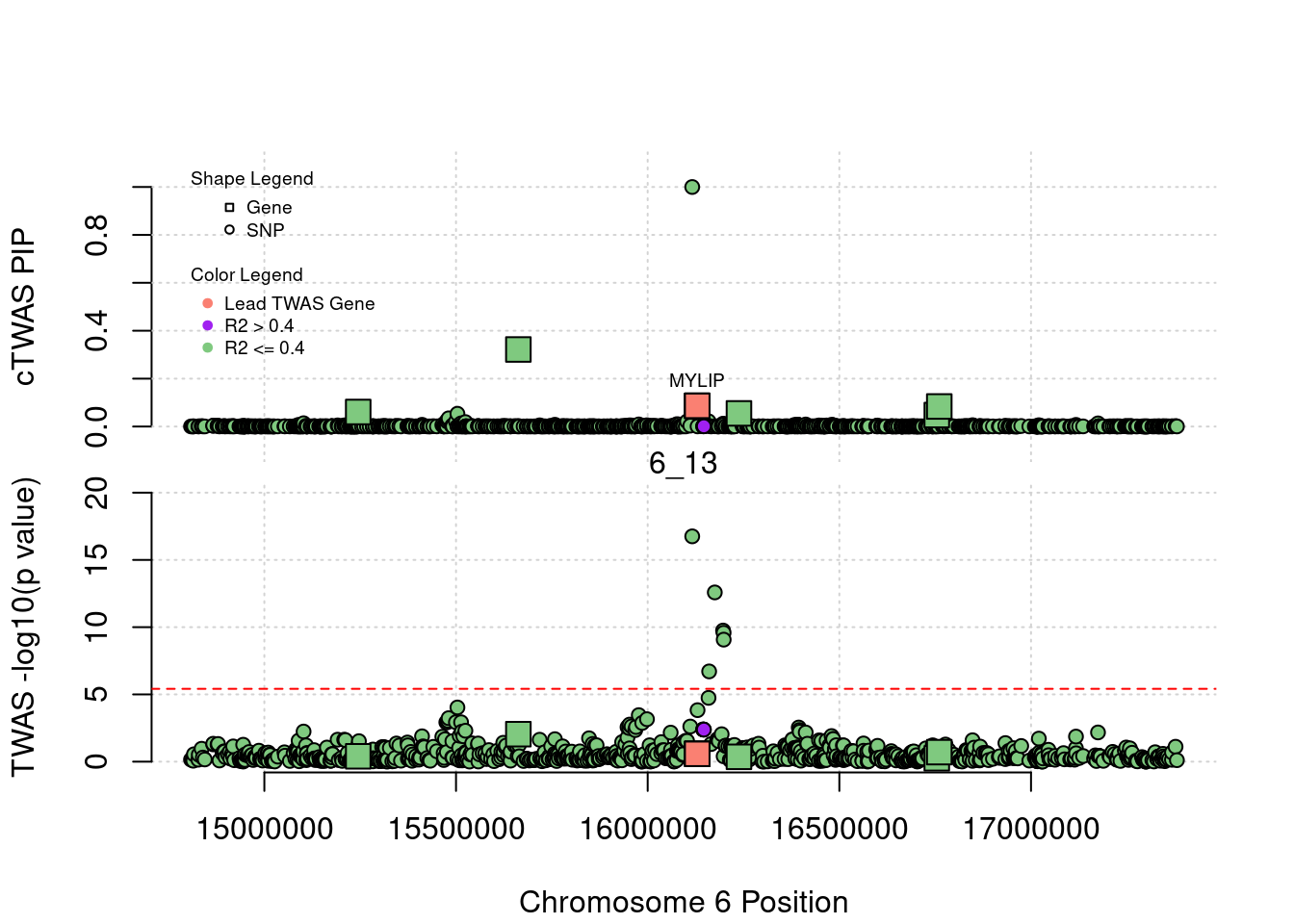
[1] "MYLIP"
[1] "6_13"
genename region_tag susie_pip mu2 PVE z
146 MYLIP 6_13 0.08653777 10.583115 2.665260e-06 1.1219050
463 DTNBP1 6_13 0.32173291 21.849708 2.045792e-05 2.6114864
4145 ATXN1 6_13 0.04786987 5.395142 7.515978e-07 0.5753436
5410 GMPR 6_13 0.05495376 6.679036 1.068148e-06 0.7796528
14194 RP11-560J1.2 6_13 0.06072175 7.243905 1.280081e-06 0.8415880
14252 RP1-151F17.2 6_13 0.08377364 10.350535 2.523426e-06 -1.2903423
num_eqtl
146 1
463 2
4145 1
5410 1
14194 2
14252 1
[1] "OSBPL5"
[1] "11_3"
genename region_tag susie_pip mu2 PVE
79 ZNF195 11_3 0.04489658 9.189417 1.200664e-06
298 OSBPL5 11_3 0.02664660 4.569047 3.543134e-07
647 TSPAN32 11_3 0.17425691 21.212698 1.075737e-05
1071 TOLLIP 11_3 0.03693407 7.443930 8.001101e-07
2819 CARS 11_3 0.07740928 14.125636 3.182155e-06
2820 SLC22A18 11_3 0.03079258 5.943437 5.326036e-07
2821 CD81 11_3 0.02696224 4.633467 3.635652e-07
2823 C11orf21 11_3 0.03069238 5.723158 5.111950e-07
4547 CDKN1C 11_3 0.19125419 22.428368 1.248329e-05
4622 LSP1 11_3 0.07511949 14.128318 3.088612e-06
4623 TNNT3 11_3 0.02719406 4.706400 3.724631e-07
6666 SYT8 11_3 0.03750832 7.572600 8.265954e-07
8708 IGF2 11_3 0.08828897 18.018558 4.629635e-06
8718 ART5 11_3 0.06505907 12.517928 2.370067e-06
9788 BRSK2 11_3 0.03458189 6.857338 6.901200e-07
10491 PHLDA2 11_3 0.33549377 27.403845 2.675570e-05
10550 MOB2 11_3 0.03264429 6.494092 6.169443e-07
10736 ASCL2 11_3 0.02875374 5.326728 4.457334e-07
10828 DUSP8 11_3 0.28864675 26.451176 2.221938e-05
12235 NAP1L4 11_3 0.16255365 20.746616 9.814412e-06
12773 LINC01150 11_3 0.13157051 18.410626 7.049323e-06
13220 IFITM10 11_3 0.03240731 6.214297 5.860779e-07
14725 PRR33 11_3 0.03815367 8.869684 9.848378e-07
2830 NUP98 11_3 0.02632474 4.454601 3.412661e-07
4546 CHRNA10 11_3 0.02743057 4.820109 3.847796e-07
4765 TRIM21 11_3 0.03208860 6.218791 5.807337e-07
8721 RRM1 11_3 0.03458420 6.885031 6.929533e-07
8722 OR51E2 11_3 0.16920407 21.212381 1.044529e-05
8723 TRIM68 11_3 0.02668311 4.574996 3.552609e-07
10415 OR51E1 11_3 0.03372023 6.672792 6.548147e-07
13487 RP11-23F23.2 11_3 0.02982610 5.565984 4.831241e-07
z num_eqtl
79 1.14786730 2
298 0.20151551 1
647 2.25278977 1
1071 -0.80841023 2
2819 -1.77831042 2
2820 -0.43733079 2
2821 0.20617703 1
2823 -0.55061372 4
4547 2.53335014 2
4622 -1.89370692 1
4623 0.20030015 1
6666 -0.86657930 1
8708 2.87768350 1
8718 1.51689797 3
9788 0.88384194 1
10491 -3.12977961 1
10550 -1.02326080 2
10736 0.57486941 2
10828 -3.08552426 1
12235 -2.48039908 2
12773 -1.99663571 1
13220 0.57719694 1
14725 -1.74750284 1
2830 -0.08259672 1
4546 0.33897462 1
4765 -0.71226005 1
8721 0.84172180 3
8722 2.36169323 2
8723 -0.20197797 2
10415 -0.76087482 3
13487 -0.57167492 2
[1] "SCARB1"
[1] "12_76"
genename region_tag susie_pip mu2 PVE
900 SCARB1 12_76 0.04497941 5.090161 6.662935e-07
1138 AACS 12_76 0.06679809 8.645254 1.680591e-06
5716 TMEM132B 12_76 0.04516820 5.075552 6.671699e-07
6801 DHX37 12_76 0.08312286 11.348157 2.745150e-06
6802 UBC 12_76 0.18059131 17.826680 9.368878e-06
14496 RP11-214K3.24 12_76 0.04312085 4.924835 6.180154e-07
z num_eqtl
900 0.4644817 2
1138 -1.1186318 2
5716 -0.3647610 2
6801 1.7052619 1
6802 -2.1757766 1
14496 -0.6831074 1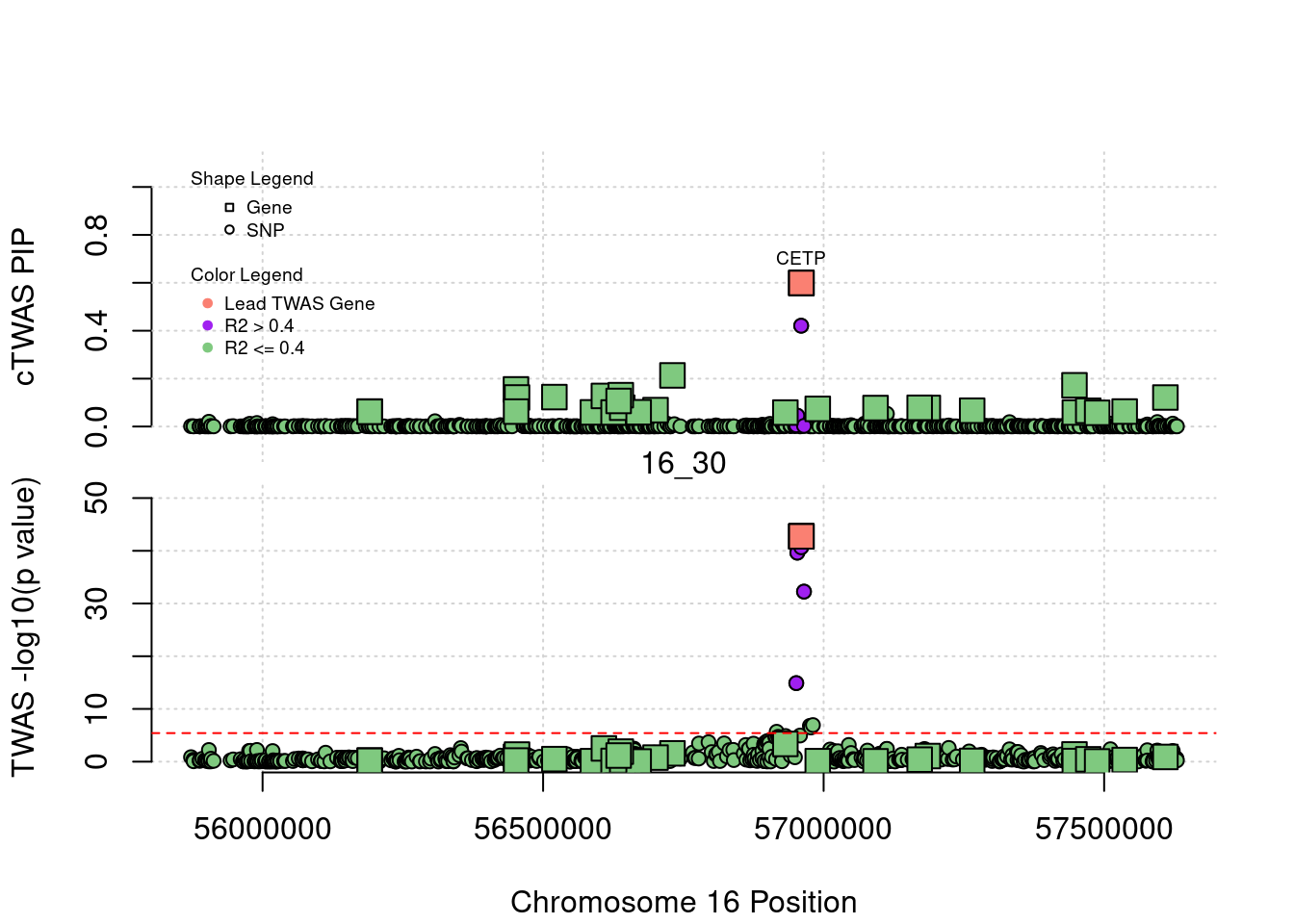
[1] "CETP"
[1] "16_30"
genename region_tag susie_pip mu2 PVE
64 CIAPIN1 16_30 0.17214874 14.897020 7.463174e-06
507 HERPUD1 16_30 0.05838811 14.363220 2.440600e-06
1287 CETP 16_30 0.59931467 157.616391 2.749012e-04
1289 MT3 16_30 0.05924333 4.707772 8.116619e-07
1291 GNAO1 16_30 0.06263732 5.222523 9.519933e-07
1292 OGFOD1 16_30 0.15510374 13.095419 5.911014e-06
1331 COQ9 16_30 0.05865500 4.637403 7.915898e-07
1987 NUP93 16_30 0.21353525 16.476560 1.023897e-05
1994 PLLP 16_30 0.06611764 5.559908 1.069807e-06
1998 POLR2C 16_30 0.06599408 6.001463 1.152610e-06
4159 BBS2 16_30 0.12288976 10.473970 3.745824e-06
4160 MT1G 16_30 0.05777613 4.753714 7.992852e-07
4161 MT2A 16_30 0.12764431 15.782956 5.862868e-06
4164 DOK4 16_30 0.05732431 4.468875 7.455167e-07
5194 CCDC102A 16_30 0.06265653 5.393224 9.834110e-07
5872 CPNE2 16_30 0.07721418 6.634346 1.490787e-06
5873 NLRC5 16_30 0.07387993 6.332294 1.361469e-06
7520 AMFR 16_30 0.12126443 12.373908 4.366773e-06
7523 RSPRY1 16_30 0.07773194 8.734951 1.975970e-06
8673 NUDT21 16_30 0.06250318 5.226823 9.507367e-07
9113 MT1E 16_30 0.05878522 4.690679 8.024615e-07
9557 FAM192A 16_30 0.07831597 7.176667 1.635661e-06
11160 MT1X 16_30 0.06861508 7.477815 1.493188e-06
11844 MT1F 16_30 0.07860421 6.798591 1.555196e-06
12219 ADGRG1 16_30 0.12120257 11.295539 3.984181e-06
12223 MT1H 16_30 0.05971606 4.770516 8.290426e-07
12224 MT1A 16_30 0.13092059 13.999055 5.333680e-06
12226 MT1M 16_30 0.10691465 10.615765 3.303002e-06
13267 RP11-461O7.1 16_30 0.06152796 5.035363 9.016201e-07
z num_eqtl
64 -2.02099466 3
507 3.52077435 2
1287 13.82423033 2
1289 0.23412884 1
1291 -0.52872064 1
1292 1.55918687 2
1331 -0.28431278 1
1987 -2.21748371 2
1994 -0.36226197 2
1998 -0.90590353 1
4159 -0.97225733 3
4160 -0.61932212 3
4161 2.95959176 2
4164 0.10732971 1
5194 0.69758013 2
5872 0.16513057 2
5873 -0.23774647 2
7520 1.99782505 1
7523 -1.71229943 3
8673 -0.49431949 1
9113 0.36161593 2
9557 0.81027778 4
11160 -1.37965021 1
11844 0.55338625 1
12219 -1.51761847 1
12223 0.07381601 1
12224 2.49797425 1
12226 1.81397194 1
13267 -0.41702961 2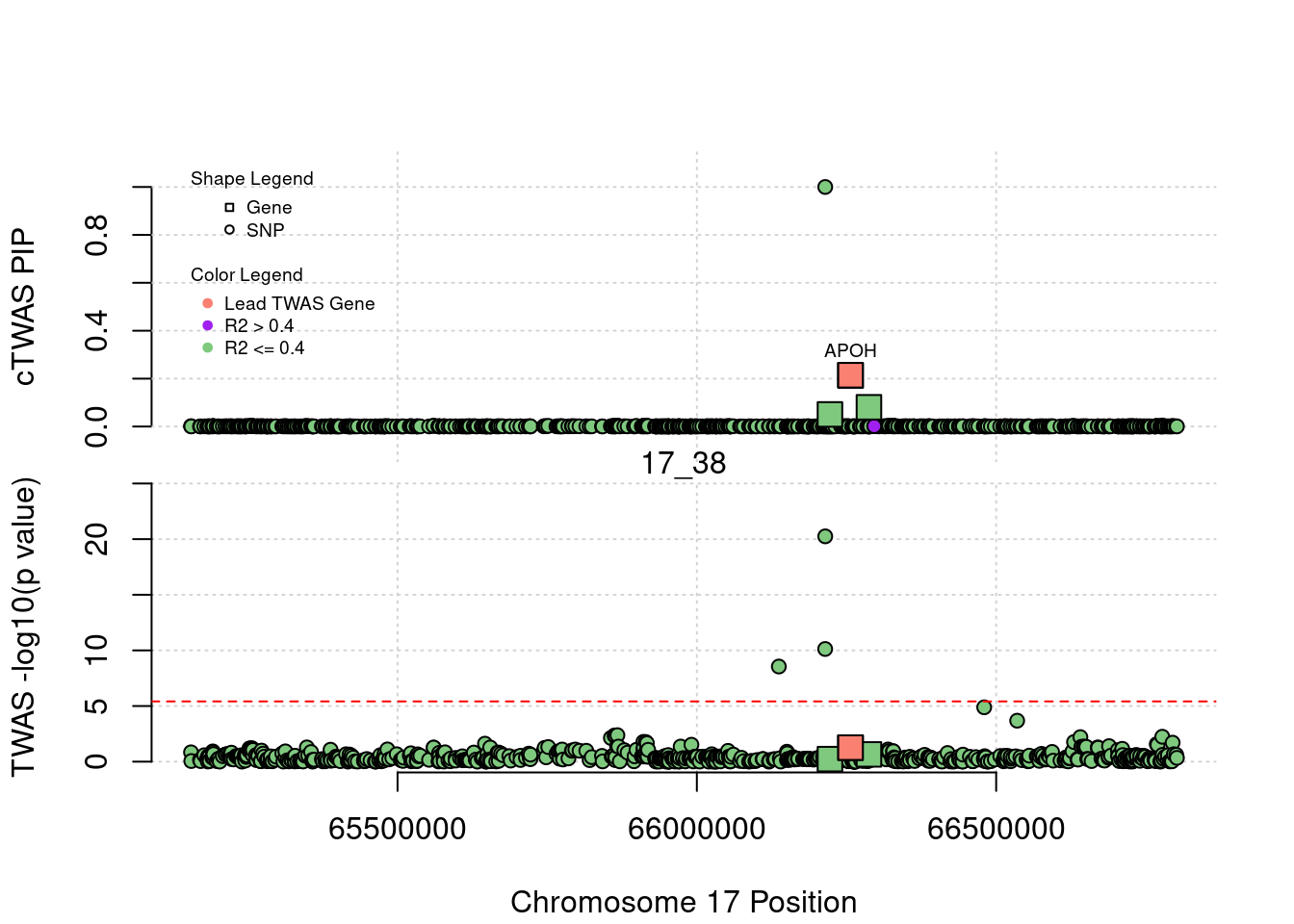
[1] "APOH"
[1] "17_38"
genename region_tag susie_pip mu2 PVE z
1453 APOH 17_38 0.21426090 17.867584 1.114113e-05 -1.9170127
7093 PRKCA 17_38 0.08046637 9.762396 2.286078e-06 -1.2170317
7095 CEP112 17_38 0.05062462 5.572178 8.209318e-07 -0.5308523
num_eqtl
1453 1
7093 3
7095 2
[1] "TSPO"
[1] "22_18"
genename region_tag susie_pip mu2 PVE z
1679 POLDIP3 22_18 0.05178187 4.633871 6.982998e-07 0.2274642
1684 CYB5R3 22_18 0.05630277 5.392417 8.835549e-07 0.5420286
1688 PACSIN2 22_18 0.07148090 7.564412 1.573568e-06 0.9678223
1689 TTLL1 22_18 0.17083194 15.676222 7.793469e-06 -2.1322245
1696 BIK 22_18 0.05092059 4.482049 6.641870e-07 0.4591806
1702 TSPO 22_18 0.05532850 5.234110 8.427758e-07 -0.4689559
1704 TTLL12 22_18 0.07272648 7.722186 1.634380e-06 -0.9795175
4423 A4GALT 22_18 0.06043456 6.035374 1.061475e-06 -0.6869080
10705 SERHL2 22_18 0.06901903 7.244560 1.455128e-06 0.8796779
11408 RRP7A 22_18 0.05433717 5.070210 8.017579e-07 -0.4501227
12780 LINC01315 22_18 0.06951375 7.309711 1.478738e-06 1.1089502
13151 ARFGAP3 22_18 0.05131251 4.551445 6.796619e-07 0.3514497
14104 RNU12 22_18 0.07141806 7.556377 1.570515e-06 0.8996741
num_eqtl
1679 1
1684 3
1688 2
1689 3
1696 1
1702 3
1704 2
4423 2
10705 4
11408 4
12780 2
13151 2
14104 1
[1] "PLTP"
[1] "20_28"
genename region_tag susie_pip mu2 PVE
332 TOMM34 20_28 0.04533655 4.445049 5.864693e-07
648 WISP2 20_28 0.04607859 4.556285 6.109847e-07
1831 PLTP 20_28 0.24925213 25.239194 1.830774e-05
1832 PCIF1 20_28 0.05235452 5.587895 8.513787e-07
1834 MMP9 20_28 0.12995649 14.283129 5.401839e-06
1841 CD40 20_28 0.37611114 23.983088 2.625074e-05
1849 PABPC1L 20_28 0.04805949 5.005387 7.000629e-07
1850 STK4 20_28 0.06987286 8.231616 1.673840e-06
1919 WFDC2 20_28 0.04573035 4.746014 6.316171e-07
1924 DNTTIP1 20_28 0.12140623 11.912726 4.208937e-06
1927 ACOT8 20_28 0.06334567 6.885678 1.269357e-06
4051 SNX21 20_28 0.04545578 5.846657 7.734229e-07
4052 SLPI 20_28 0.05020222 5.349621 7.815670e-07
4053 WFDC3 20_28 0.13959762 13.170791 5.350695e-06
4055 SLC12A5 20_28 0.12825861 19.555311 7.299138e-06
4056 SDC4 20_28 0.04597491 4.713198 6.306041e-07
4058 NCOA5 20_28 0.05691612 7.263518 1.203102e-06
4078 RBPJL 20_28 0.07347715 8.926145 1.908695e-06
4080 KCNK15 20_28 0.04809593 4.941554 6.916593e-07
4081 TP53TG5 20_28 0.05216220 6.404313 9.721846e-07
4084 NEURL2 20_28 0.24945020 26.182796 1.900729e-05
4856 SERINC3 20_28 0.08568693 10.148534 2.530685e-06
6728 JPH2 20_28 0.04662211 4.697686 6.373768e-07
8654 YWHAB 20_28 0.04679234 4.707774 6.410778e-07
8957 ZSWIM1 20_28 0.49747963 27.481750 3.978689e-05
8968 PKIG 20_28 0.04632678 4.616209 6.223545e-07
9833 UBE2C 20_28 0.06875402 8.263570 1.653431e-06
10630 WFDC10B 20_28 0.13098132 12.644729 4.819913e-06
11570 ADA 20_28 0.07021430 8.385710 1.713506e-06
11647 FITM2 20_28 0.05031676 5.386041 7.886834e-07
11781 ZNF335 20_28 0.04753503 4.821045 6.669223e-07
12040 SYS1 20_28 0.04641331 4.944932 6.679181e-07
12568 OSER1-AS1 20_28 0.08081672 9.689722 2.278940e-06
13221 DBNDD2 20_28 0.04846708 5.389257 7.601442e-07
14566 RP3-453C12.15 20_28 0.05325997 5.663392 8.778045e-07
14718 RP11-445H22.3 20_28 0.05184085 5.639257 8.507741e-07
z num_eqtl
332 -0.09477079 2
648 0.07374487 1
1831 -5.06974903 2
1832 -0.40126223 1
1834 1.76632544 1
1841 3.08821509 3
1849 -0.41033299 1
1850 -0.92647735 1
1919 0.70565748 1
1924 1.25299376 2
1927 0.50170116 2
4051 -1.04349049 2
4052 -0.52170155 2
4053 1.47955717 1
4055 3.19251988 2
4056 -0.46960970 4
4058 -1.06921473 1
4078 -1.21973824 1
4080 0.32476956 1
4081 -1.20704171 2
4084 -5.14834688 4
4856 1.31279423 2
6728 -0.28429650 4
8654 -0.26055073 2
8957 -5.21332474 2
8968 -0.27435123 2
9833 -1.29063071 1
10630 -1.43724745 1
11570 1.07627907 2
11647 0.60421286 1
11781 0.03190689 1
12040 -0.62993660 1
12568 1.23192967 3
13221 -0.82538860 2
14566 0.41494905 1
14718 0.58102262 2
[1] "LIPG"
[1] "18_27"
genename region_tag susie_pip mu2 PVE z
1942 SMAD7 18_27 0.03491073 5.878003 5.971853e-07 -0.5025531
1943 LIPG 18_27 0.04119536 8.452188 1.013299e-06 1.4047427
4967 CTIF 18_27 0.07594168 12.788218 2.826250e-06 -1.5876520
5972 DYM 18_27 0.03441476 5.662925 5.671603e-07 -0.3864952
8717 MYO5B 18_27 0.15196972 20.414857 9.028669e-06 2.7683503
8719 ACAA2 18_27 0.02985664 5.336283 4.636605e-07 -0.6137097
10091 C18orf32 18_27 0.02954518 5.592329 4.808390e-07 1.5533654
13942 RPL17 18_27 0.12896352 17.747457 6.660753e-06 -2.1255282
num_eqtl
1942 1
1943 1
4967 1
5972 3
8717 1
8719 1
10091 1
13942 2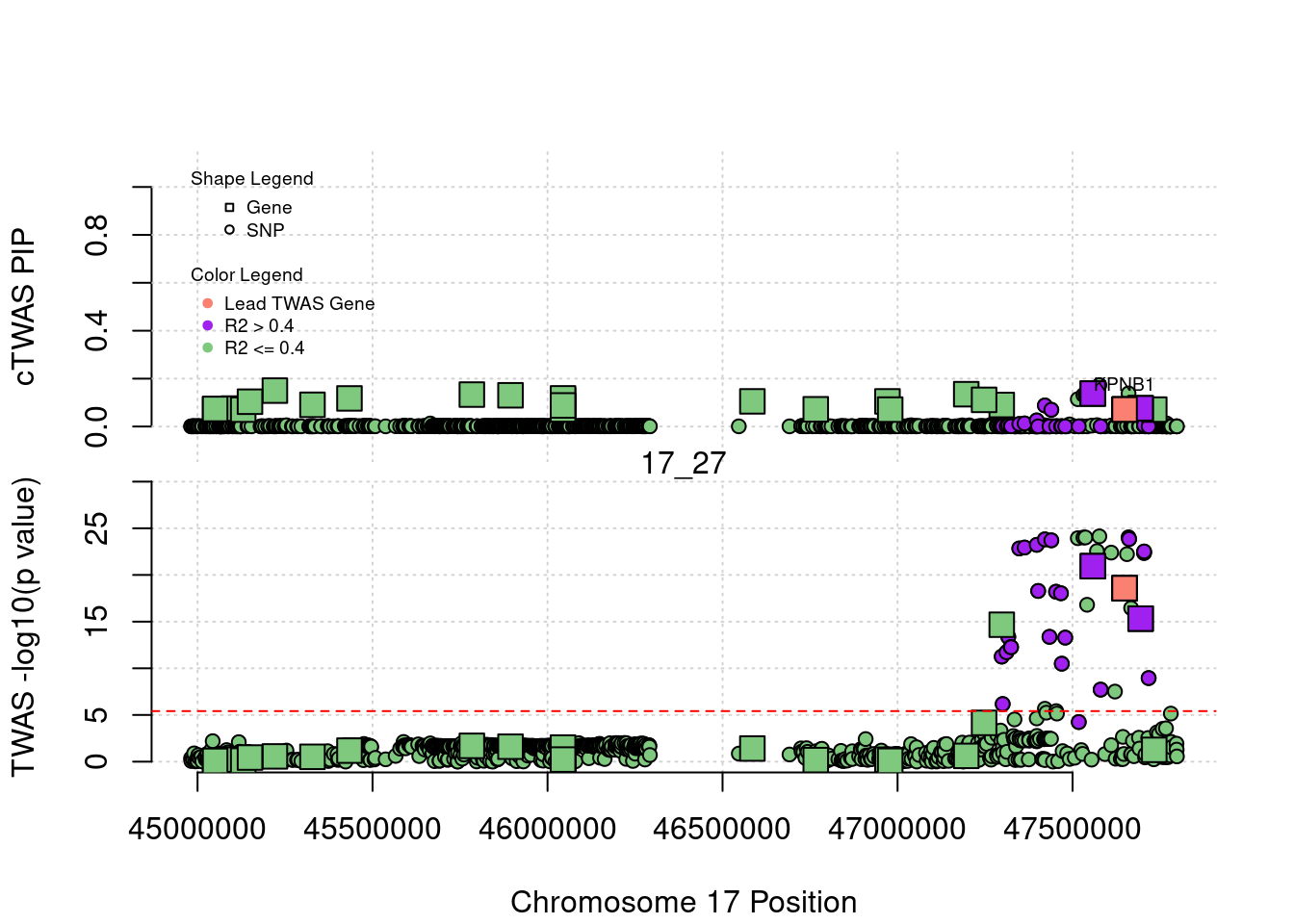
[1] "KPNB1"
[1] "17_27"
genename region_tag susie_pip mu2 PVE z
91 MAP3K14 17_27 0.09072093 6.884994 1.817738e-06 -1.0285934
920 TBX21 17_27 0.07083115 6.939598 1.430470e-06 -1.9227460
922 NSF 17_27 0.10512081 9.604572 2.938238e-06 -2.0605341
2601 WNT3 17_27 0.07205209 4.777449 1.001758e-06 0.4073694
2610 KPNB1 17_27 0.07115951 65.798026 1.362593e-05 -8.9833416
2611 GOSR2 17_27 0.10498443 7.379811 2.254709e-06 0.5031359
3740 KANSL1 17_27 0.07059257 4.726049 9.709067e-07 0.7337356
3742 CRHR1 17_27 0.13342770 9.871297 3.833015e-06 2.3227671
5301 NMT1 17_27 0.07223649 4.604850 9.680381e-07 -0.2105658
5916 NPEPPS 17_27 0.13772574 77.266226 3.096885e-05 -9.5630322
7504 ARHGAP27 17_27 0.11743928 8.577042 2.931374e-06 -1.8396376
7724 PLCD3 17_27 0.07500264 5.039775 1.100039e-06 0.5179343
9593 DCAKD 17_27 0.07339094 4.720237 1.008153e-06 0.2332407
10002 LRRC37A 17_27 0.10920883 7.720153 2.453601e-06 1.4867193
10233 EFCAB13 17_27 0.08989568 52.216423 1.366049e-05 7.9325427
10319 RPRML 17_27 0.07068060 4.598073 9.457937e-07 0.2617099
10485 ACBD4 17_27 0.07058490 4.434810 9.109765e-07 -0.3084589
10861 FMNL1 17_27 0.14994497 11.091215 4.839843e-06 1.0947893
10990 ARL17A 17_27 0.07144577 4.842763 1.006909e-06 0.5921664
11118 HEXIM1 17_27 0.10367175 7.942042 2.396144e-06 -0.9107222
11122 MAPT 17_27 0.13047166 9.704427 3.684736e-06 2.2406437
11831 MYL4 17_27 0.13504574 10.001656 3.930729e-06 -1.1719413
11984 TBKBP1 17_27 0.07623218 54.464825 1.208300e-05 -8.1169972
12749 ARL17B 17_27 0.08817019 6.058312 1.554511e-06 -0.5243534
13057 LRRC37A2 17_27 0.11749880 8.886278 3.038601e-06 2.1590825
13687 ITGB3 17_27 0.11143090 19.100830 6.194099e-06 3.9774261
13997 RP11-798G7.6 17_27 0.08817019 6.058312 1.554511e-06 -0.5243534
num_eqtl
91 1
920 1
922 1
2601 2
2610 2
2611 1
3740 2
3742 1
5301 2
5916 1
7504 2
7724 1
9593 3
10002 3
10233 5
10319 3
10485 3
10861 1
10990 2
11118 1
11122 2
11831 2
11984 1
12749 1
13057 3
13687 2
13997 1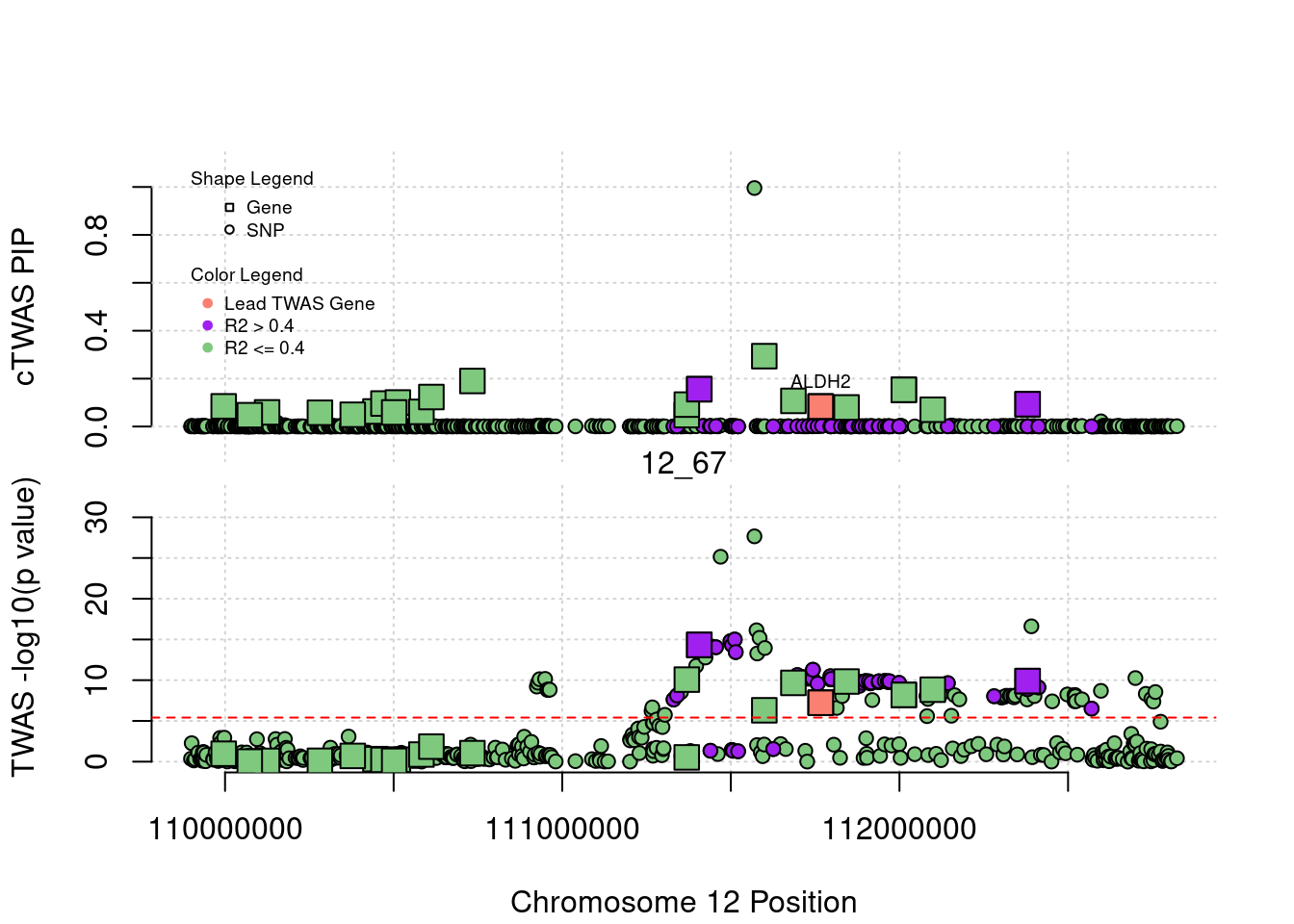
[1] "ALDH2"
[1] "12_67"
genename region_tag susie_pip mu2 PVE
1372 ERP29 12_67 0.15329765 28.289697 1.262072e-05
2872 ARPC3 12_67 0.06289760 6.513524 1.192258e-06
2873 GPN3 12_67 0.09613867 9.589092 2.682847e-06
2874 VPS29 12_67 0.10033598 9.906185 2.892567e-06
2877 SH2B3 12_67 0.15577187 44.572567 2.020584e-05
2882 ACAD10 12_67 0.10709072 30.137673 9.392515e-06
2883 ALDH2 12_67 0.08322783 24.583232 5.954261e-06
2885 NAA25 12_67 0.07014231 25.043849 5.112125e-06
3966 IFT81 12_67 0.05926969 6.067962 1.046636e-06
5723 GIT2 12_67 0.08432622 9.646757 2.367360e-06
6819 RAD9B 12_67 0.05898260 6.033048 1.035574e-06
9600 HECTD4 12_67 0.09330047 28.024117 7.609149e-06
9759 ATP2A2 12_67 0.05764606 5.868380 9.844829e-07
9763 C12orf76 12_67 0.04736602 4.432726 6.110237e-07
11056 PPP1CC 12_67 0.19006665 15.825955 8.753791e-06
11516 ANAPC7 12_67 0.04922095 5.728226 8.205223e-07
11572 PPTC7 12_67 0.06130471 7.783795 1.388691e-06
11825 TMEM116 12_67 0.15329765 28.289697 1.262072e-05
11829 FAM109A 12_67 0.04774326 5.392396 7.492283e-07
12163 ATXN2 12_67 0.29374488 28.131876 2.404857e-05
12166 TCTN1 12_67 0.12313494 14.131827 5.064073e-06
12935 MAPKAPK5-AS1 12_67 0.07979717 27.101626 6.293658e-06
13619 RP3-473L9.4 12_67 0.09147889 31.821722 8.471589e-06
z num_eqtl
1372 -5.80494471 1
2872 0.68493704 2
2873 0.51884682 1
2874 -0.55399022 1
2877 -7.83542471 1
2882 6.35151099 1
2883 -5.42199160 2
2885 6.04725215 2
3966 -0.24688650 1
5723 -1.61476918 2
6819 -0.27832495 1
9600 -6.42780377 2
9759 -0.26839506 1
9763 -0.03375633 2
11056 -1.69490241 1
11516 -1.28866259 2
11572 -1.49852195 1
11825 5.80494471 1
11829 1.03362273 1
12163 5.02733955 2
12166 2.45091897 2
12935 6.41042953 1
13619 6.49229983 2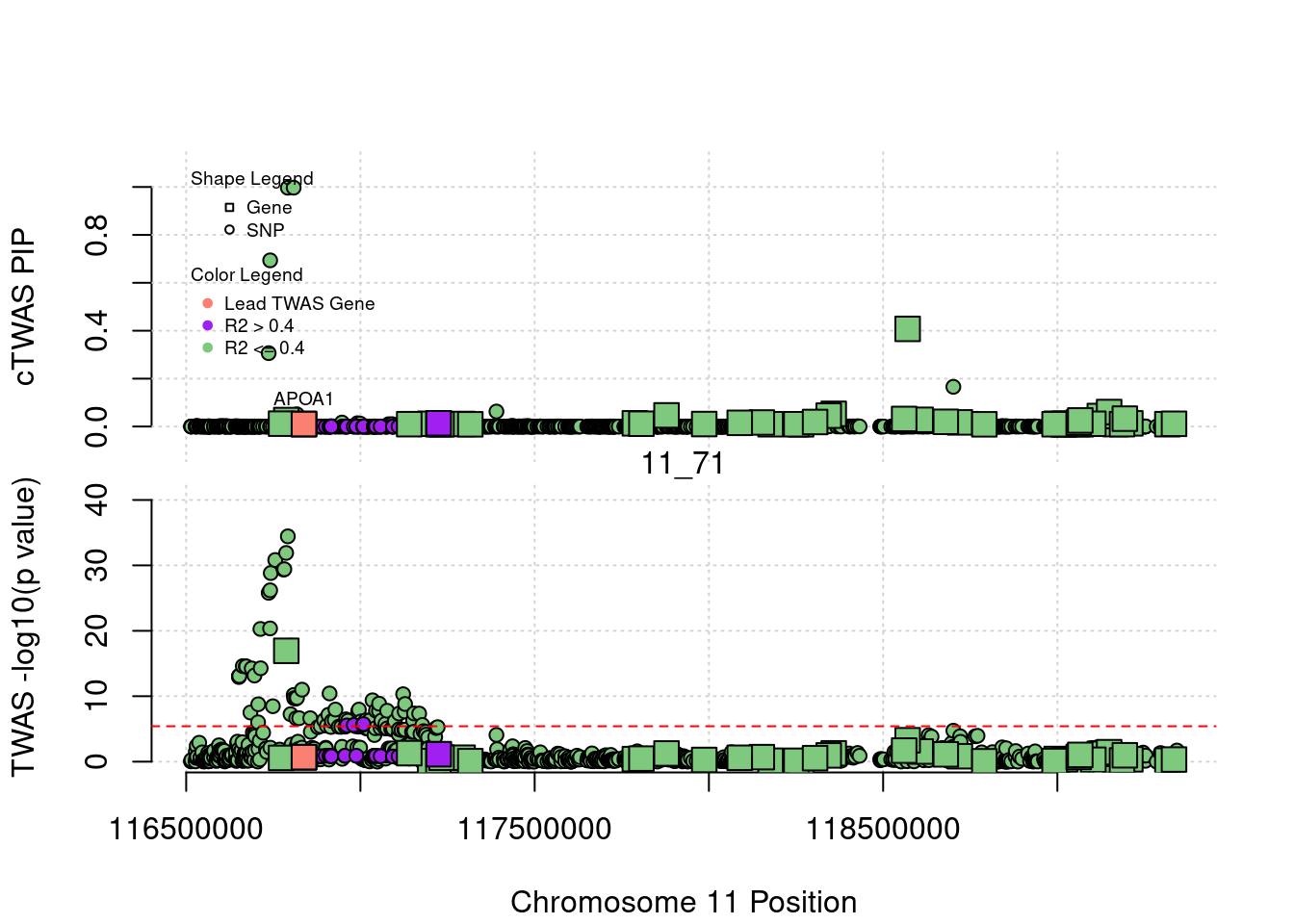
[1] "APOA1"
[1] "11_71"
genename region_tag susie_pip mu2 PVE
2758 ZPR1 11_71 0.027138501 68.532498 5.412560e-06
2796 CEP164 11_71 0.010679436 5.582893 1.735114e-07
3569 APOA1 11_71 0.010340579 6.392219 1.923609e-07
5473 BUD13 11_71 0.012132507 5.524353 1.950528e-07
5487 FXYD2 11_71 0.013365483 7.592636 2.953232e-07
6725 SIDT2 11_71 0.013582489 9.625168 3.804591e-07
6727 TAGLN 11_71 0.011561175 6.059787 2.038823e-07
7632 PCSK7 11_71 0.009501786 10.427999 2.883543e-07
8709 RNF214 11_71 0.009406343 5.546319 1.518259e-07
8876 PAFAH1B2 11_71 0.009941395 12.256689 3.546017e-07
10048 DSCAML1 11_71 0.011878776 6.836207 2.363237e-07
11059 BACE1 11_71 0.009316798 6.184582 1.676862e-07
285 PHLDB1 11_71 0.028044438 10.750712 8.774134e-07
1010 MCAM 11_71 0.009810626 4.895203 1.397616e-07
1517 ARCN1 11_71 0.407740006 22.362262 2.653502e-05
2800 IL10RA 11_71 0.010422364 5.420265 1.644020e-07
2803 UBE4A 11_71 0.053053200 18.803482 2.903155e-06
2804 DDX6 11_71 0.017482756 9.691666 4.930928e-07
2805 CBL 11_71 0.013289939 7.299399 2.823127e-07
3566 TREH 11_71 0.019744984 10.056066 5.778368e-07
3567 IFT46 11_71 0.031747840 10.423613 9.630587e-07
3571 RPS25 11_71 0.010227200 5.275859 1.570255e-07
5480 SLC37A4 11_71 0.009397659 4.537777 1.241032e-07
5485 FXYD6 11_71 0.046690588 18.268758 2.482325e-06
6703 HYOU1 11_71 0.009846593 4.836252 1.385847e-07
6723 MPZL2 11_71 0.009306304 4.443448 1.203421e-07
6724 SCN2B 11_71 0.009568653 4.688538 1.305595e-07
7629 MPZL3 11_71 0.009733494 4.844369 1.372228e-07
7633 CD3G 11_71 0.045462919 17.263010 2.283990e-06
7639 VPS11 11_71 0.009575332 4.630045 1.290207e-07
7640 NLRX1 11_71 0.010117860 5.203039 1.532026e-07
9490 HINFP 11_71 0.043989018 16.569847 2.121207e-06
9503 ABCG4 11_71 0.060640642 19.356768 3.415993e-06
9506 PDZD3 11_71 0.009938183 5.008050 1.448425e-07
9507 C2CD2L 11_71 0.011635604 6.326639 2.142310e-07
9650 RNF26 11_71 0.011399588 6.073174 2.014769e-07
10047 SCN4B 11_71 0.016975252 9.627266 4.755975e-07
11039 CCDC84 11_71 0.011845436 6.558154 2.260752e-07
11541 TRAPPC4 11_71 0.011417049 6.235715 2.071860e-07
11954 CD3E 11_71 0.018914110 10.513760 5.787144e-07
13309 CCDC153 11_71 0.032832474 14.427861 1.378561e-06
13519 RP11-110I1.12 11_71 0.009682951 4.826245 1.359995e-07
13525 TMPRSS4-AS1 11_71 0.015574798 8.944451 4.054118e-07
13574 HMBS 11_71 0.016718720 9.658456 4.699277e-07
14179 RP11-110I1.14 11_71 0.025480291 12.976492 9.622369e-07
14629 RP11-158I9.8 11_71 0.009353098 4.472246 1.217311e-07
z num_eqtl
2758 -8.558083579 3
2796 -1.151441496 2
3569 1.259896730 1
5473 1.106795181 1
5487 -0.815262931 1
6725 1.789124471 1
6727 -0.208778219 1
7632 0.427156051 3
8709 -1.149310049 2
8876 1.954200048 2
10048 -0.950975759 5
11059 -0.202145940 1
285 -2.195603226 1
1010 -0.392316857 1
1517 -3.460721439 2
2800 -0.588163124 1
2803 1.931041428 2
2804 -1.440092098 2
2805 0.768500019 1
3566 -1.769814683 1
3567 -2.321005506 2
3571 -0.553265539 2
5480 -0.175372766 3
5485 -1.905698366 1
6703 -0.149647514 1
6723 -0.007818612 2
6724 -0.347449108 1
7629 -0.333954065 2
7633 1.597891521 1
7639 0.043651363 3
7640 -0.491017849 2
9490 1.858723296 1
9503 2.097751950 2
9506 -0.444556851 1
9507 -0.651233403 1
9650 0.713961504 1
10047 1.267144486 1
11039 0.858181559 1
11541 0.798148148 1
11954 1.057119238 1
13309 1.620441106 2
13519 0.275624342 1
13525 -1.131537430 1
13574 -1.302989641 3
14179 1.677677763 3
14629 0.141389981 3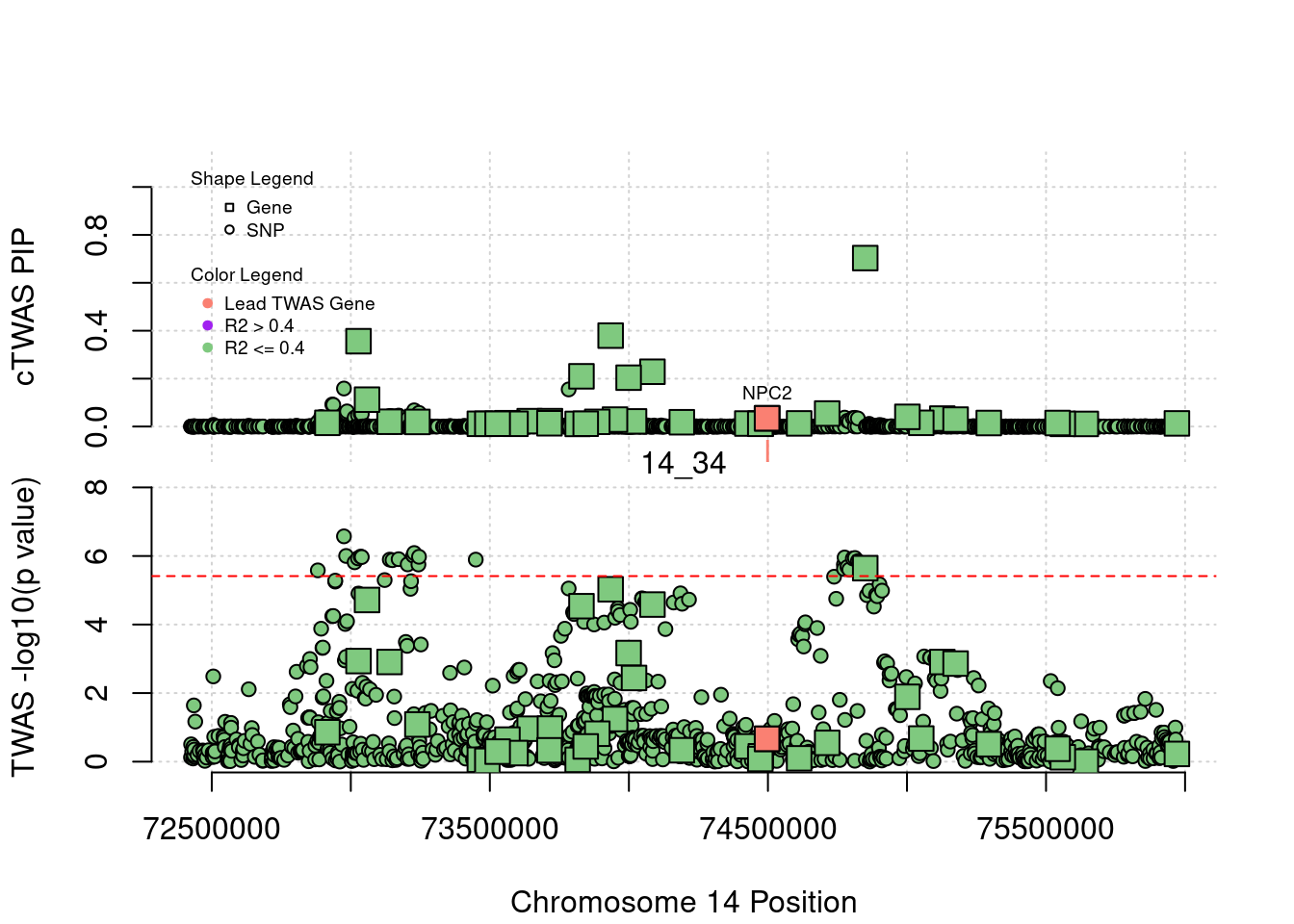
[1] "NPC2"
[1] "14_34"
genename region_tag susie_pip mu2 PVE
1114 PSEN1 14_34 0.02010397 16.375048 9.580424e-07
1804 PAPLN 14_34 0.01925153 8.438763 4.727857e-07
3677 DCAF4 14_34 0.01540989 8.667533 3.887006e-07
3678 PROX2 14_34 0.70325263 23.490338 4.807518e-05
3679 VSX2 14_34 0.01791110 8.634819 4.500864e-07
3682 BBOF1 14_34 0.02156863 9.446067 5.929170e-07
3683 NEK9 14_34 0.03309955 18.937215 1.824141e-06
3685 NPC2 14_34 0.03436382 13.214789 1.321545e-06
3686 DNAL1 14_34 0.02242416 9.885930 6.451401e-07
3688 ACOT2 14_34 0.01401022 6.851498 2.793513e-07
3689 LTBP2 14_34 0.01304209 5.766042 2.188494e-07
3690 AREL1 14_34 0.05425110 16.198697 2.557460e-06
3691 MLH3 14_34 0.01557866 8.131928 3.686752e-07
3692 TTLL5 14_34 0.01095600 4.450475 1.418988e-07
3693 FLVCR2 14_34 0.01130986 4.726730 1.555745e-07
3694 ABCD4 14_34 0.01098056 4.491665 1.435332e-07
3698 RBM25 14_34 0.11226918 20.179307 6.593061e-06
3699 ALDH6A1 14_34 0.22883768 20.398357 1.358448e-05
3702 EIF2B2 14_34 0.04052428 17.829408 2.102677e-06
3703 COQ6 14_34 0.02898938 13.818418 1.165782e-06
3704 ZNF410 14_34 0.01967664 9.342881 5.349978e-07
4958 NUMB 14_34 0.01159366 4.943189 1.667816e-07
5786 PTGR2 14_34 0.01170132 4.943757 1.683497e-07
7225 ELMSAN1 14_34 0.02257333 9.948175 6.535207e-07
7228 FAM161B 14_34 0.37956419 23.454004 2.590732e-05
7236 BATF 14_34 0.01474757 7.074562 3.036270e-07
8454 ZFYVE1 14_34 0.35634642 18.672437 1.936394e-05
9204 TMED10 14_34 0.02895004 17.858622 1.504587e-06
9227 RIOX1 14_34 0.01142021 4.994770 1.660007e-07
10019 PNMA1 14_34 0.01376277 6.108375 2.446538e-07
10080 ACOT4 14_34 0.01147450 5.176126 1.728459e-07
10684 SYNDIG1L 14_34 0.01265595 6.044141 2.226125e-07
10796 ACOT1 14_34 0.01183906 5.899872 2.032732e-07
11145 ENTPD5 14_34 0.20396123 26.614401 1.579736e-05
13651 RP5-1021I20.2 14_34 0.20971787 20.664322 1.261180e-05
13668 RP11-270M14.5 14_34 0.01309887 5.940865 2.264664e-07
13680 RP3-449M8.6 14_34 0.01196014 5.243296 1.824991e-07
13714 LINC01220 14_34 0.01465376 7.139045 3.044455e-07
14100 RP3-449M8.9 14_34 0.01144832 4.806645 1.601416e-07
z num_eqtl
1114 3.229758391 2
1804 1.737216658 1
3677 1.480577350 4
3678 4.728159457 1
3679 -0.730399204 3
3682 -2.910226590 2
3683 -3.229698770 1
3685 1.236891294 1
3686 1.601799548 1
3688 -1.206396425 4
3689 0.253509565 1
3690 -1.073394574 2
3691 -1.248354840 1
3692 -0.004986035 2
3693 -0.323831005 2
3694 -0.146051273 2
3698 4.273809776 1
3699 4.204641862 1
3702 -2.487675584 2
3703 1.903428327 2
3704 1.429488618 2
4958 0.153840456 1
5786 -0.916339410 5
7225 1.605708757 1
7228 -4.431133058 2
7236 -0.804124148 1
8454 3.247488674 1
9204 -3.199737076 1
9227 0.069112089 1
10019 0.727262838 3
10080 0.578384062 2
10684 0.918206255 1
10796 0.658176457 4
11145 -3.397736054 2
13651 4.180026519 1
13668 0.530733050 1
13680 -0.386452409 2
13714 1.016718420 3
14100 -0.204891747 3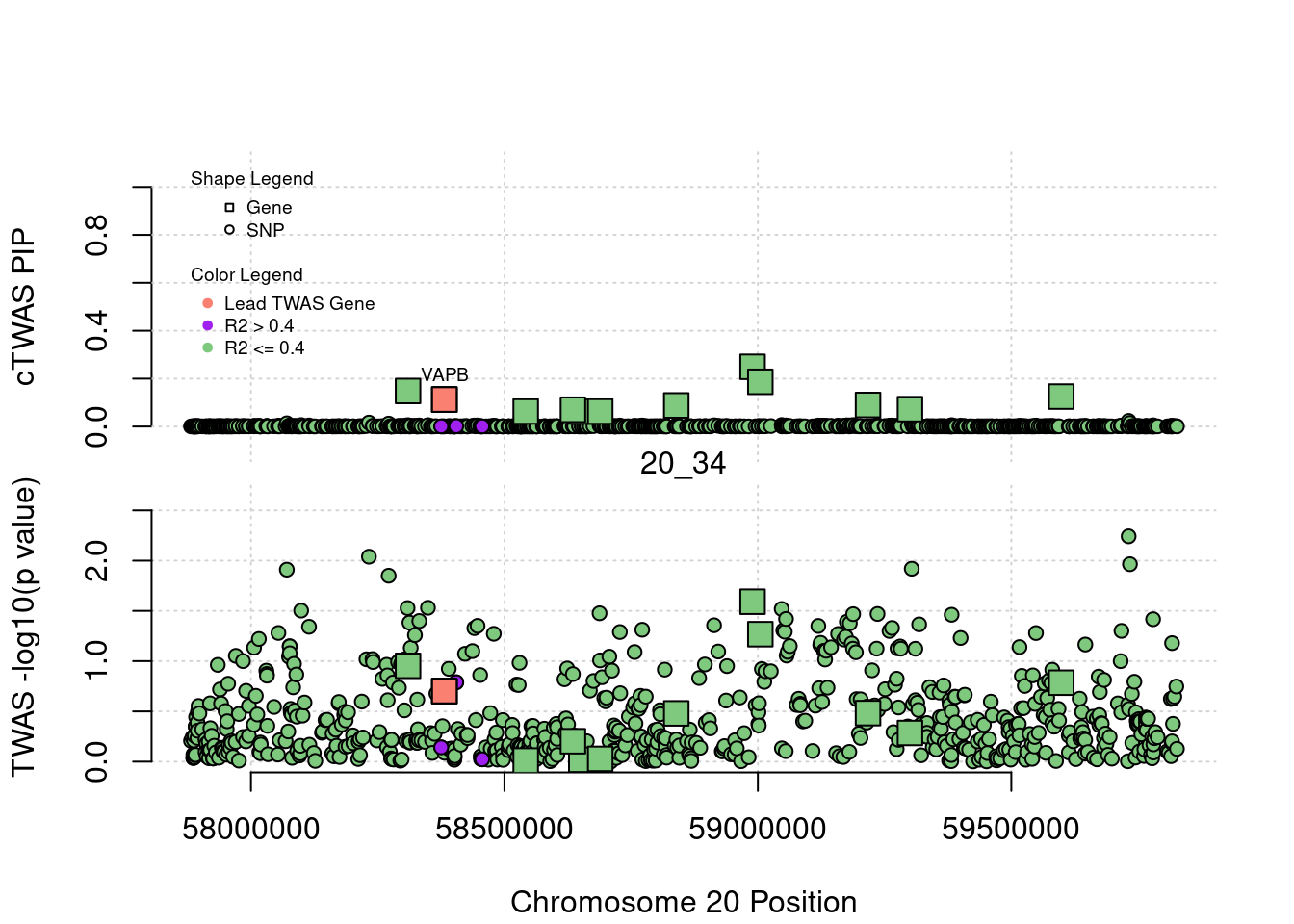
[1] "VAPB"
[1] "20_34"
genename region_tag susie_pip mu2 PVE z
1301 GNAS 20_34 0.08821990 7.538587 1.935427e-06 -0.97211018
1304 PHACTR3 20_34 0.12515137 10.780858 3.926533e-06 -1.39239344
1859 NELFCD 20_34 0.24935052 17.409189 1.263308e-05 2.23141397
1860 CTSZ 20_34 0.18660840 14.570499 7.912722e-06 -1.92748353
1862 TUBB1 20_34 0.09001034 7.723481 2.023139e-06 -0.97658919
4059 VAPB 20_34 0.11277927 9.809866 3.219680e-06 -1.28562576
4067 EDN3 20_34 0.07270281 5.765366 1.219827e-06 0.64574281
4069 RAB22A 20_34 0.14843201 12.385561 5.350121e-06 1.59196827
4075 STX16 20_34 0.06280330 4.431077 8.098640e-07 -0.05327087
11919 APCDD1L 20_34 0.06281315 4.432504 8.102517e-07 -0.03161543
12487 NPEPL1 20_34 0.06304855 4.466533 8.195321e-07 -0.07722682
14066 LINC01711 20_34 0.07021562 5.447525 1.113149e-06 -0.48678992
num_eqtl
1301 1
1304 2
1859 1
1860 2
1862 1
4059 3
4067 2
4069 1
4075 1
11919 1
12487 3
14066 1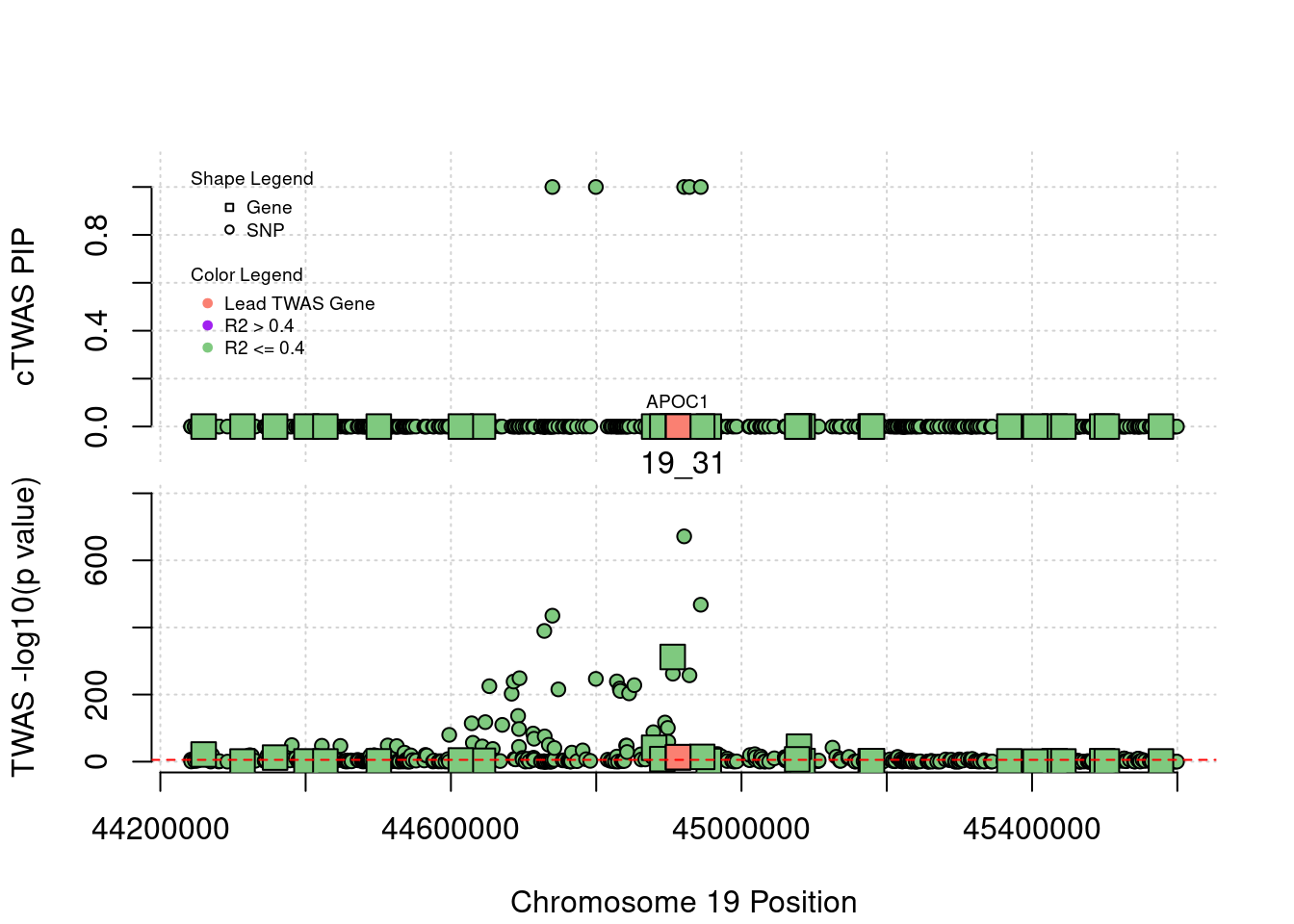
[1] "APOC1"
[1] "19_31"
genename region_tag susie_pip mu2 PVE z num_eqtl
123 MARK4 19_31 0 20.312700 0 -2.24637681 1
131 TRAPPC6A 19_31 0 16.600235 0 1.92140959 1
229 ERCC1 19_31 0 13.134793 0 -2.19401016 2
624 ZNF112 19_31 0 72.068605 0 7.18938487 3
898 PVR 19_31 0 30.112592 0 -3.01973306 1
2196 CLPTM1 19_31 0 48.802068 0 -2.57517256 1
2198 PPP1R37 19_31 0 35.405859 0 -2.37518201 2
2203 ERCC2 19_31 0 13.615522 0 1.53401174 1
3560 CD3EAP 19_31 0 16.730266 0 0.05565582 4
4222 OPA3 19_31 0 11.581430 0 1.32335471 2
4224 RTN2 19_31 0 6.803450 0 2.07007103 1
4226 VASP 19_31 0 23.808738 0 1.26035740 1
4570 NECTIN2 19_31 0 55.577675 0 -13.59762224 1
4571 APOE 19_31 0 999.967785 0 37.80754977 2
4572 TOMM40 19_31 0 56.000201 0 5.53230466 2
4573 APOC1 19_31 0 296.620171 0 7.53014709 1
6024 GEMIN7 19_31 0 157.439551 0 14.13454209 2
7552 ZNF233 19_31 0 98.228951 0 -9.63193736 1
7553 ZNF235 19_31 0 8.185909 0 -0.97877558 1
8729 ZNF180 19_31 0 25.642421 0 1.41443321 2
9263 ZNF296 19_31 0 65.538646 0 5.25423092 2
11089 CEACAM19 19_31 0 13.759203 0 3.64086985 3
11388 BLOC1S3 19_31 0 8.975124 0 2.30141189 1
12385 PPM1N 19_31 0 20.227762 0 -1.22009641 2
12950 APOC2 19_31 0 99.467017 0 7.82468739 2
14021 ZNF285 19_31 0 10.013342 0 -1.26709542 2
14628 ZNF229 19_31 0 8.630054 0 1.23011935 2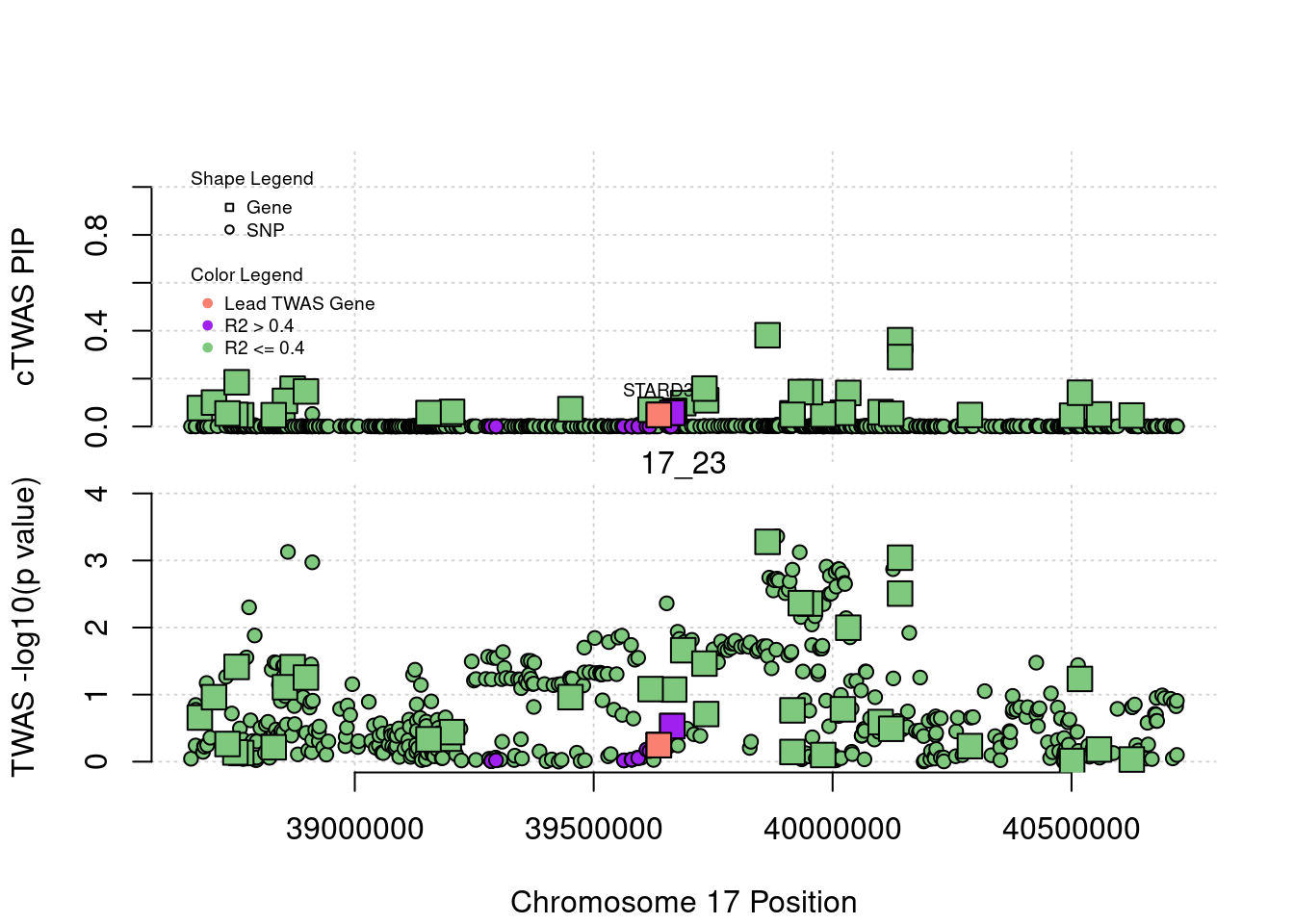
[1] "STARD3"
[1] "17_23"
genename region_tag susie_pip mu2 PVE
19 LASP1 17_23 0.15799930 15.920978 7.320575e-06
170 MED24 17_23 0.14028386 14.795945 6.040469e-06
743 CACNB1 17_23 0.05220395 5.697116 8.655232e-07
912 SMARCE1 17_23 0.04608221 4.569080 6.127486e-07
913 GSDMB 17_23 0.05519144 6.201387 9.960493e-07
1508 CDC6 17_23 0.04814919 4.965566 6.957897e-07
2596 CSF3 17_23 0.05727969 6.538324 1.089902e-06
2597 PSMD3 17_23 0.04999169 5.305080 7.718094e-07
2598 CASC3 17_23 0.36066342 24.150651 2.534844e-05
2599 RAPGEFL1 17_23 0.29020290 21.883511 1.848158e-05
4213 MED1 17_23 0.07252390 8.686548 1.833364e-06
4214 RPL23 17_23 0.10711135 12.274085 3.826000e-06
4291 CCR7 17_23 0.05066990 5.427040 8.002642e-07
4292 NR1D1 17_23 0.06016118 6.983963 1.222753e-06
4739 TNS4 17_23 0.04538777 4.431962 5.854033e-07
4740 STARD3 17_23 0.04866297 5.061517 7.168027e-07
5983 ERBB2 17_23 0.09768485 11.421362 3.246874e-06
5984 GRB7 17_23 0.10964113 12.490218 3.985326e-06
5985 MIEN1 17_23 0.15859217 15.956518 7.364448e-06
5986 PNMT 17_23 0.06528679 7.727697 1.468235e-06
5987 STAC2 17_23 0.06127600 7.150777 1.275158e-06
7700 PLXDC1 17_23 0.05617141 6.360899 1.039810e-06
7702 IKZF3 17_23 0.38108622 24.739571 2.743694e-05
8833 GSDMA 17_23 0.14346404 15.007260 6.265630e-06
9459 ORMDL3 17_23 0.14399874 15.042193 6.303622e-06
9715 TCAP 17_23 0.05755478 6.581774 1.102414e-06
11355 MSL1 17_23 0.05333864 5.891908 9.145726e-07
12444 AC087491.2 17_23 0.07034685 8.408242 1.721354e-06
13915 LINC00672 17_23 0.14645923 15.202371 6.479603e-06
13932 RP11-387H17.4 17_23 0.04908556 5.139456 7.341608e-07
13955 RP5-1028K7.2 17_23 0.14122966 14.859370 6.107263e-06
14405 CWC25 17_23 0.04729257 4.803271 6.610744e-07
14406 EPOP 17_23 0.07709694 9.245577 2.074395e-06
14476 MLLT6 17_23 0.10018332 11.654991 3.398034e-06
14540 PIP4K2B 17_23 0.04712234 4.770713 6.542300e-07
14581 PCGF2 17_23 0.04842224 5.016662 7.069358e-07
14607 PSMB3 17_23 0.18491519 17.426240 9.377705e-06
14611 CISD3 17_23 0.05458616 6.101300 9.692263e-07
z num_eqtl
19 2.06384703 1
170 2.57367225 1
743 -0.74789947 1
912 0.08854994 1
913 1.36595451 3
1508 -0.54487846 2
2596 -1.38591286 1
2597 -0.25796073 1
2598 -3.31731790 4
2599 2.95599474 2
4213 -1.60223183 2
4214 1.76434090 1
4291 -0.44220762 1
4292 1.13477197 1
4739 0.02859681 1
4740 -0.57272843 2
5983 -2.29348339 2
5984 -1.29578288 2
5985 2.11399068 1
5986 -1.72758728 2
5987 -0.90983233 1
7700 0.73156690 1
7702 3.46618563 1
8833 2.84876635 2
9459 2.85322256 1
9715 1.04770013 1
11355 0.98720359 1
12444 1.73462940 1
13915 1.91358441 1
13932 0.36181731 3
13955 1.89232102 1
14405 -0.51001116 1
14406 -1.22710237 1
14476 -1.60128295 1
14540 -0.32703052 3
14581 0.34617143 2
14607 2.06810346 2
14611 -0.60454978 1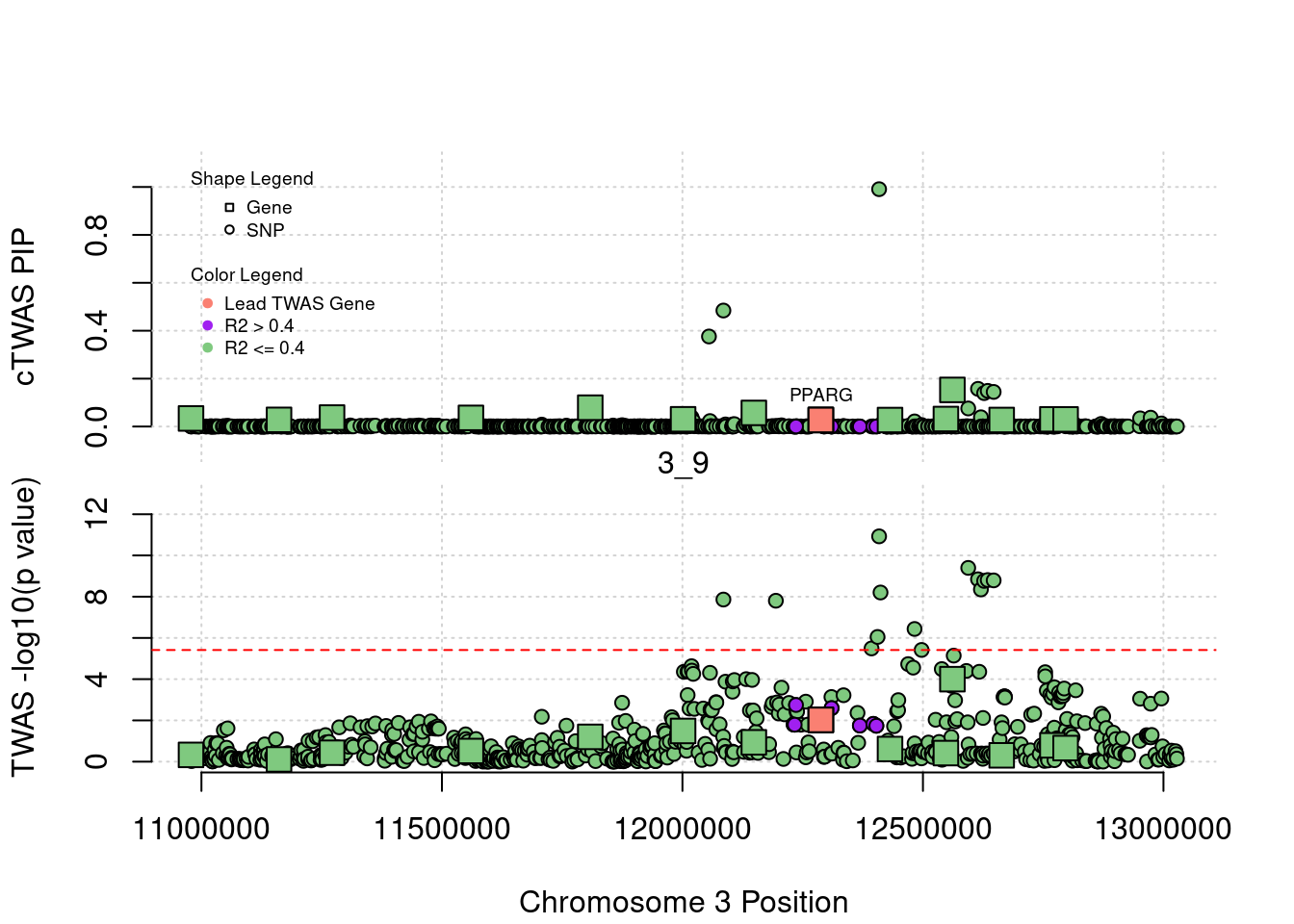
[1] "PPARG"
[1] "3_9"
genename region_tag susie_pip mu2 PVE z
988 MKRN2 3_9 0.03135400 5.430624 4.955221e-07 -0.8716686
1332 TMEM40 3_9 0.03013251 6.094506 5.344340e-07 1.3996166
4769 RAF1 3_9 0.02731073 4.897512 3.892505e-07 -0.6842182
4770 PPARG 3_9 0.02722444 8.708239 6.899373e-07 -2.5953663
6298 TAMM41 3_9 0.07878073 13.991641 3.207812e-06 1.8598576
6299 VGLL4 3_9 0.03609933 6.977527 7.330287e-07 1.0266781
6323 CAND2 3_9 0.02964105 5.796627 5.000221e-07 -1.2257763
7134 TSEN2 3_9 0.02716448 5.059164 3.999451e-07 1.1650072
7314 SLC6A1 3_9 0.03379825 6.518335 6.411375e-07 0.7212287
7319 TIMP4 3_9 0.05692187 11.262681 1.865697e-06 1.5758546
7320 SYN2 3_9 0.03018848 6.735250 5.917187e-07 2.1389711
11537 HRH1 3_9 0.02794624 4.806876 3.909368e-07 0.2808860
11684 ATG7 3_9 0.03751242 7.285704 7.953657e-07 -0.8966031
12619 MKRN2OS 3_9 0.15296674 17.188665 7.651727e-06 3.8901034
num_eqtl
988 3
1332 1
4769 1
4770 1
6298 3
6299 1
6323 3
7134 4
7314 3
7319 3
7320 2
11537 2
11684 3
12619 2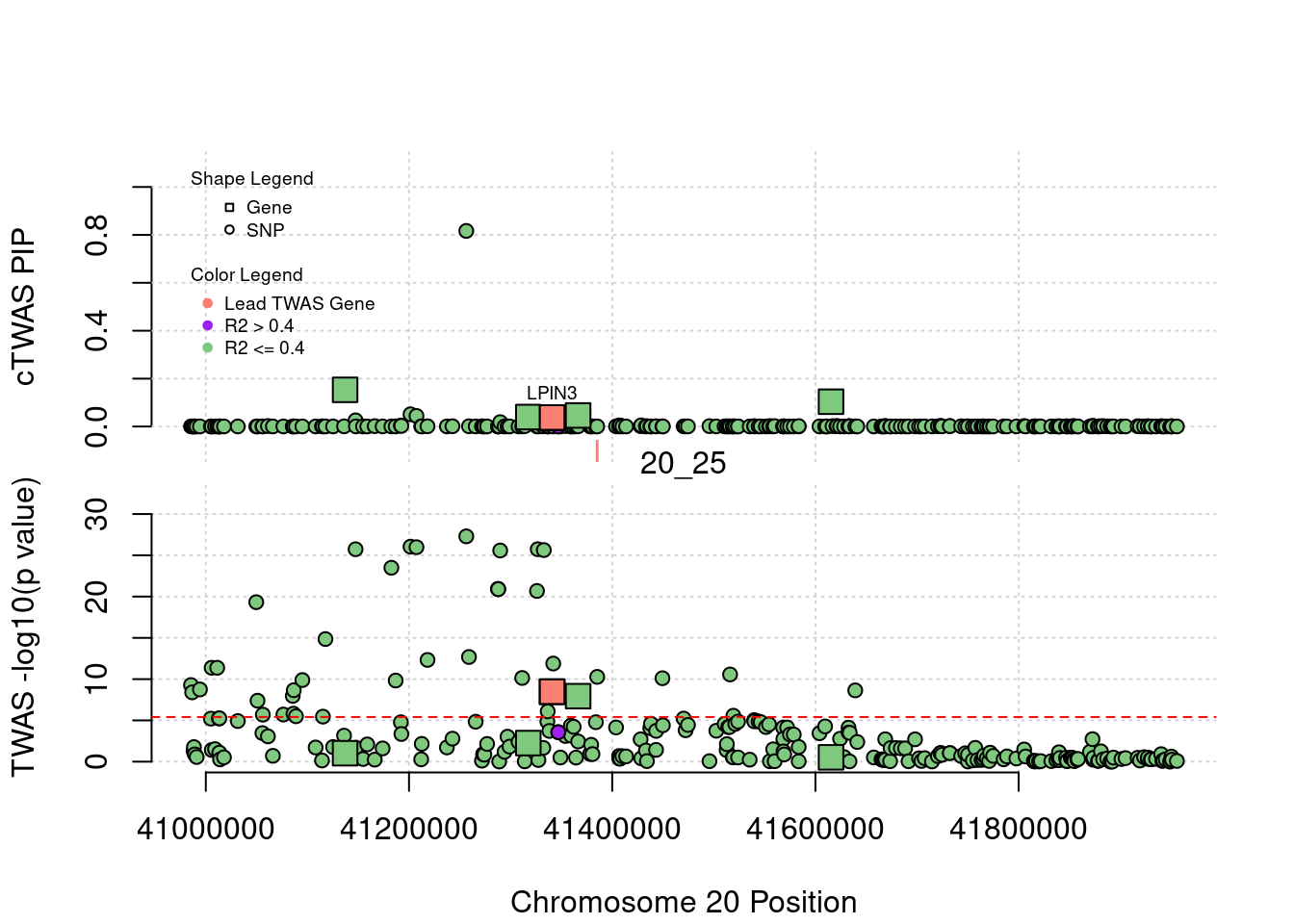
[1] "LPIN3"
[1] "20_25"
genename region_tag susie_pip mu2 PVE z
4061 CHD6 20_25 0.10371263 12.68028 3.827198e-06 1.035059
4062 PLCG1 20_25 0.15328065 16.47045 7.347050e-06 1.671800
4852 LPIN3 20_25 0.03769762 31.76804 3.485176e-06 5.915115
9747 ZHX3 20_25 0.03951900 11.71552 1.347373e-06 2.767903
10750 EMILIN3 20_25 0.04636140 31.38957 4.235086e-06 5.704566
num_eqtl
4061 2
4062 2
4852 2
9747 1
10750 2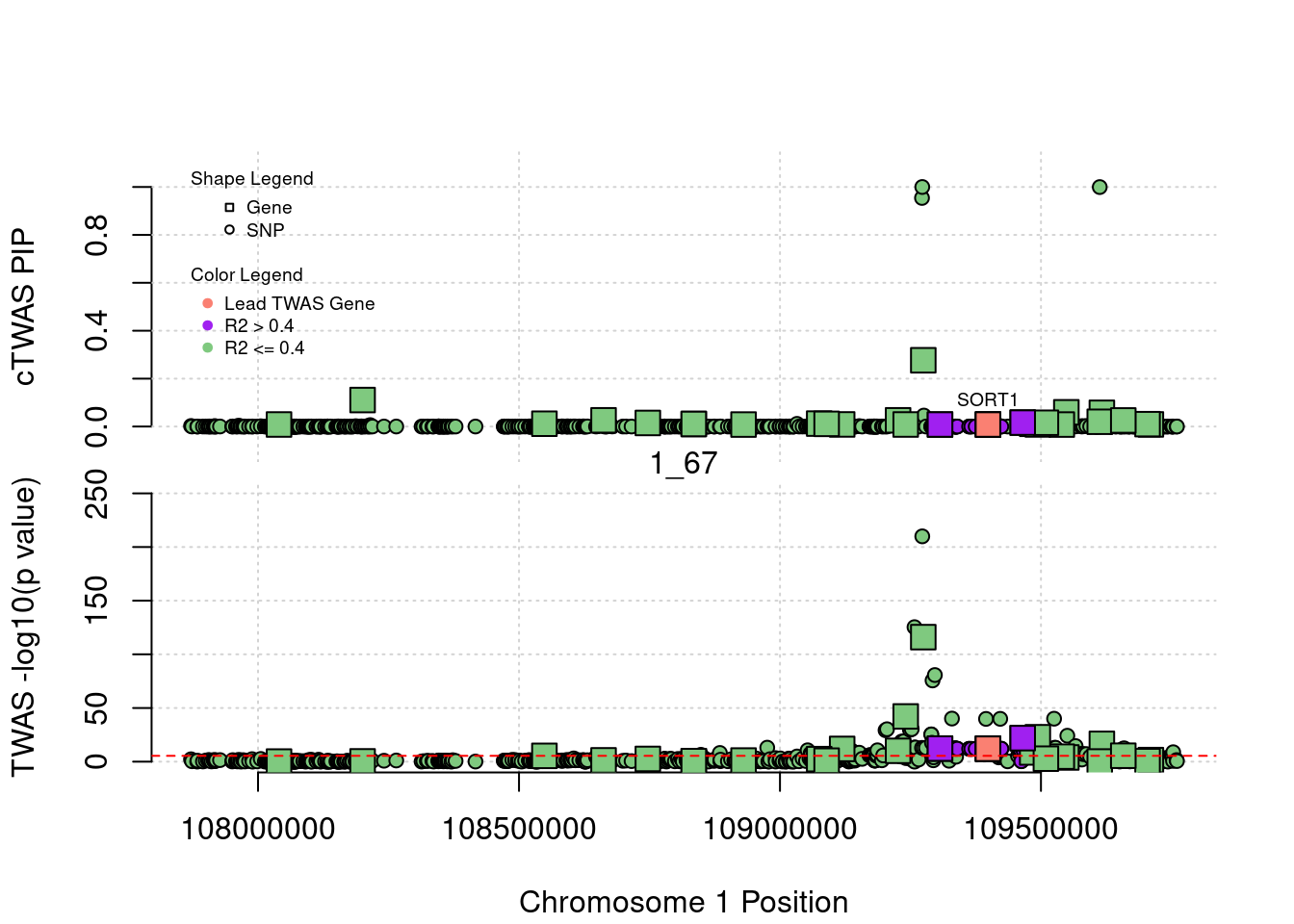
[1] "SORT1"
[1] "1_67"
genename region_tag susie_pip mu2 PVE
366 SARS 1_67 0.008842491 24.257145 6.242156e-07
680 GNAI3 1_67 0.059461335 36.597463 6.332948e-06
1230 SLC25A24 1_67 0.110623224 19.040173 6.129676e-06
3407 STXBP3 1_67 0.014885979 16.452942 7.127567e-07
3409 AMPD2 1_67 0.057412268 44.195010 7.384112e-06
3878 CLCC1 1_67 0.008350193 5.020227 1.219945e-07
3879 GPSM2 1_67 0.010770874 6.955047 2.180074e-07
4984 GSTM1 1_67 0.026030135 38.410146 2.909663e-06
4986 GSTM5 1_67 0.008934125 9.401227 2.444313e-07
4987 GSTM3 1_67 0.010048438 7.235686 2.115917e-07
4991 PSRC1 1_67 0.276724165 192.009108 1.546284e-04
4994 SORT1 1_67 0.008445869 20.739040 5.097454e-07
6084 SYPL2 1_67 0.016453770 49.423212 2.366555e-06
6093 CELSR2 1_67 0.008522725 109.890397 2.725577e-06
7232 GPR61 1_67 0.008286089 9.151074 2.206693e-07
7834 FAM102B 1_67 0.011788953 15.577365 5.344284e-07
7835 HENMT1 1_67 0.025890373 14.844196 1.118447e-06
7836 AKNAD1 1_67 0.010770874 6.955047 2.180074e-07
7840 ATXN7L2 1_67 0.011482988 14.603293 4.880070e-07
8972 GSTM4 1_67 0.019958084 16.303653 9.469435e-07
9731 CYB561D1 1_67 0.008784524 51.319596 1.311964e-06
10499 AMIGO1 1_67 0.015914284 29.501073 1.366297e-06
11729 TAF13 1_67 0.012348876 13.705360 4.925362e-07
12337 GSTM2 1_67 0.025354958 25.302853 1.867036e-06
12493 TMEM167B 1_67 0.011335261 15.309187 5.050146e-07
12546 MYBPHL 1_67 0.008420809 21.076428 5.165010e-07
12677 RP11-356N1.2 1_67 0.008511622 4.878611 1.208450e-07
z num_eqtl
366 -7.0857282 2
680 -3.8408490 1
1230 -0.6406888 2
3407 2.9914703 1
3409 8.4673955 1
3878 -1.7197469 4
3879 1.0104135 1
4984 6.5012768 6
4986 2.0991563 4
4987 -1.0142898 4
4991 -22.9681237 2
4994 -7.1373768 1
6084 9.7928029 4
6093 13.7257529 2
7232 -4.2425343 1
7834 -4.6211180 2
7835 -1.8537405 1
7836 1.0104135 1
7840 5.9371453 1
8972 -0.3531182 7
9731 9.9823505 2
10499 -3.1539161 2
11729 -2.8413870 2
12337 4.6633215 4
12493 1.5934686 1
12546 -7.1894797 1
12677 -0.2308337 2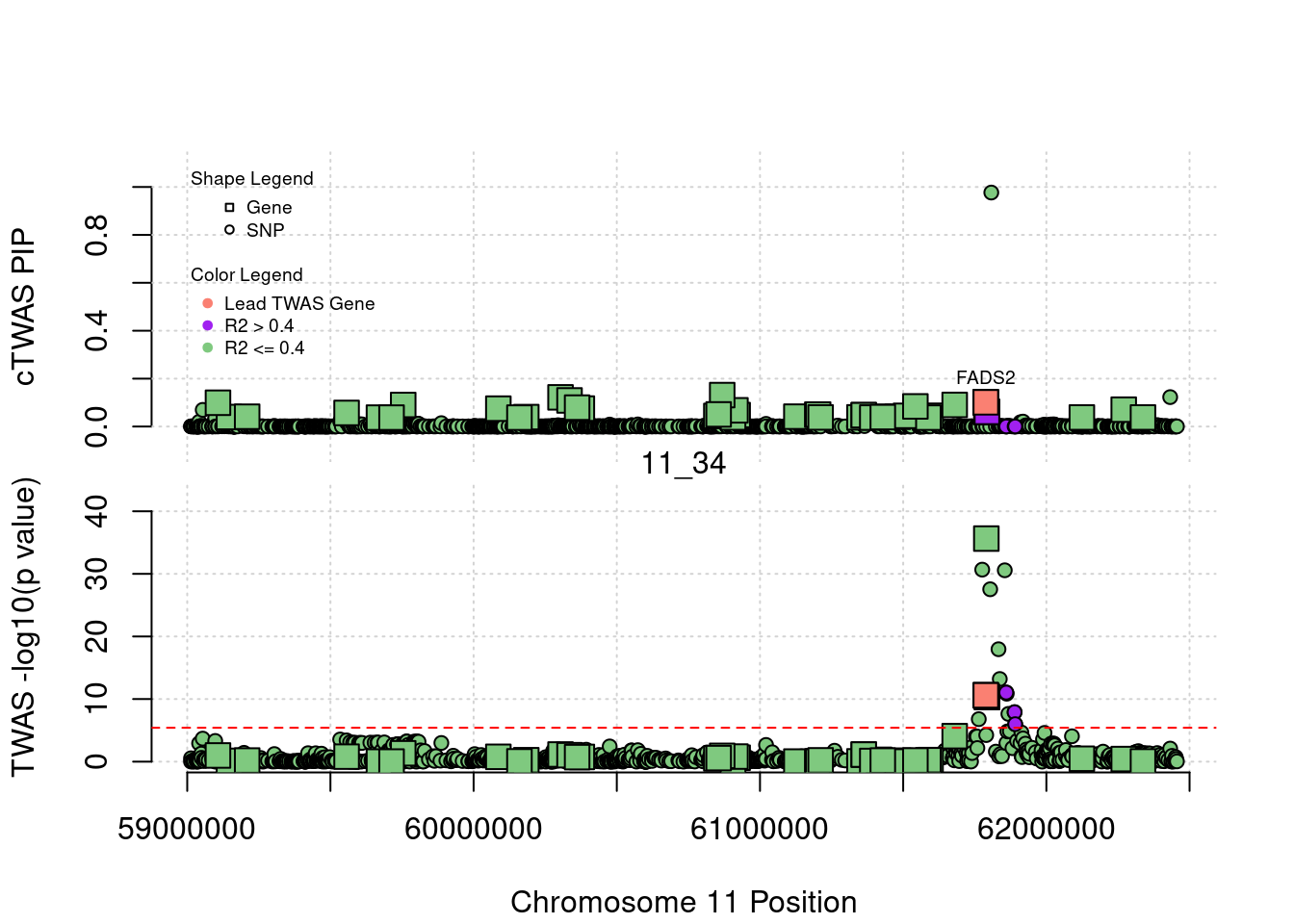
[1] "FADS2"
[1] "11_34"
genename region_tag susie_pip mu2 PVE
93 TMEM132A 11_34 0.04785366 7.015339 9.769766e-07
217 SYT7 11_34 0.04504220 6.171242 8.089327e-07
2779 MS4A6A 11_34 0.03942139 4.898568 5.619806e-07
2780 MS4A4A 11_34 0.12175706 14.565575 5.161098e-06
2784 CCDC86 11_34 0.04414017 6.348683 8.155262e-07
2785 PRPF19 11_34 0.03764336 4.544073 4.977989e-07
2786 TMEM109 11_34 0.06783652 9.977574 1.969740e-06
2812 CD5 11_34 0.04113275 5.168916 6.187390e-07
2814 SCGB2A2 11_34 0.07055845 9.376180 1.925286e-06
5063 DAGLA 11_34 0.09030423 22.360980 5.876507e-06
5068 FADS2 11_34 0.10124455 48.582684 1.431441e-05
5069 TMEM258 11_34 0.06130644 42.202763 7.529519e-06
6706 TKFC 11_34 0.03835879 4.681012 5.225466e-07
6708 TMEM138 11_34 0.04744389 8.607896 1.188496e-06
6713 INCENP 11_34 0.03775693 5.226734 5.743114e-07
6714 ZP1 11_34 0.13126658 15.042527 5.746392e-06
6716 CPSF7 11_34 0.04065351 5.418793 6.410929e-07
6717 MS4A2 11_34 0.07462519 10.413658 2.261565e-06
7760 CYB561A3 11_34 0.04744389 8.607896 1.188496e-06
7761 PPP1R32 11_34 0.04071487 5.212588 6.176276e-07
7762 ASRGL1 11_34 0.03781915 4.604150 5.067357e-07
8626 FAM111A 11_34 0.03961979 4.961783 5.720977e-07
8648 PATL1 11_34 0.03725178 4.459940 4.834998e-07
8651 STX3 11_34 0.09014207 12.338229 3.236687e-06
8658 MS4A6E 11_34 0.10910015 13.511300 4.289857e-06
8659 MS4A7 11_34 0.07506493 10.635878 2.323436e-06
8660 MS4A14 11_34 0.07895441 10.955159 2.517186e-06
8849 DDB1 11_34 0.04953782 6.511309 9.386971e-07
8941 FEN1 11_34 0.06056125 130.936081 2.307674e-05
9491 OR10V1 11_34 0.03732469 4.468866 4.854158e-07
10660 PTGDR2 11_34 0.05069792 7.427473 1.095851e-06
11140 TMEM216 11_34 0.03746280 4.562447 4.974145e-07
11381 FAM111B 11_34 0.09936200 12.979566 3.753192e-06
11705 MPEG1 11_34 0.04099168 5.378373 6.416038e-07
12177 LRRC10B 11_34 0.04467299 5.764632 7.494400e-07
12779 PGA3 11_34 0.05000776 6.578712 9.574115e-07
13357 RP11-855O10.2 11_34 0.03720880 4.688916 5.077365e-07
13511 AP001257.1 11_34 0.03715880 4.438625 4.799881e-07
13516 AP000442.4 11_34 0.05687255 8.241387 1.364028e-06
13579 RP11-794G24.1 11_34 0.08412300 10.437753 2.555301e-06
13586 RP11-286N22.8 11_34 0.03932925 4.983081 5.703402e-07
13590 PGA5 11_34 0.03801922 4.886600 5.406674e-07
z num_eqtl
93 -1.18450440 1
217 -0.57786099 1
2779 0.54425280 1
2780 1.79371022 2
2784 1.02857284 2
2785 0.26571594 1
2786 1.42183198 1
2812 -0.05673626 1
2814 -0.87430500 1
5063 -3.93195300 2
5068 -6.67154547 1
5069 -6.57743600 2
6706 -0.12983138 1
6708 -1.78280456 1
6713 -0.94777319 2
6714 -1.56544705 1
6716 -0.65925712 2
6717 -1.37362441 2
7760 -1.78280456 1
7761 -0.38265325 1
7762 -0.10311627 1
8626 0.32187672 1
8648 -0.04571303 1
8651 1.99083966 3
8658 1.67282787 1
8659 -1.35533608 3
8660 -1.30793556 3
8849 0.07077958 1
8941 12.59182931 1
9491 0.05353965 1
10660 -1.09899297 2
11140 -0.27954303 1
11381 -1.62714480 2
11705 0.44595864 2
12177 -0.07842180 1
12779 0.07975455 1
13357 -0.54787454 1
13511 -0.12178151 2
13516 1.40716759 2
13579 -0.44775309 1
13586 -0.42704781 1
13590 -0.50405613 1
[1] "CYP27A1"
[1] "2_129"
genename region_tag susie_pip mu2 PVE z
279 SLC11A1 2_129 0.04391614 4.666290 5.963705e-07 0.05100451
538 PTPRN 2_129 0.04264818 4.498007 5.582656e-07 -0.11033678
872 ASIC4 2_129 0.04271441 4.460744 5.545006e-07 0.10442281
873 SPEG 2_129 0.05207228 6.277503 9.512920e-07 0.60892801
934 BCS1L 2_129 0.04574168 7.973602 1.061419e-06 2.08166821
1080 TNS1 2_129 0.04619236 5.113780 6.874363e-07 0.37728313
3320 PLCD4 2_129 0.40823863 18.131918 2.154161e-05 4.09053549
3322 ZNF142 2_129 0.04306017 4.560569 5.714985e-07 -0.15341588
3324 PRKAG3 2_129 0.04819753 9.031493 1.266790e-06 -2.46670899
3332 CNPPD1 2_129 0.10740584 12.931062 4.041871e-06 -1.79583223
3334 ABCB6 2_129 0.08143374 9.899501 2.346054e-06 1.15422213
3335 STK16 2_129 0.14023857 15.014034 6.127526e-06 -1.90468349
4041 CHPF 2_129 0.05764271 7.202828 1.208281e-06 0.82081390
4042 DNPEP 2_129 0.10819230 13.090853 4.121778e-06 1.67585142
4043 INHA 2_129 0.04903900 5.807218 8.287624e-07 -0.65920233
4044 OBSL1 2_129 0.04671411 5.214645 7.089133e-07 0.42707562
4384 TUBA4A 2_129 0.04928770 5.660873 8.119743e-07 -0.56762321
4385 AAMP 2_129 0.14795477 14.968021 6.444863e-06 -2.20803733
5219 TTLL4 2_129 0.04395255 5.198043 6.648815e-07 0.91983273
5220 USP37 2_129 0.39622587 18.025295 2.078478e-05 -4.07816656
5225 TMBIM1 2_129 0.13034375 13.808679 5.237966e-06 -2.08113590
5226 CYP27A1 2_129 0.05326632 10.379321 1.608948e-06 -2.89315752
6301 FAM134A 2_129 0.04289250 4.514845 5.635656e-07 -0.06827746
6302 CTDSP1 2_129 0.10273155 15.123969 4.521577e-06 2.60879648
6303 CNOT9 2_129 0.05936095 7.026344 1.213810e-06 1.21362648
6305 GMPPA 2_129 0.05056185 6.010876 8.844657e-07 -0.73851578
7422 CATIP 2_129 0.09717705 14.068973 3.978748e-06 2.22430822
7438 ZFAND2B 2_129 0.10278197 11.931090 3.568760e-06 -1.58442042
7978 CXCR1 2_129 0.04612223 5.142339 6.902261e-07 0.56376400
7979 ARPC2 2_129 0.10165225 11.782469 3.485568e-06 -1.94043222
7983 RNF25 2_129 0.04488136 5.319029 6.947342e-07 -0.93528618
7984 STK36 2_129 0.10822566 14.219802 4.478619e-06 3.56927112
7988 IHH 2_129 0.04587125 5.041304 6.729825e-07 -0.61498029
7996 GLB1L 2_129 0.05075190 5.838483 8.623282e-07 -0.68615748
9836 DES 2_129 0.11064942 12.863520 4.142183e-06 -1.45556658
10338 GPBAR1 2_129 0.16656950 16.035020 7.772939e-06 -2.28666919
10422 CXCR2 2_129 0.08141085 9.681782 2.293812e-06 -1.47518963
11206 NHEJ1 2_129 0.13320177 14.824598 5.746630e-06 2.00614596
11276 RUFY4 2_129 0.13506417 15.135306 5.949105e-06 1.84473382
11333 TMEM198 2_129 0.04486642 4.873550 6.363370e-07 0.31231552
11981 ATG9A 2_129 0.04557167 5.083833 6.742276e-07 -0.37507138
12386 SLC23A3 2_129 0.04265081 4.535957 5.630106e-07 -0.13068048
12831 DIRC3 2_129 0.04511063 4.949927 6.498272e-07 0.44929498
14288 RP11-33O4.1 2_129 0.06571468 8.992541 1.719749e-06 -1.43979804
num_eqtl
279 1
538 1
872 1
873 2
934 2
1080 1
3320 1
3322 2
3324 1
3332 2
3334 1
3335 1
4041 1
4042 2
4043 2
4044 2
4384 1
4385 1
5219 1
5220 1
5225 2
5226 1
6301 1
6302 1
6303 1
6305 1
7422 1
7438 1
7978 1
7979 1
7983 1
7984 2
7988 1
7996 1
9836 1
10338 1
10422 1
11206 2
11276 2
11333 1
11981 1
12386 2
12831 2
14288 2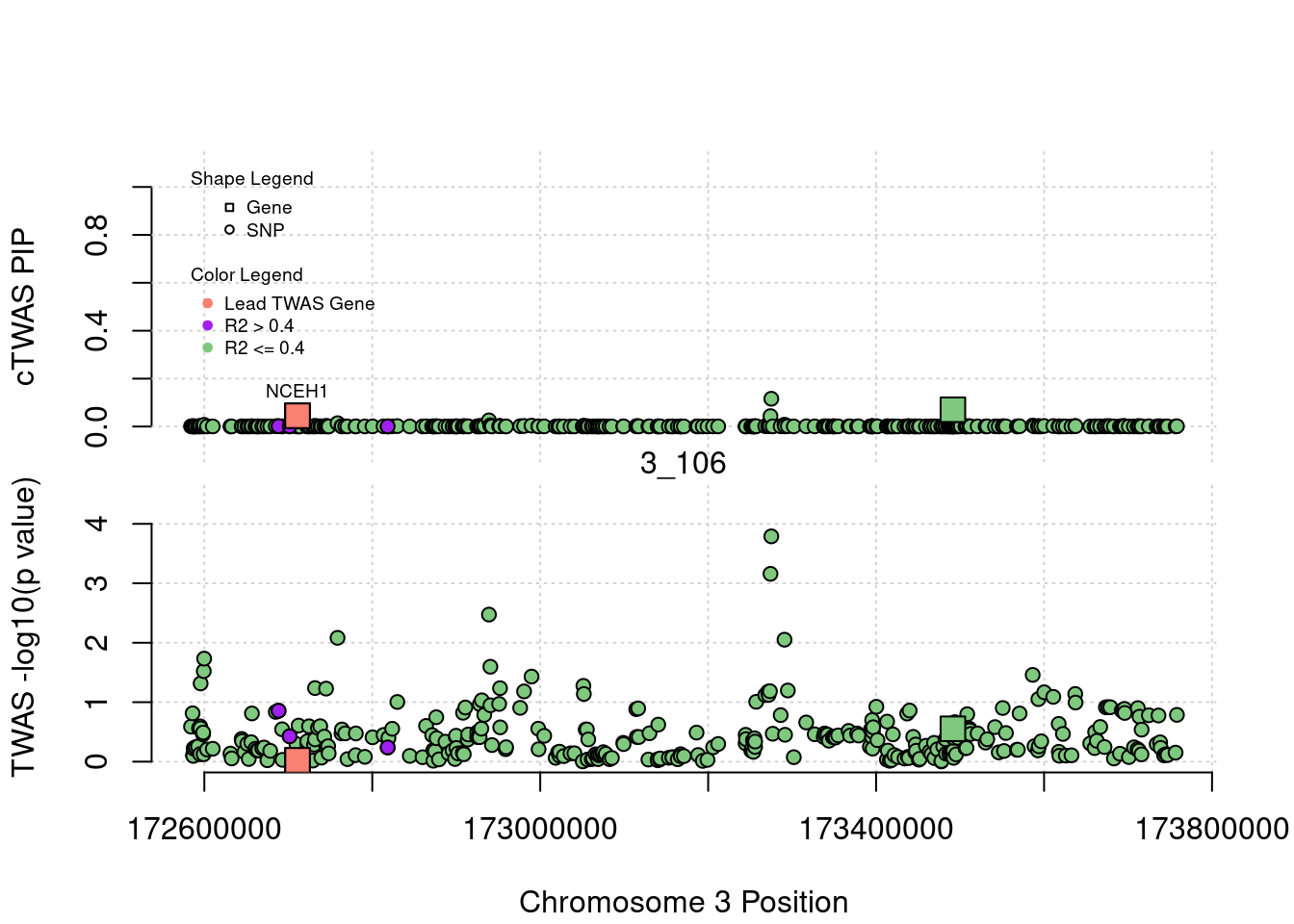
[1] "NCEH1"
[1] "3_106"
genename region_tag susie_pip mu2 PVE z
6349 NCEH1 3_106 0.04509989 4.437297 5.823904e-07 0.06081573
9120 NLGN1 3_106 0.07025444 8.458892 1.729448e-06 1.08118654
num_eqtl
6349 1
9120 1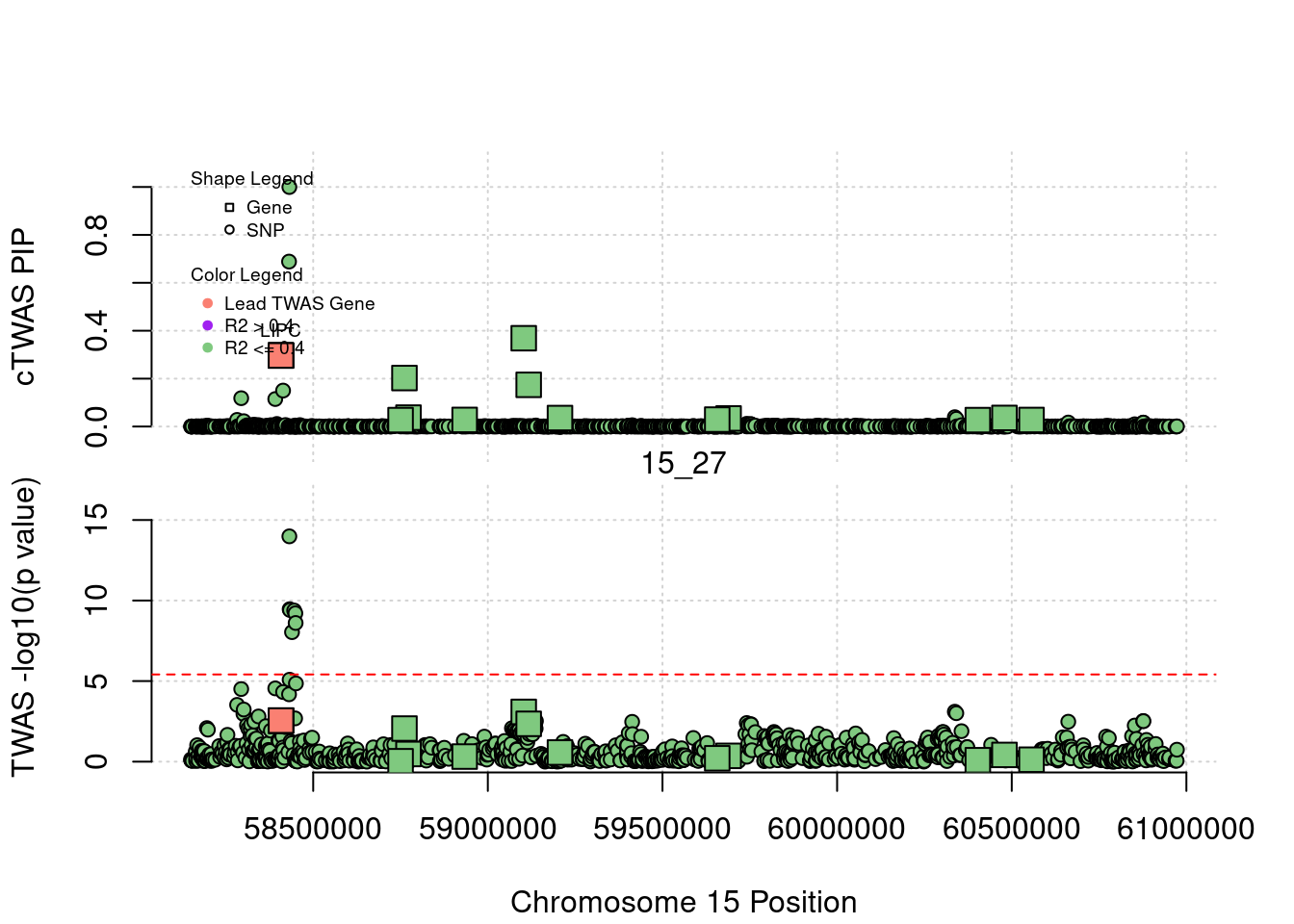
[1] "LIPC"
[1] "15_27"
genename region_tag susie_pip mu2 PVE
4474 MINDY2 15_27 0.03718240 7.533033 8.151314e-07
5493 SLTM 15_27 0.02761454 4.803092 3.859926e-07
5512 ADAM10 15_27 0.02680348 4.574160 3.567984e-07
7346 CCNB2 15_27 0.36869728 27.064346 2.903941e-05
8490 LIPC 15_27 0.29678611 25.395298 2.193397e-05
9451 LDHAL6B 15_27 0.03545826 7.491701 7.730688e-07
13260 RP11-30K9.6 15_27 0.20255558 22.862100 1.347661e-05
14571 RP11-59H7.4 15_27 0.17413663 20.722866 1.050172e-05
4473 ICE2 15_27 0.03591103 7.167848 7.490952e-07
5805 BNIP2 15_27 0.03366822 6.592942 6.459810e-07
5806 GTF2A2 15_27 0.02892651 5.238282 4.409660e-07
10608 ANXA2 15_27 0.02673418 4.537994 3.530621e-07
14422 RP11-219B17.3 15_27 0.02737735 4.754173 3.787797e-07
z num_eqtl
4474 0.9669014 1
5493 -0.6994231 2
5512 -0.0670727 2
7346 3.3483233 2
8490 2.9852576 4
9451 -1.1018855 3
13260 2.6192647 1
14571 2.8403444 2
4473 -0.8737488 2
5805 -0.7799366 1
5806 -0.4956509 1
10608 -0.2323451 1
14422 -0.3095370 1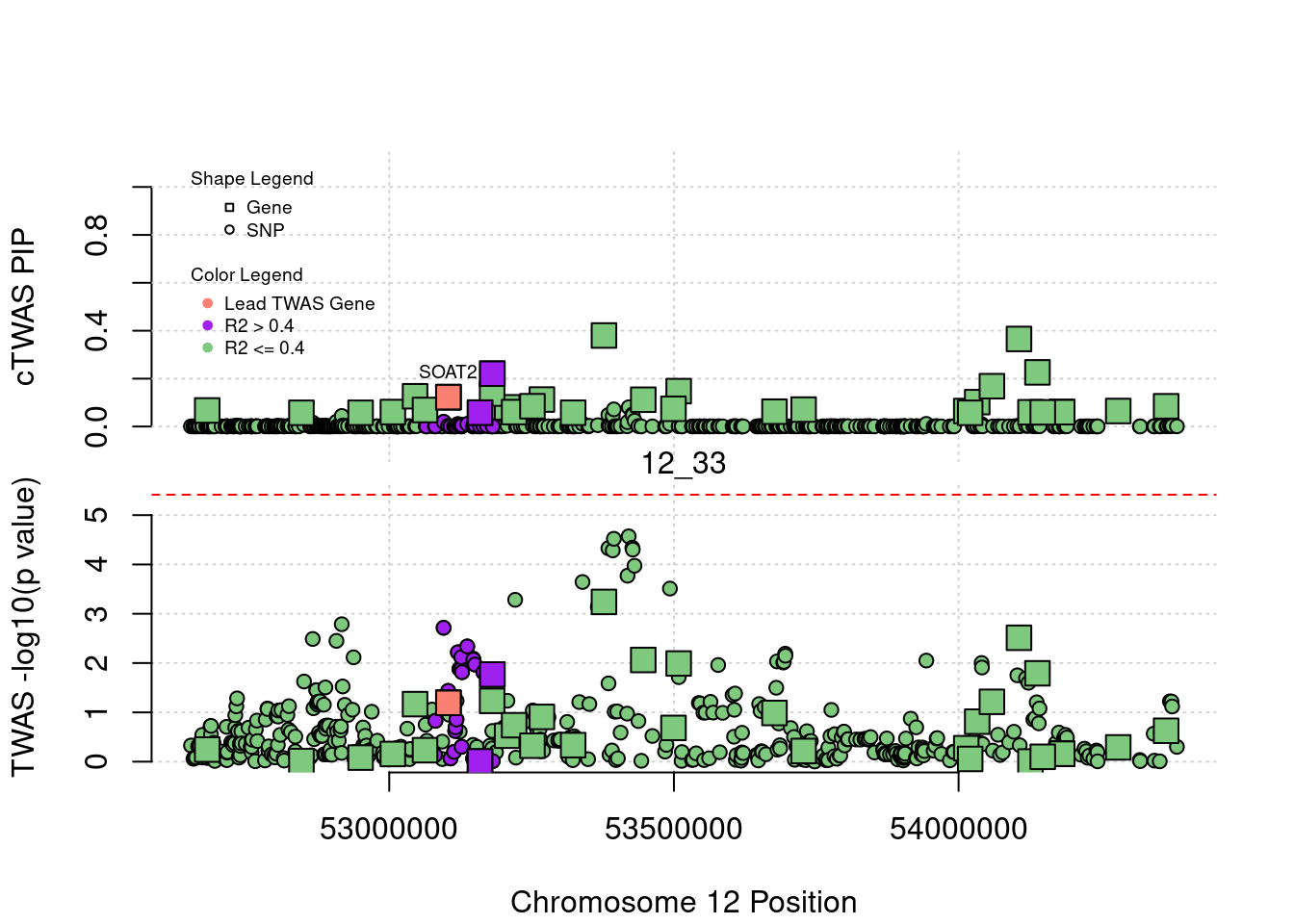
[1] "SOAT2"
[1] "12_33"
genename region_tag susie_pip mu2 PVE
236 CALCOCO1 12_33 0.07084606 6.147958 1.267555e-06
411 HOXC8 12_33 0.06493880 5.474349 1.034563e-06
630 EIF4B 12_33 0.06048567 4.934324 8.685613e-07
1509 AAAS 12_33 0.05763844 4.616415 7.743501e-07
1510 CBX5 12_33 0.06470668 5.537884 1.042830e-06
2860 KRT18 12_33 0.05745600 4.461100 7.459292e-07
2861 TNS2 12_33 0.12632401 11.858433 4.359468e-06
4006 SMUG1 12_33 0.05999442 4.845854 8.460607e-07
5145 ATP5G2 12_33 0.06210261 6.156822 1.112722e-06
5163 ESPL1 12_33 0.11286793 10.616072 3.487022e-06
5737 GPR84 12_33 0.08471970 7.900294 1.947816e-06
5738 NPFF 12_33 0.14773740 14.438155 6.207582e-06
5745 MAP3K12 12_33 0.07409543 7.091631 1.529177e-06
5746 ITGB7 12_33 0.08028431 7.545529 1.762953e-06
5748 CSAD 12_33 0.22074476 16.707851 1.073325e-05
5751 ZNF740 12_33 0.13764596 12.318065 4.934308e-06
8808 KRT1 12_33 0.06711956 5.837839 1.140306e-06
8812 SPRYD3 12_33 0.06919468 6.165285 1.241498e-06
8813 SOAT2 12_33 0.12257546 11.214208 4.000299e-06
9208 SP7 12_33 0.06038619 5.761697 1.012531e-06
9216 KRT78 12_33 0.05738783 4.444256 7.422311e-07
9560 HOXC5 12_33 0.10193040 9.489558 2.814946e-06
10587 MFSD5 12_33 0.08500710 7.540837 1.865499e-06
10952 SP1 12_33 0.37999627 15.203373 1.681278e-05
12221 PRR13 12_33 0.11204340 10.603524 3.457457e-06
13294 FLJ12825 12_33 0.16850668 14.298227 7.011640e-06
13326 RP11-834C11.6 12_33 0.36556171 21.561355 2.293808e-05
13367 LINC01598 12_33 0.05999442 4.845854 8.460607e-07
13381 RP11-834C11.4 12_33 0.05961361 4.745922 8.233534e-07
13615 RP11-834C11.10 12_33 0.05999442 4.845854 8.460607e-07
13625 RP11-1136G11.8 12_33 0.05815968 4.602031 7.789183e-07
13772 RP11-834C11.11 12_33 0.05939888 4.794122 8.287196e-07
13781 AC012531.25 12_33 0.05732387 4.434706 7.398108e-07
14706 RP11-834C11.15 12_33 0.22606324 16.893906 1.111425e-05
z num_eqtl
236 0.513018450 2
411 0.621629482 1
630 -0.385755780 2
1509 -0.735021442 1
1510 0.652766171 1
2860 -0.218426666 1
2861 -1.824491131 1
4006 0.396812394 1
5145 1.629895011 1
5163 1.538877559 2
5737 1.184293991 1
5738 -2.568783738 1
5745 -1.262287350 2
5746 -1.041271762 1
5748 -2.384777793 1
5751 1.892475105 2
8808 -0.570714701 1
8812 -0.545043625 1
8813 -1.851220053 1
9208 -1.333615935 2
9216 0.033259212 1
9560 1.423934785 1
10587 0.713409483 2
10952 3.442597770 1
12221 -2.625326238 1
13294 -1.872004676 1
13326 -2.960246455 1
13367 -0.396812394 1
13381 -0.009489289 1
13615 -0.396812394 1
13625 -0.012887459 1
13772 -0.256309447 2
13781 0.150041534 1
14706 -2.405714134 2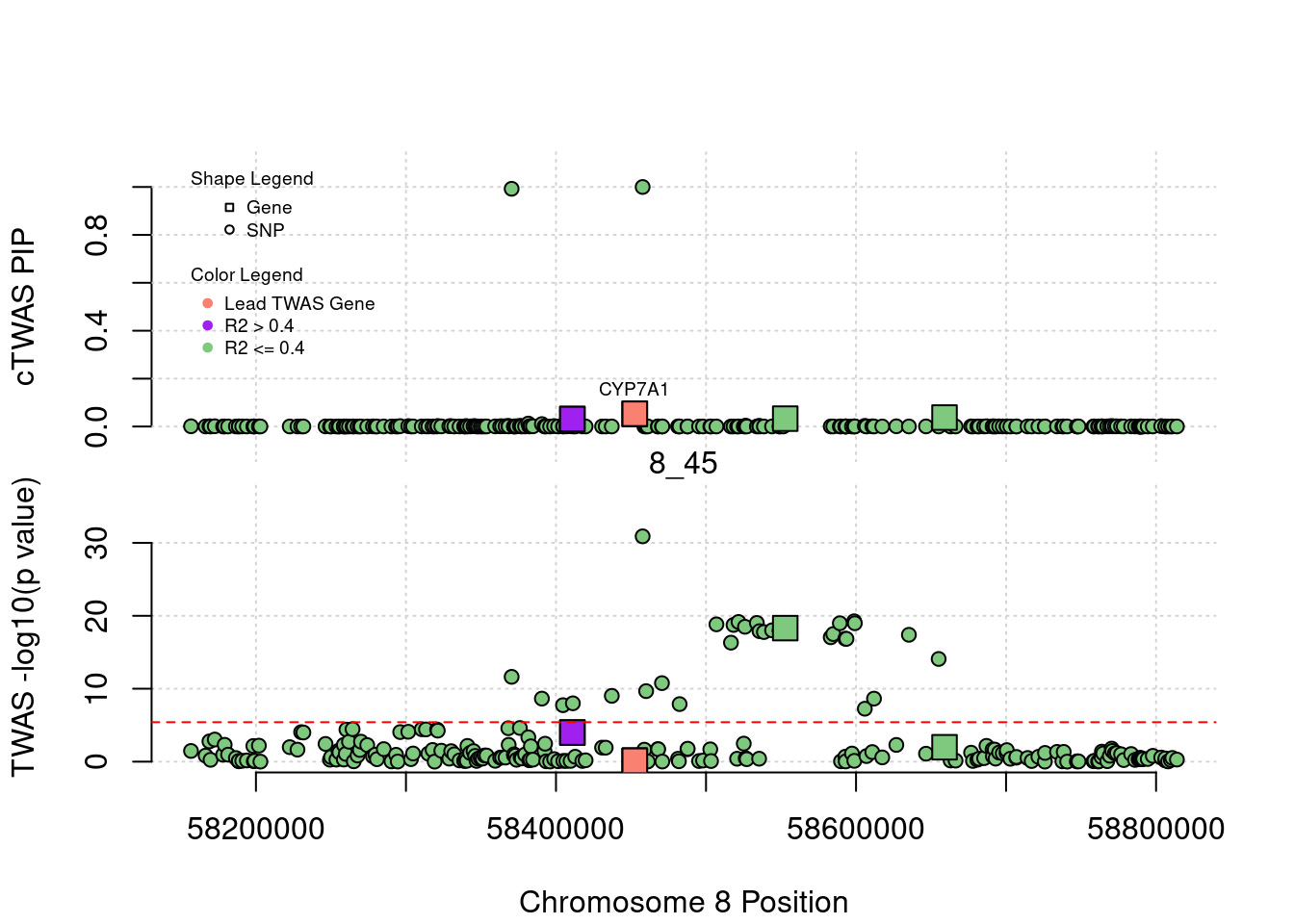
[1] "CYP7A1"
[1] "8_45"
genename region_tag susie_pip mu2 PVE z
393 NSMAF 8_45 0.03759612 10.731362 1.174135e-06 -2.5545688
5470 SDCBP 8_45 0.03299290 59.563400 5.719002e-06 8.9143332
8832 CYP7A1 8_45 0.05327798 9.516386 1.475503e-06 0.3161889
12473 UBXN2B 8_45 0.03136742 10.622745 9.696966e-07 3.8844161
num_eqtl
393 1
5470 1
8832 1
12473 3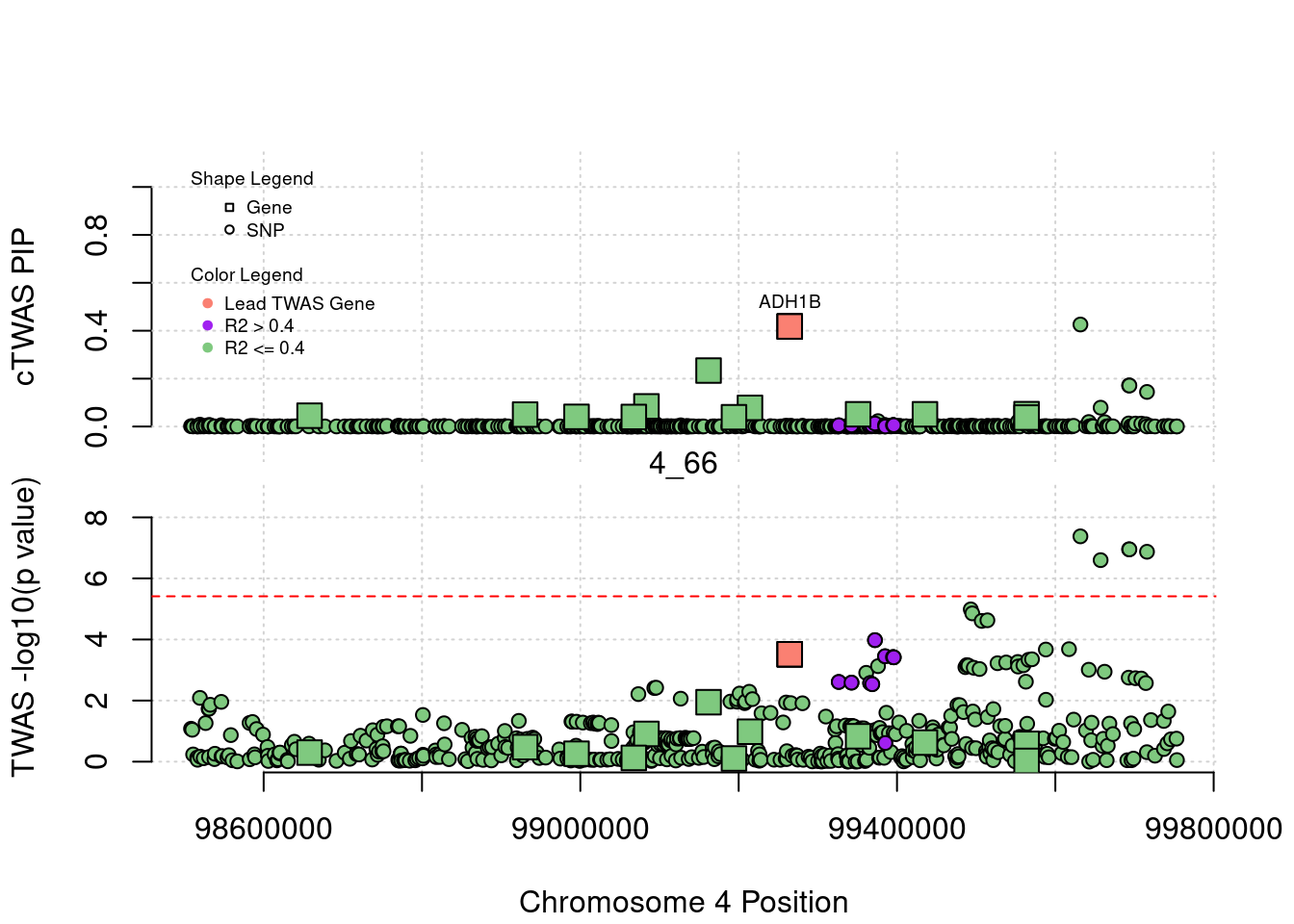
[1] "ADH1B"
[1] "4_66"
genename region_tag susie_pip mu2 PVE
5664 MTTP 4_66 0.05177806 7.559951 1.139161e-06
6378 TRMT10A 4_66 0.03740272 4.436930 4.829543e-07
6825 EIF4E 4_66 0.04995378 7.038158 1.023170e-06
8125 METAP1 4_66 0.04047155 5.165848 6.084315e-07
8978 TSPAN5 4_66 0.04530893 6.211309 8.190063e-07
9588 ADH6 4_66 0.07808769 11.029611 2.506473e-06
11210 ADH1A 4_66 0.03871596 4.778017 5.383417e-07
11474 ADH7 4_66 0.05109442 7.576550 1.126588e-06
11535 ADH1B 4_66 0.41779633 26.754222 3.252949e-05
11752 ADH5 4_66 0.08411582 11.850860 2.901001e-06
11796 ADH4 4_66 0.23368026 20.516922 1.395258e-05
13290 ADH1C 4_66 0.05128665 7.205360 1.075425e-06
14305 RP11-571L19.8 4_66 0.03997627 4.979432 5.792985e-07
z num_eqtl
5664 1.12400758 1
6378 -0.06765527 2
6825 0.96532383 1
8125 0.59927970 2
8978 -0.66850706 2
9588 1.62276202 3
11210 -0.27018216 1
11474 1.18093280 1
11535 -3.61112568 2
11752 1.54003066 3
11796 -2.53274599 1
13290 -1.44685831 2
14305 -0.31136184 4
[1] "LCAT"
[1] "16_36"
genename region_tag susie_pip mu2 PVE
759 CBFB 16_36 0.04945609 4.432840 6.380020e-07
1379 CMTM1 16_36 0.09105170 10.050709 2.663208e-06
1985 ELMO3 16_36 0.05204512 4.879790 7.390971e-07
1986 NUTF2 16_36 0.04952411 4.444004 6.404887e-07
1988 CENPT 16_36 0.04974765 4.482755 6.489898e-07
1989 TSNAXIP1 16_36 0.04957545 4.466510 6.443995e-07
1996 CTCF 16_36 0.04982033 4.503298 6.529164e-07
1999 PARD6A 16_36 0.05293053 5.105917 7.865029e-07
2013 SLC7A6OS 16_36 0.05008346 4.562238 6.649555e-07
2014 SLC7A6 16_36 0.05423927 5.234814 8.262955e-07
2015 ESRP2 16_36 0.05621634 5.555666 9.089060e-07
4047 ENKD1 16_36 0.14948598 14.483731 6.300880e-06
4158 LRRC29 16_36 0.05301326 5.033454 7.765527e-07
4162 C16orf70 16_36 0.39877735 24.355517 2.826494e-05
4826 PRMT7 16_36 0.05553741 5.552394 8.974002e-07
5193 FHOD1 16_36 0.21969237 18.249259 1.166757e-05
5195 SLC9A5 16_36 0.08324258 9.100878 2.204698e-06
5879 CMTM3 16_36 0.17133228 15.820080 7.888023e-06
5904 RANBP10 16_36 0.04957545 4.466510 6.443995e-07
5905 CTRL 16_36 0.04945904 4.433264 6.381012e-07
5906 GFOD2 16_36 0.07964234 8.873533 2.056652e-06
7526 NAE1 16_36 0.05394996 5.343806 8.390003e-07
7535 LRRC36 16_36 0.05395706 5.233319 8.217615e-07
7536 TPPP3 16_36 0.06800487 7.355783 1.455758e-06
7537 ATP6V0D1 16_36 0.06239358 6.526073 1.184983e-06
7539 C16orf86 16_36 0.04952512 4.446696 6.408898e-07
8594 BEAN1 16_36 0.07649453 8.409846 1.872142e-06
8595 TK2 16_36 0.08432855 9.338808 2.291851e-06
8710 DPEP2 16_36 0.04975082 4.506683 6.524955e-07
8902 THAP11 16_36 0.05341192 5.113641 7.948564e-07
8962 KCTD19 16_36 0.05177175 4.847883 7.304075e-07
9566 CES3 16_36 0.08275161 9.131982 2.199185e-06
9568 CES2 16_36 0.05363642 5.187902 8.097890e-07
9569 PDP2 16_36 0.05516017 5.377113 8.631675e-07
10258 EXOC3L1 16_36 0.05329745 5.080792 7.880578e-07
10734 CMTM4 16_36 0.04947520 4.437939 6.389829e-07
11248 NRN1L 16_36 0.05429687 5.401776 8.535554e-07
12209 PSMB10 16_36 0.04946207 4.434021 6.382492e-07
12212 E2F4 16_36 0.04946698 4.434273 6.383489e-07
12340 LCAT 16_36 0.04951145 4.443981 6.403215e-07
12506 CKLF 16_36 0.05817017 5.911203 1.000683e-06
13018 B3GNT9 16_36 0.05994089 6.110613 1.065929e-06
13273 RP11-61A14.4 16_36 0.06997639 7.755129 1.579286e-06
13274 LINC00920 16_36 0.08483633 9.286053 2.292627e-06
13839 CTD-2012K14.6 16_36 0.08414722 9.215129 2.256636e-06
14494 RP11-615I2.6 16_36 0.10659383 11.658928 3.616688e-06
z num_eqtl
759 -0.252876085 2
1379 1.756325429 2
1985 0.510149510 2
1986 -0.034587389 1
1988 -0.005954879 1
1989 -0.067069628 1
1996 0.240048062 1
1999 0.496802872 2
2013 -0.257953484 1
2014 0.467985675 1
2015 0.620211670 1
4047 -1.748584122 1
4158 -0.607413397 1
4162 3.161389249 1
4826 0.645370435 3
5193 2.386229548 2
5195 1.427810548 2
5879 2.158846432 2
5904 0.067069628 1
5905 0.084186643 2
5906 1.359729574 1
7526 -0.703973566 1
7535 0.550976590 1
7536 0.844633083 2
7537 -1.037404231 2
7539 -0.151633954 1
8594 1.501374372 3
8595 1.663854673 2
8710 -0.164482155 1
8902 0.295264078 1
8962 0.415312501 2
9566 1.189325464 2
9568 -0.624495208 2
9569 -0.115955712 1
10258 0.621774861 1
10734 0.178201387 1
11248 0.727341908 2
12209 -0.080306770 2
12212 0.083523326 2
12340 0.055498649 1
12506 0.855232520 1
13018 -0.679554308 1
13273 -1.396847840 1
13274 1.043513113 2
13839 1.285399757 1
14494 -1.992338697 1
[1] "VDAC1"
[1] "5_80"
genename region_tag susie_pip mu2 PVE
121 CDKL3 5_80 0.09633617 5.995324 1.680824e-06
447 JADE2 5_80 0.08431510 4.768195 1.169983e-06
519 FSTL4 5_80 0.08345060 4.673574 1.135008e-06
877 AFF4 5_80 0.08397072 4.730614 1.156021e-06
3126 SKP1 5_80 0.13079595 8.839815 3.364788e-06
3127 PPP2CA 5_80 0.10072360 6.406905 1.878018e-06
3129 C5orf15 5_80 0.19807264 12.792830 7.374141e-06
3135 TXNDC15 5_80 0.08592546 4.941989 1.235788e-06
4822 PCBD2 5_80 0.10695089 6.962635 2.167097e-06
6969 SAR1B 5_80 0.09489964 5.856664 1.617466e-06
8223 SHROOM1 5_80 0.11262349 7.442699 2.439381e-06
8224 GDF9 5_80 0.15457784 10.415628 4.685468e-06
8225 UQCRQ 5_80 0.11747974 7.835795 2.678961e-06
8226 LEAP2 5_80 0.08878525 5.243042 1.354704e-06
8256 CAMLG 5_80 0.08128820 4.432698 1.048615e-06
9247 HSPA4 5_80 0.08146390 4.452498 1.055575e-06
10517 C5orf24 5_80 0.08173711 4.483208 1.066420e-06
12355 VDAC1 5_80 0.13426487 9.085562 3.550050e-06
13019 CDKN2AIPNL 5_80 0.15403528 10.382270 4.654069e-06
13394 LINC01843 5_80 0.10362695 6.669961 2.011483e-06
14117 CTD-2410N18.3 5_80 0.08858876 5.222657 1.346451e-06
z num_eqtl
121 0.7946318 1
447 -0.3248676 1
519 -0.2767465 1
877 0.3132544 2
3126 -1.2408018 1
3127 0.8605539 1
3129 1.7000678 1
3135 -0.4386475 1
4822 0.8755768 2
6969 0.6871367 3
8223 1.0107556 2
8224 -1.4862194 2
8225 -1.1575811 1
8226 0.6903155 1
8256 0.1049360 1
9247 0.0232656 2
10517 0.2016299 1
12355 1.2027849 1
13019 -1.3298699 2
13394 0.8436844 2
14117 0.4989330 1
[1] "APOC2"
[1] "19_31"
genename region_tag susie_pip mu2 PVE z num_eqtl
123 MARK4 19_31 0 20.312700 0 -2.24637681 1
131 TRAPPC6A 19_31 0 16.600235 0 1.92140959 1
229 ERCC1 19_31 0 13.134793 0 -2.19401016 2
624 ZNF112 19_31 0 72.068605 0 7.18938487 3
898 PVR 19_31 0 30.112592 0 -3.01973306 1
2196 CLPTM1 19_31 0 48.802068 0 -2.57517256 1
2198 PPP1R37 19_31 0 35.405859 0 -2.37518201 2
2203 ERCC2 19_31 0 13.615522 0 1.53401174 1
3560 CD3EAP 19_31 0 16.730266 0 0.05565582 4
4222 OPA3 19_31 0 11.581430 0 1.32335471 2
4224 RTN2 19_31 0 6.803450 0 2.07007103 1
4226 VASP 19_31 0 23.808738 0 1.26035740 1
4570 NECTIN2 19_31 0 55.577675 0 -13.59762224 1
4571 APOE 19_31 0 999.967785 0 37.80754977 2
4572 TOMM40 19_31 0 56.000201 0 5.53230466 2
4573 APOC1 19_31 0 296.620171 0 7.53014709 1
6024 GEMIN7 19_31 0 157.439551 0 14.13454209 2
7552 ZNF233 19_31 0 98.228951 0 -9.63193736 1
7553 ZNF235 19_31 0 8.185909 0 -0.97877558 1
8729 ZNF180 19_31 0 25.642421 0 1.41443321 2
9263 ZNF296 19_31 0 65.538646 0 5.25423092 2
11089 CEACAM19 19_31 0 13.759203 0 3.64086985 3
11388 BLOC1S3 19_31 0 8.975124 0 2.30141189 1
12385 PPM1N 19_31 0 20.227762 0 -1.22009641 2
12950 APOC2 19_31 0 99.467017 0 7.82468739 2
14021 ZNF285 19_31 0 10.013342 0 -1.26709542 2
14628 ZNF229 19_31 0 8.630054 0 1.23011935 2#run APOE locus again using full SNPs
# focus <- "APOE"
# region_tag <- ctwas_res$region_tag[which(ctwas_res$genename==focus)]
#
# locus_plot(region_tag, label="TWAS", rerun_ctwas = T)
#
# mtext(text=region_tag)
#
# print(focus)
# print(region_tag)
# print(ctwas_gene_res[ctwas_gene_res$region_tag==region_tag,report_cols,])Locus Plots - False positives
This section produces locus plots for all bystander genes with PIP>0.8 (false positives). The highlighted gene at each region is the false positive gene.
false_positives <- ctwas_gene_res$genename[ctwas_gene_res$genename %in% unrelated_genes & ctwas_gene_res$susie_pip>0.8]
for (i in 1:length(false_positives)){
focus <- false_positives[i]
region_tag <- ctwas_res$region_tag[which(ctwas_res$genename==focus)]
locus_plot3(region_tag, focus=focus)
mtext(text=region_tag)
print(focus)
print(region_tag)
print(ctwas_gene_res[ctwas_gene_res$region_tag==region_tag,report_cols,])
#genes at this locus that are in known annotations
ctwas_gene_res$genename[ctwas_gene_res$region_tag==region_tag][ctwas_gene_res$genename[ctwas_gene_res$region_tag==region_tag] %in% known_annotations]
}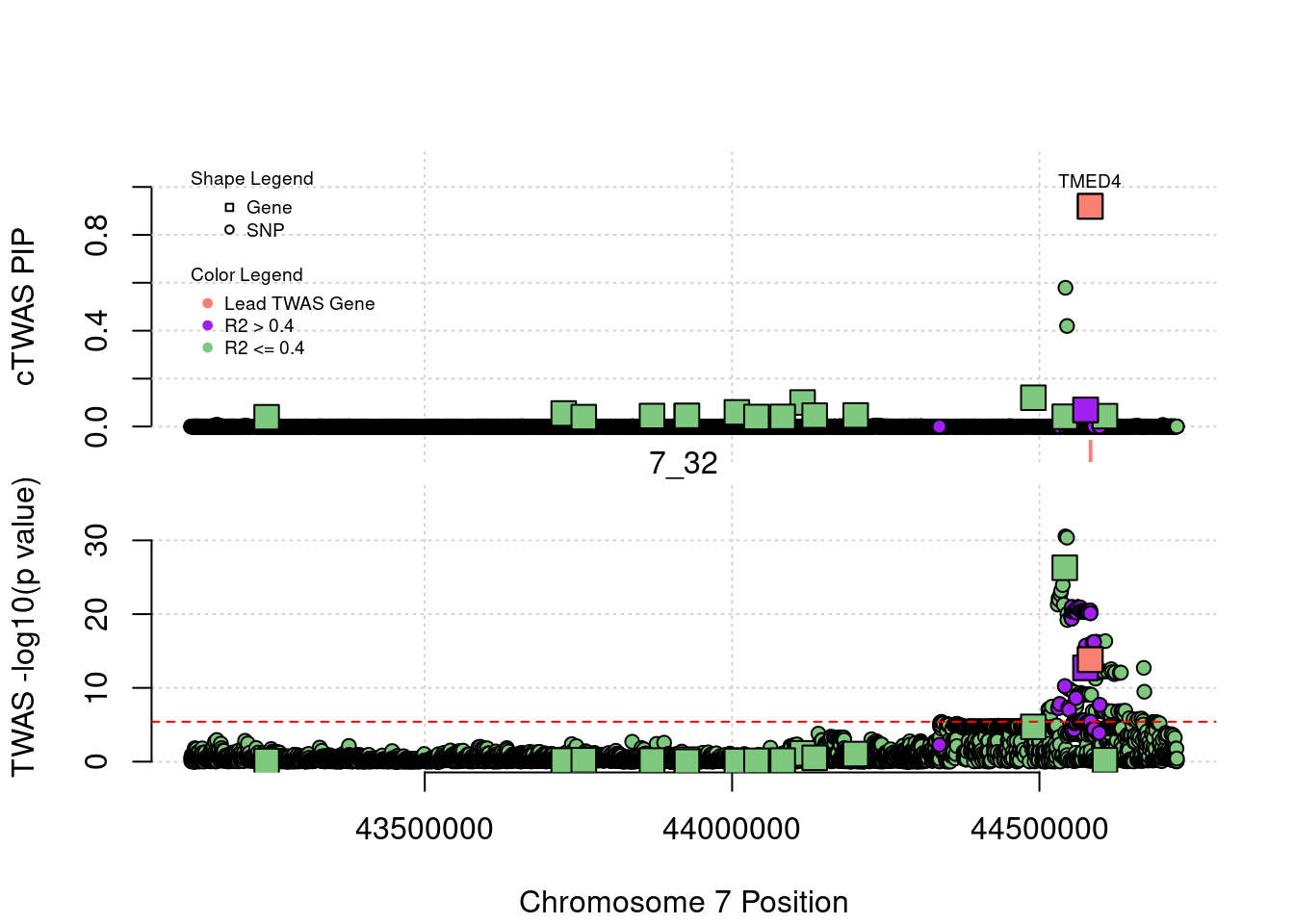
[1] "TMED4"
[1] "7_32"
genename region_tag susie_pip mu2 PVE z
17 HECW1 7_32 0.03879622 4.502064 5.083015e-07 -0.23376042
268 NPC1L1 7_32 0.04099715 82.556624 9.849767e-06 10.76193109
270 NUDCD3 7_32 0.12016967 17.520565 6.127217e-06 4.27589199
627 MRPS24 7_32 0.04400621 5.502620 7.046992e-07 0.38278180
1072 UBE2D4 7_32 0.04316907 5.512225 6.925003e-07 0.62909522
2384 OGDH 7_32 0.04345606 22.901957 2.896298e-06 0.47417198
2465 COA1 7_32 0.05470517 6.827556 1.086961e-06 -0.37405767
2466 BLVRA 7_32 0.03930484 4.699292 5.375251e-07 0.46600524
2467 URGCP 7_32 0.04456617 5.287322 6.857430e-07 0.03508092
2472 POLD2 7_32 0.10048790 13.317824 3.894640e-06 1.78074584
2474 GCK 7_32 0.04566065 6.311854 8.387246e-07 1.02935564
2476 YKT6 7_32 0.04642044 7.365875 9.950706e-07 1.67948623
3938 POLM 7_32 0.04055106 4.857444 5.732319e-07 -0.36899829
5285 DDX56 7_32 0.06932067 30.497390 6.152417e-06 7.35708820
7441 TMED4 7_32 0.91919811 36.878293 9.865071e-05 7.68827409
12733 AC004951.6 7_32 0.05933237 7.272867 1.255792e-06 0.22091509
12967 LINC00957 7_32 0.03984318 4.700790 5.450609e-07 0.29011976
num_eqtl
17 3
268 1
270 1
627 1
1072 1
2384 2
2465 2
2466 1
2467 2
2472 4
2474 2
2476 1
3938 3
5285 2
7441 2
12733 1
12967 1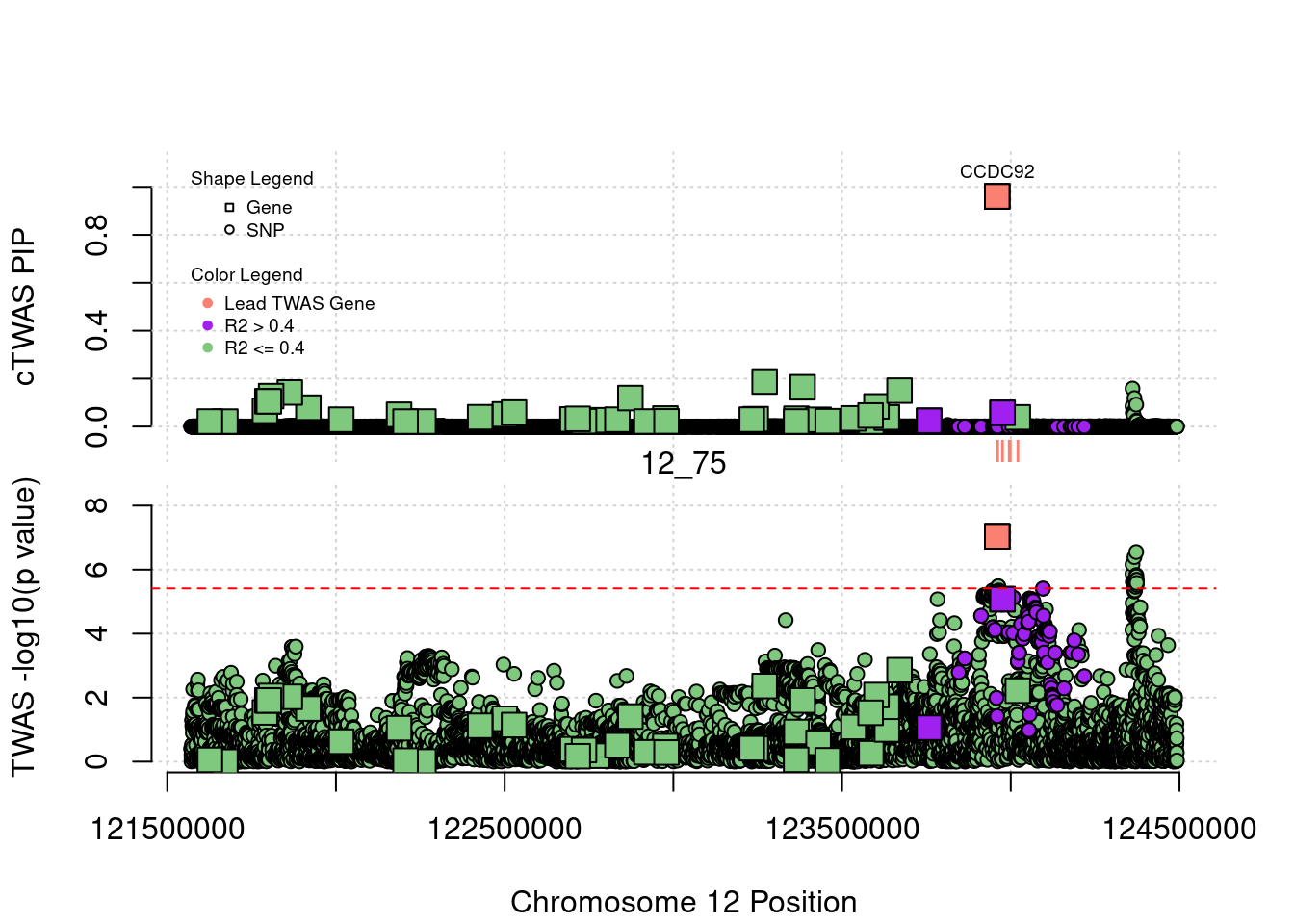
[1] "CCDC92"
[1] "12_75"
genename region_tag susie_pip mu2 PVE
372 ZCCHC8 12_75 0.05064241 12.817141 1.888973e-06
515 MPHOSPH9 12_75 0.03087760 7.425851 6.672831e-07
1268 TMED2 12_75 0.02079112 4.585181 2.774308e-07
2857 BCL7A 12_75 0.02875572 7.171329 6.001284e-07
2888 OGFOD2 12_75 0.03356930 8.204507 8.015212e-07
2889 CDK2AP1 12_75 0.18773826 23.330727 1.274681e-05
2895 GTF2H3 12_75 0.03792096 10.244128 1.130511e-06
2896 EIF2B1 12_75 0.03806846 11.612525 1.286507e-06
2897 DDX55 12_75 0.08396178 17.893757 4.372235e-06
3647 CCDC92 12_75 0.96048401 26.340340 7.362610e-05
4656 CLIP1 12_75 0.03878883 10.315993 1.164496e-06
4657 CCDC62 12_75 0.02630153 6.215464 4.757457e-07
4658 HIP1R 12_75 0.02863272 7.637638 6.364173e-07
4668 C12orf65 12_75 0.02883123 6.956820 5.837061e-07
5756 SBNO1 12_75 0.03272792 9.237419 8.798108e-07
5757 MORN3 12_75 0.02074617 4.471989 2.699970e-07
5758 VPS33A 12_75 0.02089689 4.519520 2.748491e-07
5759 VPS37B 12_75 0.11881371 19.999490 6.915217e-06
5760 RHOF 12_75 0.06582808 13.414260 2.569793e-06
6799 ABCB9 12_75 0.02100541 4.900367 2.995574e-07
6800 RILPL2 12_75 0.02826604 7.418349 6.102286e-07
7391 WDR66 12_75 0.07868177 15.872314 3.634416e-06
7398 HPD 12_75 0.14212649 21.541090 8.909699e-06
7401 LRRC43 12_75 0.04922749 11.939856 1.710516e-06
8975 TCTN2 12_75 0.14984380 24.210330 1.055747e-05
9970 B3GNT4 12_75 0.02074467 4.462980 2.694337e-07
10235 RFLNA 12_75 0.05720051 21.048278 3.503780e-06
10282 ZNF664 12_75 0.03824348 14.877484 1.655798e-06
10547 ARL6IP4 12_75 0.02135634 4.793908 2.979455e-07
10616 HCAR2 12_75 0.02966372 7.197248 6.213158e-07
10768 KMT5A 12_75 0.16388338 21.329940 1.017290e-05
10792 SNRNP35 12_75 0.02233216 4.959506 3.223216e-07
10819 KNTC1 12_75 0.05837394 13.415466 2.279004e-06
11244 RILPL1 12_75 0.03397789 9.373187 9.268383e-07
11582 HCAR1 12_75 0.02235176 5.203103 3.384499e-07
11709 DNAH10 12_75 0.02411867 7.560196 5.306483e-07
12301 LINC01089 12_75 0.12695826 19.015800 7.025801e-06
13281 RP11-486O12.2 12_75 0.04620271 12.506896 1.681656e-06
13533 HCAR3 12_75 0.03208497 7.773161 7.258044e-07
13565 RP13-942N8.1 12_75 0.02079414 4.593501 2.779746e-07
14318 RP11-347I19.8 12_75 0.10577689 17.432702 5.366310e-06
14442 RP11-347I19.7 12_75 0.10511020 17.378625 5.315946e-06
14526 ORAI1 12_75 0.02084106 4.481764 2.718248e-07
z num_eqtl
372 -1.997330862 1
515 -0.850400109 1
1268 0.601043736 1
2857 1.181541124 1
2888 -0.977970651 1
2889 2.851959720 1
2895 1.649373878 2
2896 2.339753799 3
2897 -2.641060332 3
3647 -5.343007339 5
4656 -1.775873434 1
4657 -0.453713661 1
4658 -1.021315410 1
4668 0.875821894 2
5756 -1.561826949 2
5757 0.002443962 4
5758 -0.014751572 1
5759 -2.064935470 2
5760 2.184756664 1
6799 0.656964218 1
6800 -1.133101871 3
7391 2.293784845 2
7398 2.595685436 1
7401 1.694167023 1
8975 3.196517420 3
9970 0.092723463 1
10235 -4.455405839 1
10282 -2.745109828 2
10547 -0.647121985 1
10616 -0.826359203 2
10768 -2.504311853 2
10792 0.111175630 1
10819 -1.799641224 1
11244 -1.735085267 1
11582 -0.779399534 1
11709 -1.713076506 3
12301 2.616204262 1
13281 -2.173324783 2
13533 0.415374217 2
13565 0.152862225 3
14318 2.506680919 1
14442 2.503442974 1
14526 -0.150022733 1Genes with many eQTL
#distribution of number of eQTL for all imputed genes (after dropping ambiguous variants)
table(ctwas_gene_res$num_eqtl)
1 2 3 4 5 6 7 8
6881 4360 1307 321 72 14 4 2 #all genes with 4+ eQTL
ctwas_gene_res[ctwas_gene_res$num_eqtl>3,] chrom id pos type region_tag1 region_tag2
540 1 ENSG00000054523.17 10210344 gene 1 7
3427 1 ENSG00000116663.10 11663463 gene 1 9
12643 1 ENSG00000226029.1 16458366 gene 1 11
7459 1 ENSG00000158825.5 20588679 gene 1 14
1421 1 ENSG00000090686.15 21782050 gene 1 15
10125 1 ENSG00000177868.11 42805362 gene 1 26
3556 1 ENSG00000117834.12 48212526 gene 1 30
7782 1 ENSG00000162383.11 53085621 gene 1 33
13102 1 ENSG00000240563.1 62194418 gene 1 39
7796 1 ENSG00000162437.14 64734373 gene 1 41
3449 1 ENSG00000116783.14 74215526 gene 1 47
3452 1 ENSG00000116791.13 74702349 gene 1 47
14321 1 ENSG00000272864.1 74701793 gene 1 47
5522 1 ENSG00000137944.17 88979282 gene 1 54
7116 1 ENSG00000154511.11 92934792 gene 1 56
9301 1 ENSG00000170989.8 101154806 gene 1 62
3878 1 ENSG00000121940.15 108930191 gene 1 67
4984 1 ENSG00000134184.12 109226267 gene 1 67
4986 1 ENSG00000134201.10 109710593 gene 1 67
4987 1 ENSG00000134202.10 109704237 gene 1 67
6084 1 ENSG00000143028.8 109465030 gene 1 67
8972 1 ENSG00000168765.16 109612653 gene 1 67
12337 1 ENSG00000213366.12 109657738 gene 1 67
11807 1 ENSG00000198162.12 117367233 gene 1 72
11756 1 ENSG00000197915.5 152219919 gene 1 74
11240 1 ENSG00000188004.9 159838179 gene 1 79
6101 1 ENSG00000143162.7 167544347 gene 1 82
3535 1 ENSG00000117533.14 171741914 gene 1 84
12590 1 ENSG00000224687.1 178093586 gene 1 87
12555 1 ENSG00000223396.2 201474507 gene 1 101
4884 1 ENSG00000133059.16 205210869 gene 1 104
6911 1 ENSG00000152104.11 214371230 gene 1 109
6195 1 ENSG00000143653.9 246720005 gene 1 131
13729 1 ENSG00000259865.1 247179932 gene 1 131
13065 1 ENSG00000238243.3 247638887 gene 1 131
3339 2 ENSG00000115705.20 1095735 gene 2 1
11076 2 ENSG00000186453.12 24119662 gene 2 14
12939 2 ENSG00000234690.6 47336895 gene 2 30
5540 2 ENSG00000138039.14 48685277 gene 2 31
7881 2 ENSG00000162869.15 48428760 gene 2 31
6244 2 ENSG00000144026.11 95142476 gene 2 57
3333 2 ENSG00000115652.14 106194127 gene 2 63
3642 2 ENSG00000119147.9 106024112 gene 2 63
4203 2 ENSG00000125629.14 118088243 gene 2 69
4205 2 ENSG00000125633.10 118013438 gene 2 69
863 2 ENSG00000071967.11 171522308 gene 2 104
4455 2 ENSG00000128652.11 176136417 gene 2 107
5596 2 ENSG00000138400.12 206765180 gene 2 122
6293 2 ENSG00000144488.14 238099977 gene 2 141
8927 2 ENSG00000168427.8 238127296 gene 2 141
12043 2 ENSG00000204104.11 238316563 gene 2 141
10005 2 ENSG00000176720.4 241550210 gene 2 144
10430 2 ENSG00000180902.17 241734689 gene 2 144
12958 2 ENSG00000235151.1 241816853 gene 2 144
6289 3 ENSG00000144455.13 4368850 gene 3 4
7134 3 ENSG00000154743.17 12431546 gene 3 9
3157 3 ENSG00000113812.13 53858803 gene 3 36
10381 3 ENSG00000180376.16 56545510 gene 3 39
9498 3 ENSG00000172340.14 67365898 gene 3 45
3202 3 ENSG00000114544.16 126082224 gene 3 78
11415 3 ENSG00000189366.9 125925826 gene 3 78
8085 3 ENSG00000163864.15 139674109 gene 3 85
6960 3 ENSG00000152580.8 151457865 gene 3 93
13112 3 ENSG00000240875.5 156751124 gene 3 97
10026 3 ENSG00000176945.16 195719309 gene 3 120
3645 3 ENSG00000119227.7 196947388 gene 3 121
4357 4 ENSG00000127415.12 971932 gene 4 2
8107 4 ENSG00000163945.15 1347151 gene 4 2
10958 4 ENSG00000185619.18 706823 gene 4 2
9729 4 ENSG00000174137.12 1676701 gene 4 3
9279 4 ENSG00000170846.16 6673665 gene 4 7
13412 4 ENSG00000251580.1 6673694 gene 4 7
13415 4 ENSG00000251615.3 8355960 gene 4 10
10163 4 ENSG00000178163.7 10456862 gene 4 11
2737 4 ENSG00000109689.15 26826816 gene 4 22
11860 4 ENSG00000198515.13 47955024 gene 4 37
4408 4 ENSG00000128039.10 55346203 gene 4 40
277 4 ENSG00000018189.12 70704417 gene 4 49
5643 4 ENSG00000138744.14 75918159 gene 4 51
11814 4 ENSG00000198189.10 87298285 gene 4 59
14305 4 ENSG00000272777.1 99067562 gene 4 66
5654 4 ENSG00000138777.19 105472284 gene 4 69
2744 4 ENSG00000109771.15 185383595 gene 4 119
6397 4 ENSG00000145476.15 186191240 gene 4 120
8216 4 ENSG00000164344.15 186191475 gene 4 120
13385 4 ENSG00000250829.2 186890499 gene 4 120
14237 4 ENSG00000272218.1 186839668 gene 4 120
8219 5 ENSG00000164366.3 188803 gene 5 1
13411 5 ENSG00000251532.1 1515420 gene 5 2
13370 5 ENSG00000250490.1 6308511 gene 5 6
13383 5 ENSG00000250786.1 9541581 gene 5 8
3079 5 ENSG00000112977.15 10675380 gene 5 9
8188 5 ENSG00000164237.8 10281437 gene 5 9
7191 5 ENSG00000155542.11 56909530 gene 5 33
11737 5 ENSG00000197822.10 69279407 gene 5 42
9006 5 ENSG00000168938.5 122859139 gene 5 74
8307 5 ENSG00000164904.17 126543217 gene 5 77
12336 5 ENSG00000213347.10 177313243 gene 5 106
9863 5 ENSG00000175309.14 178231895 gene 5 107
10183 5 ENSG00000178338.10 178836120 gene 5 107
14272 6 ENSG00000272463.1 710558 gene 6 1
5426 6 ENSG00000137275.13 3064015 gene 6 3
6445 6 ENSG00000145949.9 2677719 gene 6 3
13215 6 ENSG00000244041.7 2981538 gene 6 3
7012 6 ENSG00000153157.12 10829086 gene 6 10
6446 6 ENSG00000145979.17 13267390 gene 6 12
2948 6 ENSG00000111801.15 26391163 gene 6 20
11608 6 ENSG00000197062.11 28259100 gene 6 22
5430 6 ENSG00000137312.14 30718130 gene 6 26
12119 6 ENSG00000204516.9 31493994 gene 6 26
12121 6 ENSG00000204525.16 31262189 gene 6 26
12122 6 ENSG00000204531.17 31113082 gene 6 26
12123 6 ENSG00000204536.13 31150242 gene 6 26
12277 6 ENSG00000206344.7 31172291 gene 6 26
12376 6 ENSG00000213780.10 30902222 gene 6 26
14238 6 ENSG00000272221.1 31393306 gene 6 26
12077 6 ENSG00000204301.6 32205480 gene 6 26
12068 6 ENSG00000204256.12 32968664 gene 6 27
11916 6 ENSG00000198755.10 35461005 gene 6 28
450 6 ENSG00000044090.8 43045924 gene 6 33
11465 6 ENSG00000196284.15 45377511 gene 6 34
7023 6 ENSG00000153291.15 46630562 gene 6 35
9518 6 ENSG00000172469.15 95541495 gene 6 65
9531 6 ENSG00000172594.12 122788734 gene 6 82
3599 6 ENSG00000118507.15 131135842 gene 6 87
2988 6 ENSG00000112110.9 159789152 gene 6 103
3902 6 ENSG00000122335.13 158151578 gene 6 103
14208 6 ENSG00000272047.1 158167424 gene 6 103
13165 7 ENSG00000242611.2 58458 gene 7 1
3937 7 ENSG00000122674.11 5887685 gene 7 10
14257 7 ENSG00000272361.2 16584790 gene 7 16
840 7 ENSG00000070882.12 24860542 gene 7 21
11457 7 ENSG00000196247.11 64665395 gene 7 43
11782 7 ENSG00000198039.11 64869717 gene 7 43
13122 7 ENSG00000241258.6 66095787 gene 7 43
8358 7 ENSG00000165171.10 73842285 gene 7 47
4394 7 ENSG00000127947.15 77503532 gene 7 49
74 7 ENSG00000005471.15 87472091 gene 7 54
2363 7 ENSG00000105793.15 90334836 gene 7 55
828 7 ENSG00000070669.16 97873010 gene 7 60
8671 7 ENSG00000166997.7 100086039 gene 7 61
12216 7 ENSG00000205307.11 100478874 gene 7 61
5125 7 ENSG00000135269.17 116210408 gene 7 70
4452 7 ENSG00000128617.2 128771566 gene 7 78
7190 7 ENSG00000155530.2 134043283 gene 7 81
21 7 ENSG00000002933.7 150777235 gene 7 94
2461 7 ENSG00000106565.17 150788995 gene 7 94
11166 7 ENSG00000187260.15 151404454 gene 7 94
13842 7 ENSG00000261455.1 152420372 gene 7 94
13463 8 ENSG00000253958.1 8667057 gene 8 11
2120 8 ENSG00000104213.12 17575683 gene 8 19
11633 8 ENSG00000197217.12 23359473 gene 8 24
9737 8 ENSG00000174226.8 100646399 gene 8 69
9305 8 ENSG00000171045.14 142307501 gene 8 93
258 8 ENSG00000014164.6 143534573 gene 8 94
6574 8 ENSG00000147813.15 143578019 gene 8 94
2505 9 ENSG00000107099.15 205428 gene 9 1
9626 9 ENSG00000173253.15 1045596 gene 9 1
6576 9 ENSG00000147853.16 4718165 gene 9 5
3749 9 ENSG00000120217.13 5452560 gene 9 6
8332 9 ENSG00000164989.16 15531752 gene 9 13
12586 9 ENSG00000224549.1 22767062 gene 9 17
5390 9 ENSG00000137074.18 33024919 gene 9 25
8329 9 ENSG00000164978.17 34151465 gene 9 27
13808 9 ENSG00000260947.1 33612306 gene 9 27
2485 9 ENSG00000106733.20 75087456 gene 9 35
2490 9 ENSG00000106789.12 98122860 gene 9 49
5359 9 ENSG00000136870.10 101399660 gene 9 50
2486 9 ENSG00000106771.12 109112805 gene 9 56
9627 9 ENSG00000173258.12 111524995 gene 9 56
3664 9 ENSG00000119431.9 113343432 gene 9 58
1524 9 ENSG00000095397.13 114424432 gene 9 59
4619 9 ENSG00000130560.8 135926392 gene 9 73
7616 9 ENSG00000160360.12 136327342 gene 9 73
2577 10 ENSG00000107959.15 3148307 gene 10 4
12883 10 ENSG00000233117.2 4211392 gene 10 5
5031 10 ENSG00000134463.14 11742089 gene 10 10
6948 10 ENSG00000152465.17 15170861 gene 10 13
12157 10 ENSG00000204740.10 19490064 gene 10 16
9876 10 ENSG00000175395.15 37974942 gene 10 28
4930 10 ENSG00000133661.15 79970366 gene 10 51
3905 10 ENSG00000122375.11 86653675 gene 10 56
10862 10 ENSG00000184923.12 87206818 gene 10 56
12445 10 ENSG00000214562.14 87220987 gene 10 56
11898 10 ENSG00000198682.12 87660153 gene 10 56
6650 10 ENSG00000148690.11 93699055 gene 10 60
7173 10 ENSG00000155265.10 97822001 gene 10 62
8536 10 ENSG00000166275.15 102853979 gene 10 66
2543 10 ENSG00000107672.14 121974763 gene 10 76
6866 10 ENSG00000151640.12 132186469 gene 10 83
9420 10 ENSG00000171813.13 132365124 gene 10 83
6009 11 ENSG00000142102.15 288845 gene 11 1
10901 11 ENSG00000185201.16 306862 gene 11 1
12411 11 ENSG00000214063.10 842775 gene 11 1
13526 11 ENSG00000255284.1 728487 gene 11 1
14141 11 ENSG00000270972.1 330061 gene 11 1
2823 11 ENSG00000110665.11 2295490 gene 11 3
3829 11 ENSG00000121236.20 5580472 gene 11 4
4945 11 ENSG00000133816.13 12054282 gene 11 9
2808 11 ENSG00000110427.14 33378145 gene 11 22
3857 11 ENSG00000121691.4 34434584 gene 11 23
6670 11 ENSG00000149089.12 34916054 gene 11 23
6834 11 ENSG00000151348.13 44066439 gene 11 27
7775 11 ENSG00000162341.16 69030673 gene 11 38
9895 11 ENSG00000175581.13 73786901 gene 11 41
7439 11 ENSG00000158555.14 75504376 gene 11 42
13261 11 ENSG00000246067.7 83072772 gene 11 46
4035 11 ENSG00000123892.11 88175178 gene 11 49
2794 11 ENSG00000110218.8 94128854 gene 11 53
6690 11 ENSG00000149289.10 110092976 gene 11 65
5482 11 ENSG00000137713.15 111764842 gene 11 66
6691 11 ENSG00000149292.16 113289048 gene 11 67
10385 11 ENSG00000180425.11 114399882 gene 11 68
10048 11 ENSG00000177103.13 117806324 gene 11 71
2774 11 ENSG00000110060.8 125898452 gene 11 77
3771 11 ENSG00000120451.10 130909773 gene 11 81
13536 11 ENSG00000255455.2 130866376 gene 11 81
2866 12 ENSG00000111186.12 1507541 gene 12 2
13577 12 ENSG00000256433.2 6384185 gene 12 7
13765 12 ENSG00000260423.1 9313517 gene 12 9
13543 12 ENSG00000255621.1 13000839 gene 12 12
162 12 ENSG00000008394.12 16242555 gene 12 14
474 12 ENSG00000048540.14 16368510 gene 12 14
14514 12 ENSG00000275764.1 27036619 gene 12 18
2846 12 ENSG00000110888.17 30737069 gene 12 21
12982 12 ENSG00000235884.3 30762606 gene 12 21
4351 12 ENSG00000127337.6 69358729 gene 12 42
4348 12 ENSG00000127328.21 69698495 gene 12 43
5243 12 ENSG00000136014.11 95546940 gene 12 56
5718 12 ENSG00000139372.14 103957554 gene 12 62
12178 12 ENSG00000204954.9 103957554 gene 12 62
14626 12 ENSG00000278266.1 127113737 gene 12 77
10866 12 ENSG00000184967.6 132083372 gene 12 81
10897 12 ENSG00000185163.9 132118203 gene 12 81
10970 12 ENSG00000185684.12 132083996 gene 12 81
13585 12 ENSG00000256576.2 132189573 gene 12 81
10413 13 ENSG00000180776.15 21459023 gene 13 2
1963 13 ENSG00000102683.7 23157008 gene 13 4
10316 13 ENSG00000179630.10 43873996 gene 13 17
5268 13 ENSG00000136161.12 48532925 gene 13 20
5256 13 ENSG00000136111.12 75311388 gene 13 37
4166 13 ENSG00000125246.15 99606547 gene 13 50
10909 14 ENSG00000185271.7 20365494 gene 14 1
7334 14 ENSG00000157326.18 23953662 gene 14 3
7340 14 ENSG00000157379.13 24291558 gene 14 3
4318 14 ENSG00000126790.11 59444736 gene 14 27
5780 14 ENSG00000139998.14 64972366 gene 14 30
3677 14 ENSG00000119599.16 72915777 gene 14 34
3688 14 ENSG00000119673.14 73563190 gene 14 34
5786 14 ENSG00000140043.11 73844725 gene 14 34
10796 14 ENSG00000184227.7 73528024 gene 14 34
180 14 ENSG00000009830.11 77318589 gene 14 36
639 14 ENSG00000063761.15 77800264 gene 14 36
546 14 ENSG00000054983.16 87948531 gene 14 44
265 14 ENSG00000015133.18 91324116 gene 14 46
13650 14 ENSG00000258572.1 95515367 gene 14 49
10872 14 ENSG00000184990.12 104747641 gene 14 55
10533 15 ENSG00000182117.5 34254250 gene 15 10
12482 15 ENSG00000215252.11 34377560 gene 15 10
4466 15 ENSG00000128829.11 39933819 gene 15 13
12768 15 ENSG00000229474.6 44705554 gene 15 17
5612 15 ENSG00000138600.9 50733462 gene 15 20
8490 15 ENSG00000166035.10 58407738 gene 15 27
5503 15 ENSG00000137819.13 69291095 gene 15 32
2094 15 ENSG00000103811.15 78937302 gene 15 37
10526 15 ENSG00000182054.9 90102033 gene 15 42
5833 15 ENSG00000140470.13 100047379 gene 15 50
2021 16 ENSG00000103148.15 89636 gene 16 1
1798 16 ENSG00000100726.14 1472291 gene 16 2
2061 16 ENSG00000103381.11 12803575 gene 16 13
10689 16 ENSG00000183426.16 14873824 gene 16 15
10749 16 ENSG00000183793.13 15036083 gene 16 15
10704 16 ENSG00000183549.10 20409166 gene 16 19
9557 16 ENSG00000172775.16 57171173 gene 16 30
13817 16 ENSG00000261079.1 74350208 gene 16 39
691 16 ENSG00000065457.10 75617776 gene 16 40
10827 16 ENSG00000184517.11 75144532 gene 16 40
2018 16 ENSG00000103111.14 77190635 gene 16 42
5882 16 ENSG00000140943.16 84070873 gene 16 48
13826 16 ENSG00000261175.5 86638764 gene 16 51
7450 16 ENSG00000158717.10 88700532 gene 16 53
25 16 ENSG00000003249.13 89958562 gene 16 54
4697 16 ENSG00000131165.14 89628212 gene 16 54
10449 17 ENSG00000181031.15 216884 gene 17 1
11665 17 ENSG00000197417.7 3608661 gene 17 3
10622 17 ENSG00000182853.11 4769728 gene 17 4
2621 17 ENSG00000108509.20 4942750 gene 17 5
4499 17 ENSG00000129204.16 5029546 gene 17 5
2690 17 ENSG00000109063.14 10657024 gene 17 9
4325 17 ENSG00000126858.16 32105418 gene 17 19
4768 17 ENSG00000132141.13 34935404 gene 17 21
8617 17 ENSG00000166750.9 35243449 gene 17 21
9543 17 ENSG00000172716.16 35294108 gene 17 21
2598 17 ENSG00000108349.16 40141200 gene 17 23
9689 17 ENSG00000173801.16 41778026 gene 17 25
7716 17 ENSG00000161653.10 43961022 gene 17 26
10233 17 ENSG00000178852.15 47297628 gene 17 27
5919 17 ENSG00000141295.13 47838029 gene 17 28
8529 17 ENSG00000166260.10 54942351 gene 17 32
13895 17 ENSG00000263004.1 57078726 gene 17 33
13973 17 ENSG00000266714.6 75590244 gene 17 42
8828 17 ENSG00000167889.12 76870349 gene 17 43
10585 17 ENSG00000182534.13 76678775 gene 17 43
5952 17 ENSG00000141519.14 80036451 gene 17 45
10599 17 ENSG00000182612.10 81636805 gene 17 46
13891 17 ENSG00000262877.4 81389871 gene 17 46
5970 17 ENSG00000141580.15 82610870 gene 17 47
1936 18 ENSG00000101577.9 2994799 gene 18 3
13913 18 ENSG00000263753.6 5237163 gene 18 4
5932 18 ENSG00000141401.11 11939159 gene 18 9
977 18 ENSG00000075643.5 36187019 gene 18 19
8705 18 ENSG00000167216.16 46917629 gene 18 26
4909 18 ENSG00000133313.14 74495925 gene 18 44
8598 18 ENSG00000166573.5 77241685 gene 18 46
2316 19 ENSG00000105556.11 289498 gene 19 1
4553 19 ENSG00000129946.10 417714 gene 19 1
684 19 ENSG00000065268.10 982793 gene 19 3
1593 19 ENSG00000099817.11 1088640 gene 19 3
4580 19 ENSG00000130270.16 1807137 gene 19 3
11723 19 ENSG00000197766.7 859368 gene 19 3
12702 19 ENSG00000227500.9 1905406 gene 19 3
4220 19 ENSG00000125734.15 6731755 gene 19 7
10448 19 ENSG00000181029.8 7682243 gene 19 7
13217 19 ENSG00000244165.1 10111179 gene 19 9
13989 19 ENSG00000267100.1 10653111 gene 19 9
8736 19 ENSG00000167461.11 16040922 gene 19 13
2327 19 ENSG00000105639.18 17826295 gene 19 14
10335 19 ENSG00000179913.10 17780011 gene 19 14
659 19 ENSG00000064607.16 18981625 gene 19 15
2345 19 ENSG00000105699.16 35248017 gene 19 24
7697 19 ENSG00000161281.10 36149867 gene 19 25
3560 19 ENSG00000117877.10 45404602 gene 19 31
2308 19 ENSG00000105501.12 51621059 gene 19 37
13483 19 ENSG00000254415.3 51548567 gene 19 37
7612 19 ENSG00000160336.14 53429981 gene 19 37
8806 19 ENSG00000167766.18 52615913 gene 19 37
9296 19 ENSG00000170949.17 53080949 gene 19 37
9297 19 ENSG00000170954.11 53087254 gene 19 37
11497 19 ENSG00000196417.12 53372867 gene 19 37
11996 19 ENSG00000203326.11 53365182 gene 19 37
302 19 ENSG00000022556.15 54955709 gene 19 37
1189 19 ENSG00000083828.15 57740129 gene 19 39
4747 19 ENSG00000131845.14 57351684 gene 19 39
11850 19 ENSG00000198466.11 57761718 gene 19 39
11510 20 ENSG00000196476.11 276391 gene 20 1
4255 20 ENSG00000125895.5 1171679 gene 20 2
1344 20 ENSG00000088882.7 2763521 gene 20 4
4254 20 ENSG00000125885.13 5950827 gene 20 5
11696 20 ENSG00000197586.12 25195465 gene 20 18
6729 20 ENSG00000149599.15 31870423 gene 20 19
13751 20 ENSG00000260257.2 32854832 gene 20 19
4056 20 ENSG00000124145.6 45319581 gene 20 28
4084 20 ENSG00000124257.6 45891103 gene 20 28
6728 20 ENSG00000149596.6 44111668 gene 20 28
11675 20 ENSG00000197496.5 46705549 gene 20 29
12581 20 ENSG00000224397.5 50267397 gene 20 31
6740 20 ENSG00000149679.11 62392095 gene 20 37
1869 20 ENSG00000101190.12 62846860 gene 20 37
4200 20 ENSG00000125534.9 63520461 gene 20 37
6012 21 ENSG00000142166.12 33300656 gene 21 14
7479 21 ENSG00000159110.19 33229937 gene 21 14
10932 21 ENSG00000185437.13 39446004 gene 21 19
7355 21 ENSG00000157617.16 41887791 gene 21 23
10696 21 ENSG00000183486.12 41266758 gene 21 23
7580 21 ENSG00000160200.17 42972160 gene 21 23
6015 21 ENSG00000142178.7 43629777 gene 21 23
10847 21 ENSG00000184787.18 44775644 gene 21 23
14416 21 ENSG00000273796.1 45359863 gene 21 23
10570 21 ENSG00000182362.13 46272884 gene 21 24
14366 22 ENSG00000273203.1 17013110 gene 22 1
1612 22 ENSG00000099956.18 23775338 gene 22 6
1635 22 ENSG00000100036.12 30631127 gene 22 10
10920 22 ENSG00000185339.8 30606651 gene 22 10
1667 22 ENSG00000100191.5 32274318 gene 22 12
1708 22 ENSG00000100316.15 39319603 gene 22 16
1669 22 ENSG00000100197.20 42121632 gene 22 17
11989 22 ENSG00000198951.11 42047904 gene 22 17
10705 22 ENSG00000183569.17 42571264 gene 22 18
11408 22 ENSG00000189306.10 42510050 gene 22 18
1497 22 ENSG00000093000.18 45102024 gene 22 20
1729 22 ENSG00000100376.11 45272131 gene 22 20
12659 22 ENSG00000226328.6 45105543 gene 22 20
13130 22 ENSG00000241484.9 44727745 gene 22 20
169 22 ENSG00000008735.13 50595411 gene 22 24
1701 22 ENSG00000100299.17 50604850 gene 22 24
521 1 ENSG00000053371.12 19310497 gene 1 13
4362 1 ENSG00000127463.13 19227605 gene 1 13
9138 1 ENSG00000169914.5 19878843 gene 1 13
10806 1 ENSG00000184313.19 54566589 gene 1 34
7879 1 ENSG00000162836.11 147646379 gene 1 73
9028 3 ENSG00000169087.10 122792659 gene 3 76
2472 7 ENSG00000106628.10 44114687 gene 7 32
2417 7 ENSG00000106178.6 75823266 gene 7 48
11287 7 ENSG00000188372.14 76397141 gene 7 48
5352 9 ENSG00000136856.17 127348293 gene 9 66
2558 10 ENSG00000107819.13 101032154 gene 10 64
5563 10 ENSG00000138152.8 122252762 gene 10 77
2871 12 ENSG00000111224.13 3803806 gene 12 4
4562 12 ENSG00000130038.9 3744539 gene 12 4
3647 12 ENSG00000119242.8 123959862 gene 12 75
5757 12 ENSG00000139714.12 121672174 gene 12 75
4278 13 ENSG00000126217.20 112894196 gene 13 62
14576 13 ENSG00000277159.1 112604083 gene 13 62
7037 13 ENSG00000153531.13 113448962 gene 13 62
12926 19 ENSG00000234465.10 43569117 gene 19 30
2302 19 ENSG00000105479.15 48319272 gene 19 33
2307 19 ENSG00000105499.13 48101216 gene 19 33
10447 19 ENSG00000181027.10 46745328 gene 19 33
2186 19 ENSG00000104805.15 48899921 gene 19 34
4615 19 ENSG00000130529.15 49158325 gene 19 34
7712 19 ENSG00000161618.9 49453017 gene 19 34
1639 22 ENSG00000100056.11 19144548 gene 22 4
10823 22 ENSG00000184470.20 19876115 gene 22 4
cs_index susie_pip mu2 region_tag PVE
540 0 4.962198e-02 6.151463 1_7 8.883268e-07
3427 0 1.459724e-01 12.226166 1_9 5.193752e-06
12643 0 6.108426e-02 4.452795 1_11 7.915573e-07
7459 0 5.712227e-02 5.788403 1_14 9.622426e-07
1421 0 6.199786e-02 5.085586 1_15 9.175676e-07
10125 0 2.479930e-01 17.926730 1_26 1.293781e-05
3556 0 4.620811e-02 4.443790 1_30 5.975746e-07
7782 0 7.147293e-02 5.136337 1_33 1.068355e-06
13102 0 6.293373e-02 8.561895 1_39 1.568100e-06
7796 0 3.921595e-01 24.142355 1_41 2.755260e-05
3449 0 5.969453e-02 5.001651 1_47 8.688969e-07
3452 0 2.259012e-01 17.462696 1_47 1.148022e-05
14321 0 1.546989e-01 13.809065 1_47 6.216869e-06
5522 0 6.265668e-02 6.255446 1_54 1.140633e-06
7116 0 4.230440e-02 9.489285 1_56 1.168259e-06
9301 0 5.882386e-02 6.507252 1_62 1.113965e-06
3878 0 8.350193e-03 5.020227 1_67 1.219945e-07
4984 0 2.603014e-02 38.410146 1_67 2.909663e-06
4986 0 8.934125e-03 9.401227 1_67 2.444313e-07
4987 0 1.004844e-02 7.235686 1_67 2.115917e-07
6084 0 1.645377e-02 49.423212 1_67 2.366555e-06
8972 0 1.995808e-02 16.303653 1_67 9.469435e-07
12337 0 2.535496e-02 25.302853 1_67 1.867036e-06
11807 0 9.559667e-02 5.511477 1_72 1.533314e-06
11756 0 6.914879e-02 4.936152 1_74 9.933296e-07
11240 0 3.692451e-02 6.626546 1_79 7.120694e-07
6101 0 1.021405e-01 6.824831 1_82 2.028664e-06
3535 0 1.231723e-01 13.164119 1_84 4.718730e-06
12590 0 9.983088e-02 12.507379 1_87 3.633720e-06
12555 0 6.523429e-02 6.277605 1_101 1.191764e-06
4884 0 5.413492e-02 5.056409 1_104 7.965994e-07
6911 0 9.449775e-02 6.506331 1_109 1.789278e-06
6195 0 3.983107e-01 22.463017 1_131 2.603816e-05
13729 0 6.580479e-02 5.037900 1_131 9.647779e-07
13065 0 6.178501e-02 4.464281 1_131 8.027031e-07
3339 0 5.928944e-02 4.431071 2_1 7.645508e-07
11076 0 4.402040e-02 5.437825 2_14 6.966257e-07
12939 0 4.631808e-02 4.501695 2_30 6.068019e-07
5540 0 5.469097e-02 4.829381 2_31 7.686478e-07
7881 0 1.016970e-01 10.504292 2_31 3.108817e-06
6244 0 8.507865e-02 4.636932 2_57 1.148079e-06
3333 0 1.738982e-01 16.052970 2_63 8.124016e-06
3642 0 4.951368e-02 4.435019 2_63 6.390590e-07
4203 0 3.245491e-02 8.580922 2_69 8.104657e-07
4205 0 7.452446e-02 11.589147 2_69 2.513452e-06
863 0 8.447531e-02 7.279747 2_104 1.789643e-06
4455 0 7.410507e-02 4.528827 2_107 9.766836e-07
5596 0 6.333036e-02 4.740234 2_122 8.736390e-07
6293 0 9.372274e-02 9.808610 2_141 2.675302e-06
8927 0 8.689394e-02 9.112061 2_141 2.304233e-06
12043 0 6.638940e-02 6.649341 2_141 1.284688e-06
10005 0 5.361436e-02 6.899845 2_144 1.076566e-06
10430 0 4.832261e-02 5.960935 2_144 8.382723e-07
12958 0 4.228788e-02 4.587183 2_144 5.645239e-07
6289 0 1.330990e-01 7.352716 3_4 2.848019e-06
7134 0 2.716448e-02 5.059164 3_9 3.999451e-07
3157 0 5.057829e-02 4.450069 3_36 6.550150e-07
10381 0 8.825056e-02 9.233335 3_39 2.371354e-06
9498 0 5.848364e-02 4.641471 3_45 7.899694e-07
3202 0 8.019696e-02 10.060856 3_78 2.348082e-06
11415 0 1.976205e-01 18.284667 3_78 1.051573e-05
8085 0 6.681521e-02 4.446657 3_85 8.646280e-07
6960 0 7.018250e-02 7.175796 3_93 1.465613e-06
13112 0 5.213648e-02 4.752577 3_97 7.210928e-07
10026 0 5.298066e-02 5.318083 3_120 8.199602e-07
3645 0 9.494103e-02 10.022379 3_121 2.769141e-06
4357 0 7.348521e-02 7.531999 4_2 1.610759e-06
8107 0 6.026888e-02 5.725442 4_2 1.004205e-06
10958 0 5.314359e-02 4.583951 4_2 7.089426e-07
9729 0 6.106321e-02 5.888569 4_3 1.046429e-06
9279 0 9.531015e-02 9.529912 4_7 2.643312e-06
13412 0 1.522725e-01 13.906550 4_7 6.162559e-06
13415 0 6.381428e-02 8.388830 4_10 1.557900e-06
10163 0 5.805510e-02 5.268586 4_11 8.901328e-07
2737 0 6.443088e-02 7.408764 4_22 1.389185e-06
11860 0 1.214173e-01 13.722318 4_37 4.848733e-06
4408 0 8.222100e-02 8.556922 4_40 2.047485e-06
277 0 5.872617e-02 5.706183 4_49 9.752090e-07
5643 0 7.673055e-02 4.666961 4_51 1.042132e-06
11814 0 6.246114e-02 6.582809 4_59 1.196579e-06
14305 0 3.997627e-02 4.979432 4_66 5.792985e-07
5654 0 5.882682e-02 4.876210 4_69 8.347915e-07
2744 0 8.094222e-02 4.979169 4_119 1.172877e-06
6397 0 9.483345e-02 8.922516 4_120 2.462460e-06
8216 0 6.777560e-02 5.842907 4_120 1.152451e-06
13385 0 6.051281e-02 4.811263 4_120 8.472795e-07
14237 0 5.928961e-02 4.625734 4_120 7.981408e-07
8219 0 7.288928e-02 9.810360 5_1 2.080985e-06
13411 0 6.465297e-02 6.496067 5_2 1.222248e-06
13370 0 5.361669e-02 4.458000 5_6 6.956013e-07
13383 0 4.898161e-02 4.431700 5_8 6.317187e-07
3079 0 1.547413e-01 14.603423 5_9 6.576293e-06
8188 0 6.629082e-02 6.746521 5_9 1.301528e-06
7191 0 6.029925e-02 7.885593 5_33 1.383778e-06
11737 0 9.752168e-02 9.425459 5_42 2.675001e-06
9006 0 6.431821e-02 5.387417 5_74 1.008405e-06
8307 0 4.938261e-02 4.468748 5_77 6.422148e-07
12336 0 8.369397e-02 4.654048 5_106 1.133562e-06
9863 0 6.310943e-02 5.615066 5_107 1.031263e-06
10183 0 8.769038e-02 8.622963 5_107 2.200538e-06
14272 0 5.119606e-02 4.510033 6_1 6.719494e-07
5426 0 5.875755e-02 4.661196 6_3 7.970422e-07
6445 0 6.367771e-02 5.392188 6_3 9.992468e-07
13215 0 5.820621e-02 4.575597 6_3 7.750636e-07
7012 0 5.159617e-02 4.645635 6_10 6.975621e-07
6446 0 7.096637e-02 4.748818 6_12 9.807502e-07
2948 0 1.744862e-02 7.945247 6_20 4.034491e-07
11608 0 3.043622e-02 9.024496 6_22 7.993445e-07
5430 0 4.597292e-06 43.678102 6_26 5.843676e-10
12119 0 1.815565e-05 39.048802 6_26 2.063193e-09
12121 0 1.006660e-07 4.939213 6_26 1.446974e-12
12122 0 1.290769e-07 11.790210 6_26 4.428844e-12
12123 0 3.472355e-07 10.867848 6_26 1.098217e-11
12277 0 2.767221e-07 15.995949 6_26 1.288173e-11
12376 0 1.007762e-07 4.487520 6_26 1.316087e-12
14238 0 1.492781e-06 42.215525 6_26 1.833955e-10
12077 0 3.162212e-07 20.375684 6_26 1.875096e-11
12068 0 3.744613e-02 7.874679 6_27 8.581438e-07
11916 0 1.597898e-02 5.300136 6_28 2.464656e-07
450 0 7.777504e-02 6.188843 6_33 1.400780e-06
11465 0 7.734628e-02 6.871423 6_34 1.546701e-06
7023 0 8.210109e-02 6.964793 6_35 1.664092e-06
9518 0 5.836155e-02 4.456243 6_65 7.568607e-07
9531 0 5.661882e-02 4.506577 6_82 7.425538e-07
3599 0 7.209793e-02 5.985630 6_87 1.255894e-06
2988 0 1.253084e-02 11.698907 6_103 4.266246e-07
3902 0 5.767690e-03 4.500919 6_103 7.554807e-08
14208 0 5.847242e-03 4.589372 6_103 7.809525e-08
13165 0 4.996746e-02 4.711955 7_1 6.851863e-07
3937 0 1.750491e-01 16.459022 7_10 8.384637e-06
14257 0 8.854947e-02 7.573181 7_16 1.951572e-06
840 0 5.963248e-02 4.525519 7_21 7.853650e-07
11457 0 8.739968e-02 6.294777 7_43 1.601071e-06
11782 0 8.618212e-02 6.165812 7_43 1.546421e-06
13122 0 1.093557e-01 8.365447 7_43 2.662262e-06
8358 0 1.593654e-01 16.876074 7_47 7.826826e-06
4394 0 5.791046e-02 4.434545 7_49 7.473541e-07
74 0 1.953289e-01 16.351823 7_54 9.295078e-06
2363 0 7.152440e-02 4.434585 7_55 9.230548e-07
828 0 1.076902e-01 11.501663 7_60 3.604599e-06
8671 0 2.317753e-02 5.392645 7_61 3.637385e-07
12216 0 2.467731e-02 5.868715 7_61 4.214646e-07
5125 0 6.459907e-02 5.297491 7_70 9.959024e-07
4452 0 1.144931e-01 7.702405 7_78 2.566409e-06
7190 0 6.577891e-02 6.893642 7_81 1.319641e-06
21 0 1.115392e-01 14.565704 7_94 4.728021e-06
2461 0 8.965804e-02 12.546033 7_94 3.273527e-06
11166 0 3.191695e-01 24.776896 7_94 2.301381e-05
13842 0 3.956681e-02 5.109374 7_94 5.883274e-07
13463 0 3.907697e-02 7.557771 8_11 8.594784e-07
2120 0 1.492037e-01 15.428678 8_19 6.699286e-06
11633 0 1.344084e-01 12.791543 8_24 5.003450e-06
9737 0 7.766489e-02 4.431132 8_69 1.001520e-06
9305 0 8.894617e-02 7.511646 8_93 1.944387e-06
258 0 3.349737e-02 5.311013 8_94 5.177360e-07
6574 0 3.246077e-02 4.890865 8_94 4.620242e-07
2505 0 5.017175e-02 4.462162 9_1 6.515157e-07
9626 0 5.744776e-02 5.689267 9_1 9.511516e-07
6576 0 5.188202e-02 6.761704 9_5 1.020924e-06
3749 0 4.850107e-02 4.886942 9_6 6.897772e-07
8332 0 4.698164e-02 4.491708 9_13 6.141295e-07
12586 0 9.362888e-02 9.144406 9_17 2.491642e-06
5390 0 7.359709e-02 7.090790 9_25 1.518712e-06
8329 0 6.401293e-02 5.562218 9_27 1.036182e-06
13808 0 7.975355e-02 7.569837 9_27 1.756940e-06
2485 0 4.046784e-02 5.551293 9_35 6.537692e-07
2490 0 6.393012e-02 4.895959 9_49 9.108851e-07
5359 0 9.186241e-02 5.846452 9_50 1.562970e-06
2486 0 4.823332e-02 4.852722 9_56 6.811658e-07
9627 0 4.755173e-02 4.777804 9_56 6.611727e-07
3664 0 6.134900e-02 4.537313 9_58 8.100775e-07
1524 0 2.418645e-01 21.637752 9_59 1.523016e-05
4619 0 7.191431e-02 4.433594 9_73 9.278795e-07
7616 0 7.285912e-02 4.707852 9_73 9.982217e-07
2577 0 7.285774e-02 7.978241 10_4 1.691621e-06
12883 0 5.055248e-02 4.457466 10_5 6.557688e-07
5031 0 1.067489e-01 8.998420 10_10 2.795440e-06
6948 0 5.407575e-02 5.423068 10_13 8.534300e-07
12157 0 5.808927e-02 4.451971 10_16 7.526075e-07
9876 0 6.760057e-02 4.786728 10_28 9.416931e-07
4930 0 8.191094e-02 6.266143 10_51 1.493697e-06
3905 0 8.057165e-02 9.374366 10_56 2.198085e-06
10862 0 6.704642e-02 7.695141 10_56 1.501456e-06
12445 0 6.651215e-02 7.622273 10_56 1.475386e-06
11898 0 2.946381e-01 21.762928 10_56 1.866064e-05
6650 0 2.091865e-01 19.133611 10_60 1.164799e-05
7173 0 2.106864e-01 14.506682 10_62 8.894567e-06
8536 0 7.573597e-02 7.112368 10_66 1.567605e-06
2543 0 4.546087e-01 27.965271 10_76 3.699790e-05
6866 0 7.546753e-02 4.681919 10_83 1.028263e-06
9420 0 7.359364e-02 4.452022 10_83 9.534939e-07
6009 0 6.621158e-02 5.997767 11_1 1.155697e-06
10901 0 5.963631e-02 5.046318 11_1 8.758016e-07
12411 0 7.083342e-02 6.612905 11_1 1.363172e-06
13526 0 1.190789e-01 11.401767 11_1 3.951183e-06
14141 0 6.734565e-02 6.152498 11_1 1.205817e-06
2823 0 3.069238e-02 5.723158 11_3 5.111950e-07
3829 0 1.006951e-01 14.498486 11_4 4.248653e-06
4945 0 6.083487e-02 5.509095 11_9 9.753335e-07
2808 0 6.121774e-02 4.650116 11_22 8.284408e-07
3857 0 5.794818e-02 4.997260 11_23 8.427370e-07
6670 0 6.119944e-02 5.493139 11_23 9.783367e-07
6834 0 1.414444e-01 11.104757 11_27 4.571042e-06
7775 0 4.757130e-02 10.231065 11_38 1.416401e-06
9895 0 1.077713e-01 9.923706 11_41 3.112415e-06
7439 0 1.097952e-01 8.034023 11_42 2.567065e-06
13261 0 1.063917e-01 6.738380 11_46 2.086333e-06
4035 0 8.026508e-02 8.310418 11_49 1.941198e-06
2794 0 8.702610e-02 9.532550 11_53 2.414232e-06
6690 0 4.685023e-02 4.579029 11_65 6.243174e-07
5482 0 1.505083e-01 14.574095 11_66 6.383550e-06
6691 0 8.854745e-02 13.248003 11_67 3.413868e-06
10385 1 8.291038e-02 10.228953 11_68 2.468087e-06
10048 0 1.187878e-02 6.836207 11_71 2.363237e-07
2774 0 4.713120e-02 4.617144 11_77 6.332894e-07
3771 0 4.943787e-02 5.000034 11_81 7.193711e-07
13536 0 5.096424e-02 5.275147 11_81 7.823847e-07
2866 0 8.051852e-02 7.783980 12_2 1.823971e-06
13577 0 1.608206e-01 14.142104 12_7 6.618752e-06
13765 0 5.166516e-02 5.982148 12_9 8.994463e-07
13543 0 7.868682e-02 5.490205 12_12 1.257219e-06
162 0 6.576507e-02 6.587485 12_14 1.260768e-06
474 0 7.475655e-02 7.756932 12_14 1.687561e-06
14514 0 1.329510e-01 12.043368 12_18 4.659722e-06
2846 0 6.760241e-02 4.443052 12_21 8.741057e-07
12982 0 1.210897e-01 9.813933 12_21 3.458362e-06
4351 0 6.944753e-02 5.440086 12_42 1.099469e-06
4348 0 5.026103e-02 4.499966 12_43 6.582046e-07
5243 0 6.408990e-02 5.089866 12_56 9.493279e-07
5718 0 4.995516e-02 4.496906 12_62 6.537542e-07
12178 0 8.680689e-02 9.531941 12_62 2.407997e-06
14626 0 5.412173e-02 4.556771 12_77 7.177102e-07
10866 0 5.496139e-02 5.827674 12_81 9.321231e-07
10897 0 5.613339e-02 6.019008 12_81 9.832558e-07
10970 0 4.745017e-02 4.497614 12_81 6.210696e-07
13585 0 5.254676e-02 5.420581 12_81 8.289189e-07
10413 0 7.652726e-02 6.472837 13_2 1.441555e-06
1963 0 5.290293e-02 4.480018 13_4 6.897310e-07
10316 0 5.549466e-02 4.503128 13_17 7.272535e-07
5268 0 1.007927e-01 8.924099 13_20 2.617664e-06
5256 0 6.460352e-02 6.447870 13_37 1.212252e-06
4166 0 9.171074e-02 9.273224 13_50 2.474978e-06
10909 0 7.309326e-02 8.116696 14_1 1.726541e-06
7334 0 3.138852e-02 5.003863 14_3 4.570845e-07
7340 0 6.998099e-02 12.425296 14_3 2.530504e-06
4318 0 6.407755e-02 4.897846 14_27 9.133376e-07
5780 0 5.111519e-02 4.432401 14_30 6.593400e-07
3677 0 1.540989e-02 8.667533 14_34 3.887006e-07
3688 0 1.401022e-02 6.851498 14_34 2.793513e-07
5786 0 1.170132e-02 4.943757 14_34 1.683497e-07
10796 0 1.183906e-02 5.899872 14_34 2.032732e-07
180 0 6.796402e-02 4.888867 14_36 9.669579e-07
639 0 7.664378e-02 5.986676 14_36 1.335313e-06
546 0 6.871550e-02 6.265298 14_44 1.252901e-06
265 0 6.321513e-02 4.475653 14_46 8.233752e-07
13650 0 9.746358e-02 9.124874 14_49 2.588151e-06
10872 0 7.400890e-02 7.144219 14_55 1.538718e-06
10533 0 1.123047e-01 6.081570 15_10 1.987623e-06
12482 0 1.001055e-01 5.015204 15_10 1.461055e-06
4466 0 5.796269e-02 4.435662 15_13 7.482165e-07
12768 0 6.491627e-02 4.835227 15_17 9.134626e-07
5612 0 5.894637e-02 4.473329 15_20 7.673760e-07
8490 0 2.967861e-01 25.395298 15_27 2.193397e-05
5503 0 6.375021e-02 7.061282 15_32 1.310043e-06
2094 0 7.242486e-01 18.424674 15_37 3.883361e-05
10526 0 5.386119e-02 6.030730 15_42 9.452924e-07
5833 0 2.338924e-01 16.303247 15_50 1.109713e-05
2021 0 1.118614e-01 9.425998 16_1 3.068513e-06
1798 0 4.684180e-02 4.661097 16_2 6.353924e-07
2061 0 5.028406e-02 5.294908 16_13 7.748347e-07
10689 0 8.561826e-02 10.025994 16_15 2.498125e-06
10749 0 8.553850e-02 10.017500 16_15 2.493683e-06
10704 0 6.551777e-02 5.633390 16_19 1.074111e-06
9557 0 7.831597e-02 7.176667 16_30 1.635661e-06
13817 0 5.561881e-02 4.571053 16_39 7.398747e-07
691 0 1.332788e-01 15.577328 16_40 6.041911e-06
10827 0 1.294159e-01 15.604637 16_40 5.877081e-06
2018 0 9.232796e-02 4.764566 16_42 1.280197e-06
5882 0 4.872495e-02 5.218431 16_48 7.399658e-07
13826 0 3.876962e-02 4.768236 16_51 5.379843e-07
7450 0 4.644543e-02 4.438984 16_53 5.999940e-07
25 0 1.612424e-01 13.108651 16_54 6.151167e-06
4697 0 9.403704e-02 8.064270 16_54 2.206908e-06
10449 0 7.195120e-02 5.745481 17_1 1.203053e-06
11665 0 6.867295e-02 4.670162 17_3 9.333359e-07
10622 0 2.768906e-01 21.989342 17_4 1.771906e-05
2621 0 6.182728e-02 4.501095 17_5 8.098762e-07
4499 0 7.774076e-02 6.590311 17_5 1.490991e-06
2690 0 6.453747e-02 6.479287 17_9 1.216913e-06
4325 0 6.089621e-02 5.226389 17_19 9.262160e-07
4768 0 7.146983e-02 4.491532 17_21 9.341950e-07
8617 0 7.224344e-02 4.589854 17_21 9.649783e-07
9543 0 7.104578e-02 4.437199 17_21 9.174184e-07
2598 0 3.606634e-01 24.150651 17_23 2.534844e-05
9689 0 4.068807e-02 5.027927 17_25 5.953555e-07
7716 0 1.159428e-01 11.301158 17_26 3.813178e-06
10233 0 8.989568e-02 52.216423 17_27 1.366049e-05
5919 0 2.619212e-01 22.572042 17_28 1.720529e-05
8529 0 8.654918e-02 9.971425 17_32 2.511542e-06
13895 0 5.549062e-02 4.850018 17_33 7.832190e-07
13973 0 1.091120e-01 11.427713 17_42 3.628708e-06
8828 0 1.916633e-01 12.267487 17_43 6.842501e-06
10585 0 8.376221e-02 4.500947 17_43 1.097166e-06
5952 0 1.726286e-01 14.831638 17_45 7.451129e-06
10599 0 8.089401e-02 4.789408 17_46 1.127505e-06
13891 0 7.802561e-02 4.458599 17_46 1.012409e-06
5970 0 5.774971e-02 5.833462 17_47 9.803847e-07
1936 0 5.976521e-02 4.970231 18_3 8.644609e-07
13913 0 6.136664e-02 4.928011 18_4 8.800844e-07
5932 0 1.027418e-01 6.213652 18_9 1.857867e-06
977 0 7.493622e-02 5.888977 18_19 1.284257e-06
8705 0 7.215816e-02 6.640385 18_26 1.394437e-06
4909 0 6.079921e-02 6.419650 18_44 1.135872e-06
8598 0 5.154203e-02 4.693079 18_46 7.039466e-07
2316 0 4.827033e-02 5.040011 19_1 7.079981e-07
4553 0 4.750850e-02 4.896218 19_1 6.769434e-07
684 0 5.403014e-02 10.028716 19_3 1.576891e-06
1593 0 1.090855e-01 16.366819 19_3 5.195793e-06
4580 0 2.566234e-01 24.152998 19_3 1.803797e-05
11723 0 3.112734e-02 5.148867 19_3 4.664166e-07
12702 0 3.036951e-02 4.920810 19_3 4.349052e-07
4220 0 6.634561e-02 4.486721 19_7 8.662865e-07
10448 0 2.658225e-01 17.606759 19_7 1.362045e-05
13217 0 0.000000e+00 11.300737 19_9 0.000000e+00
13989 0 0.000000e+00 41.555232 19_9 0.000000e+00
8736 0 1.369393e-01 17.474782 19_13 6.964021e-06
2327 0 2.229788e-02 4.713721 19_14 3.058776e-07
10335 0 1.262573e-01 19.462223 19_14 7.151038e-06
659 0 2.898426e-02 20.965941 19_15 1.768467e-06
2345 0 5.185812e-02 7.325244 19_24 1.105501e-06
7697 0 6.425510e-02 5.196142 19_25 9.716479e-07
3560 0 0.000000e+00 16.730266 19_31 0.000000e+00
2308 0 3.850122e-01 33.095022 19_37 3.708151e-05
13483 0 2.159035e-02 6.009246 19_37 3.775723e-07
7612 0 2.402062e-02 6.963331 19_37 4.867675e-07
8806 0 1.821347e-02 4.489784 19_37 2.379789e-07
9296 0 1.888176e-02 4.811532 19_37 2.643907e-07
9297 0 2.623274e-02 7.752043 19_37 5.918070e-07
11497 0 1.825871e-02 4.511931 19_37 2.397468e-07
11996 0 1.877551e-02 4.761145 19_37 2.601498e-07
302 0 2.278848e-02 6.492222 19_37 4.305554e-07
1189 0 4.951397e-02 9.969522 19_39 1.436555e-06
4747 0 4.450368e-02 7.996791 19_39 1.035695e-06
11850 0 3.338469e-02 5.124183 19_39 4.978429e-07
11510 0 8.925609e-02 10.472400 20_1 2.720222e-06
4255 0 7.555796e-02 4.953577 20_2 1.089230e-06
1344 0 4.894765e-02 4.764979 20_4 6.787551e-07
4254 0 4.008864e-02 5.078698 20_5 5.925076e-07
11696 0 3.959287e-02 7.749467 20_18 8.929130e-07
6729 0 5.120928e-02 6.503883 20_19 9.692632e-07
13751 0 8.154104e-02 9.582896 20_19 2.274015e-06
4056 0 4.597491e-02 4.713198 20_28 6.306041e-07
4084 1 2.494502e-01 26.182796 20_28 1.900729e-05
6728 0 4.662211e-02 4.697686 20_28 6.373768e-07
11675 0 7.024505e-02 5.789737 20_29 1.183573e-06
12581 0 3.561318e-02 5.176869 20_31 5.365351e-07
6740 0 1.292605e-01 10.113235 20_37 3.804313e-06
1869 0 7.031657e-02 4.490480 20_37 9.189053e-07
4200 0 7.670387e-02 5.285063 20_37 1.179744e-06
6012 0 3.353109e-01 24.162518 21_14 2.357817e-05
7479 0 4.192593e-02 4.501676 21_14 5.492590e-07
10932 0 2.208200e-02 9.153284 21_19 5.882143e-07
7355 0 7.571556e-02 13.423468 21_23 2.957809e-06
10696 0 4.611006e-02 8.917912 21_23 1.196683e-06
7580 0 5.416612e-02 10.374689 21_23 1.635397e-06
6015 0 2.848489e-02 4.584989 21_23 3.800784e-07
10847 0 2.863881e-02 4.633308 21_23 3.861593e-07
14416 0 3.021118e-02 5.112689 21_23 4.495079e-07
10570 0 2.443258e-01 19.716303 21_24 1.401894e-05
14366 0 5.958036e-02 6.637191 22_1 1.150821e-06
1612 0 1.823335e-01 14.186677 22_6 7.527790e-06
1635 0 1.011679e-01 9.233762 22_10 2.718578e-06
10920 0 9.683034e-02 8.828665 22_10 2.487865e-06
1667 0 6.941895e-02 5.402849 22_12 1.091494e-06
1708 0 5.746443e-02 5.509720 22_16 9.214015e-07
1669 0 5.467582e-02 4.601589 22_17 7.321894e-07
11989 0 5.633310e-02 4.935735 22_17 8.091626e-07
10705 0 6.901903e-02 7.244560 22_18 1.455128e-06
11408 0 5.433717e-02 5.070210 22_18 8.017579e-07
1497 0 1.879331e-01 15.857129 22_20 8.672575e-06
1729 0 2.136961e-01 17.104694 22_20 1.063732e-05
12659 0 6.465651e-02 5.914065 22_20 1.112804e-06
13130 0 6.898421e-02 6.504445 22_20 1.305811e-06
169 0 7.001428e-02 5.717285 22_24 1.164922e-06
1701 0 6.228559e-02 4.758866 22_24 8.626037e-07
521 0 1.693736e-01 14.330277 1_13 7.063512e-06
4362 0 9.143968e-02 8.798896 1_13 2.341441e-06
9138 0 9.319030e-02 9.001120 1_13 2.441111e-06
10806 0 1.868505e-12 45.488781 1_34 2.473540e-16
7879 1 9.932775e-01 21.778099 1_73 6.295220e-05
9028 0 3.734024e-02 4.907991 3_76 5.333363e-07
2472 0 1.004879e-01 13.317824 7_32 3.894640e-06
2417 0 2.323409e-01 17.612682 7_48 1.190889e-05
11287 0 6.890716e-02 6.653633 7_48 1.334269e-06
5352 0 9.000403e-02 13.339162 9_66 3.493903e-06
2558 0 1.465765e-01 13.418411 10_64 5.723818e-06
5563 0 4.245584e-02 4.461662 10_77 5.512574e-07
2871 0 3.542435e-01 26.121923 12_4 2.692944e-05
4562 0 7.406565e-02 12.054995 12_4 2.598389e-06
3647 1 9.604840e-01 26.340340 12_75 7.362610e-05
5757 0 2.074617e-02 4.471989 12_75 2.699970e-07
4278 0 2.829055e-01 21.425725 13_62 1.763994e-05
14576 0 3.781685e-02 4.533875 13_62 4.989709e-07
7037 0 9.517772e-02 12.243474 13_62 3.391254e-06
12926 0 6.526748e-03 18.826025 19_30 3.575821e-07
2302 0 3.483748e-02 14.182171 19_33 1.437837e-06
2307 0 1.455655e-02 9.600340 19_33 4.066919e-07
10447 3 9.482623e-01 20.617877 19_33 5.689744e-05
2186 0 1.045121e-03 12.380242 19_34 3.765442e-08
4615 0 1.659913e-03 24.346541 19_34 1.176096e-07
7712 5 9.241005e-03 51.337514 19_34 1.380621e-06
1639 0 1.315866e-01 19.033222 22_4 7.288603e-06
10823 0 2.500688e-01 24.769840 22_4 1.802615e-05
genename gene_type z num_eqtl
540 KIF1B protein_coding -0.420023025 4
3427 FBXO6 protein_coding -1.572219800 4
12643 LINC01772 lincRNA -0.398779133 4
7459 CDA protein_coding 0.585809577 4
1421 USP48 protein_coding 0.398466350 4
10125 SVBP protein_coding -2.392855228 4
3556 SLC5A9 protein_coding 0.063362895 4
7782 SLC1A7 protein_coding -0.504506046 4
13102 L1TD1 protein_coding -1.234862240 5
7796 RAVER2 protein_coding 3.020571751 4
3449 TNNI3K protein_coding -0.474773503 4
3452 CRYZ protein_coding -2.282589321 5
14321 RP11-17E13.2 lincRNA -1.942794637 4
5522 KYAT3 protein_coding 1.036122617 4
7116 FAM69A protein_coding 2.551431698 4
9301 S1PR1 protein_coding -0.748495522 5
3878 CLCC1 protein_coding -1.719746875 4
4984 GSTM1 protein_coding 6.501276770 6
4986 GSTM5 protein_coding 2.099156257 4
4987 GSTM3 protein_coding -1.014289761 4
6084 SYPL2 protein_coding 9.792802919 4
8972 GSTM4 protein_coding -0.353118235 7
12337 GSTM2 protein_coding 4.663321525 4
11807 MAN1A2 protein_coding -0.552900612 4
11756 HRNR protein_coding 0.388616850 5
11240 C1orf204 protein_coding 0.762346726 4
6101 CREG1 protein_coding -0.853956131 4
3535 VAMP4 protein_coding -2.021000926 4
12590 RASAL2-AS1 lincRNA 1.577768381 4
12555 RP11-134G8.5 lincRNA -0.771680303 4
4884 DSTYK protein_coding -0.819870882 4
6911 PTPN14 protein_coding 0.657855506 4
6195 SCCPDH protein_coding -3.048942646 4
13729 RP11-488L18.10 lincRNA -0.373514274 5
13065 OR2W3 protein_coding -0.065549661 4
3339 TPO protein_coding 0.008096188 5
11076 FAM228A protein_coding 0.940387821 5
12939 AC073283.4 lincRNA -0.112657370 4
5540 LHCGR protein_coding 0.382961327 4
7881 PPP1R21 protein_coding -1.214145067 4
6244 ZNF514 protein_coding -0.291985227 4
3333 UXS1 protein_coding 1.974871040 4
3642 C2orf40 protein_coding 0.071374113 4
4203 INSIG2 protein_coding 1.427561022 4
4205 CCDC93 protein_coding -0.578522282 4
863 CYBRD1 protein_coding 0.923509271 4
4455 HOXD3 protein_coding 0.002308690 4
5596 MDH1B protein_coding 0.304853191 4
6293 ESPNL protein_coding -1.417367242 4
8927 KLHL30 protein_coding -1.242112550 6
12043 TRAF3IP1 protein_coding -0.865308184 5
10005 BOK protein_coding 0.928550024 4
10430 D2HGDH protein_coding 0.739406811 4
12958 AC114730.2 lincRNA -0.030084372 4
6289 SUMF1 protein_coding -0.959777571 4
7134 TSEN2 protein_coding 1.165007194 4
3157 ACTR8 protein_coding -0.104181026 4
10381 CCDC66 protein_coding -1.319878876 4
9498 SUCLG2 protein_coding 0.222588098 4
3202 SLC41A3 protein_coding 1.320648871 4
11415 ALG1L protein_coding 2.174161853 5
8085 NMNAT3 protein_coding -0.058455493 4
6960 IGSF10 protein_coding -0.884441643 4
13112 LINC00886 lincRNA 0.334099160 4
10026 MUC20 protein_coding -0.548228111 4
3645 PIGZ protein_coding 1.385192240 4
4357 IDUA protein_coding -1.261859716 4
8107 UVSSA protein_coding -0.861322805 5
10958 PCGF3 protein_coding -0.260635278 4
9729 FAM53A protein_coding 0.669441330 4
9279 AC093323.3 lincRNA -1.392536686 5
13412 RP11-539L10.3 lincRNA -1.834959651 4
13415 RP11-774O3.3 lincRNA 0.974762445 5
10163 ZNF518B protein_coding 0.571070739 4
2737 STIM2 protein_coding 0.916001158 4
11860 CNGA1 protein_coding -1.923053579 4
4408 SRD5A3 protein_coding -1.170589307 4
277 RUFY3 protein_coding 0.567301477 4
5643 NAAA protein_coding -0.203875610 4
11814 HSD17B11 protein_coding 0.829426820 5
14305 RP11-571L19.8 lincRNA -0.311361835 4
5654 PPA2 protein_coding -0.255177151 4
2744 LRP2BP protein_coding 0.401078316 4
6397 CYP4V2 protein_coding -1.282357327 4
8216 KLKB1 protein_coding 0.752919864 5
13385 RP11-11N5.1 lincRNA 0.317906005 4
14237 RP11-11N5.3 lincRNA 0.126423375 4
8219 CCDC127 protein_coding -1.286290946 4
13411 CTD-2245E15.3 lincRNA 0.760792106 4
13370 LINC02145 lincRNA -0.085938722 5
13383 SNHG18 lincRNA 0.081069368 5
3079 DAP protein_coding -1.836357379 5
8188 CMBL protein_coding 0.833093909 4
7191 SETD9 protein_coding -0.882862786 4
11737 OCLN protein_coding -1.211458994 4
9006 PPIC protein_coding -0.564727535 4
8307 ALDH7A1 protein_coding -0.027854047 4
12336 MXD3 protein_coding -0.276558915 4
9863 PHYKPL protein_coding -0.582546796 7
10183 ZNF354B protein_coding -1.086779855 5
14272 RP11-532F6.3 lincRNA 0.175732225 4
5426 RIPK1 protein_coding -0.267420987 4
6445 MYLK4 protein_coding 0.509598061 6
13215 LINC01011 lincRNA -0.245955915 4
7012 SYCP2L protein_coding -0.442810060 4
6446 TBC1D7 protein_coding 0.296434275 4
2948 BTN3A3 protein_coding 0.015487617 5
11608 ZSCAN26 protein_coding 0.229080872 4
5430 FLOT1 protein_coding -3.971254534 4
12119 MICB protein_coding -3.059399101 6
12121 HLA-C protein_coding 0.183194817 4
12122 POU5F1 protein_coding -2.393737307 5
12123 CCHCR1 protein_coding 2.931589024 4
12277 HCG27 lincRNA 1.864693269 8
12376 GTF2H4 protein_coding -0.174362404 4
14238 XXbac-BPG181B23.7 lincRNA -4.476510146 5
12077 NOTCH4 protein_coding 3.165455071 4
12068 BRD2 protein_coding 1.894966403 4
11916 RPL10A protein_coding 0.898562553 4
450 CUL7 protein_coding 0.747906425 4
11465 SUPT3H protein_coding -0.904083359 4
7023 SLC25A27 protein_coding 0.889313319 4
9518 MANEA protein_coding 0.065091049 4
9531 SMPDL3A protein_coding -0.142989824 5
3599 AKAP7 protein_coding -0.616835011 4
2988 MRPL18 protein_coding -2.249761834 4
3902 SERAC1 protein_coding 0.021599445 4
14208 GTF2H5 protein_coding 0.146671647 4
13165 AC093627.8 lincRNA 0.292277061 5
3937 CCZ1 protein_coding 1.926156679 5
14257 GS1-166A23.2 lincRNA -0.926818846 4
840 OSBPL3 protein_coding 0.218366418 4
11457 ZNF107 protein_coding -0.694440454 5
11782 ZNF273 protein_coding 0.689406338 4
13122 CRCP protein_coding 1.236269262 4
8358 WBSCR27 protein_coding -2.596799408 4
4394 PTPN12 protein_coding -0.068707659 4
74 ABCB4 protein_coding 1.847895928 4
2363 GTPBP10 protein_coding 0.103843671 4
828 ASNS protein_coding 1.769924731 4
8671 CNPY4 protein_coding -0.020060388 4
12216 SAP25 protein_coding 0.068927392 4
5125 TES protein_coding -0.519405918 4
4452 OPN1SW protein_coding 1.024844827 4
7190 LRGUK protein_coding 0.889078139 5
21 TMEM176A protein_coding -1.826630114 4
2461 TMEM176B protein_coding -1.669675814 4
11166 WDR86 protein_coding -2.895133691 4
13842 LINC01003 lincRNA -0.425451886 6
13463 CLDN23 protein_coding 1.625196493 4
2120 PDGFRL protein_coding 1.866632709 4
11633 ENTPD4 protein_coding 1.932973905 4
9737 SNX31 protein_coding 0.009455059 5
9305 TSNARE1 protein_coding 1.012418419 5
258 ZC3H3 protein_coding -0.615685951 4
6574 NAPRT protein_coding 0.438990829 5
2505 DOCK8 protein_coding -0.050483179 4
9626 DMRT2 protein_coding 0.641670575 5
6576 AK3 protein_coding -0.848931759 7
3749 CD274 protein_coding -0.470739120 4
8332 CCDC171 protein_coding -0.157121597 4
12586 RP11-370B11.3 lincRNA 1.203288096 4
5390 APTX protein_coding -0.671241425 4
8329 NUDT2 protein_coding 0.550071297 4
13808 RP11-384P7.7 lincRNA 0.923319969 4
2485 NMRK1 protein_coding 0.645405159 4
2490 CORO2A protein_coding 0.290834207 4
5359 ZNF189 protein_coding 0.670394596 4
2486 TMEM245 protein_coding 0.242908835 4
9627 ZNF483 protein_coding -0.328936474 4
3664 HDHD3 protein_coding 0.189012099 7
1524 WHRN protein_coding 2.752308321 4
4619 UBAC1 protein_coding 0.030488086 4
7616 GPSM1 protein_coding -0.505102970 4
2577 PITRM1 protein_coding -1.007937847 4
12883 LINC00702 lincRNA -0.082658134 4
5031 ECHDC3 protein_coding 1.393594958 4
6948 NMT2 protein_coding -0.515306167 4
12157 MALRD1 protein_coding 0.026142064 4
9876 ZNF25 protein_coding 0.333750519 4
4930 SFTPD protein_coding -0.818569148 5
3905 OPN4 protein_coding -1.102800039 4
10862 NUTM2A protein_coding 1.246248487 4
12445 NUTM2D protein_coding 1.215417899 4
11898 PAPSS2 protein_coding -2.916812770 4
6650 FRA10AC1 protein_coding -2.261837100 5
7173 GOLGA7B protein_coding -1.895166814 4
8536 BORCS7 protein_coding 1.464598160 4
2543 NSMCE4A protein_coding 3.456734448 4
6866 DPYSL4 protein_coding 0.304520368 5
9420 PWWP2B protein_coding 0.066112686 4
6009 PGGHG protein_coding 0.675771275 4
10901 IFITM2 protein_coding -0.410119901 6
12411 TSPAN4 protein_coding 0.941077857 4
13526 AP006621.5 lincRNA -2.255614909 4
14141 RP11-326C3.15 lincRNA 0.748207679 4
2823 C11orf21 protein_coding -0.550613719 4
3829 TRIM6 protein_coding -2.456361327 4
4945 MICAL2 protein_coding 0.504432935 4
2808 KIAA1549L protein_coding -0.335820831 4
3857 CAT protein_coding -0.409668221 4
6670 APIP protein_coding -0.618278593 4
6834 EXT2 protein_coding -1.459667131 5
7775 TPCN2 protein_coding -1.237753184 4
9895 MRPL48 protein_coding 1.190168138 4
7439 GDPD5 protein_coding -1.147220793 4
13261 RAB30-AS1 lincRNA 0.929715236 4
4035 RAB38 protein_coding 1.073954861 4
2794 PANX1 protein_coding 1.243733119 4
6690 ZC3H12C protein_coding -0.176249865 4
5482 PPP2R1B protein_coding 2.737449710 4
6691 TTC12 protein_coding -1.696940513 4
10385 C11orf71 protein_coding 1.293467398 4
10048 DSCAML1 protein_coding -0.950975759 5
2774 PUS3 protein_coding -0.580011271 4
3771 SNX19 protein_coding 0.419111598 4
13536 RP11-890B15.3 lincRNA 0.501667838 4
2866 WNT5B protein_coding -1.012958275 5
13577 RP1-102E24.8 lincRNA 1.794614984 5
13765 RP13-735L24.1 lincRNA 0.161045988 6
13543 RP11-377D9.3 lincRNA -0.583178672 4
162 MGST1 protein_coding -0.837298544 4
474 LMO3 protein_coding 0.957135074 4
14514 RP11-582E3.6 lincRNA 2.303870106 4
2846 CAPRIN2 protein_coding 0.001507103 4
12982 LINC00941 lincRNA 1.284727799 4
4351 YEATS4 protein_coding 0.575990296 4
4348 RAB3IP protein_coding -0.077277878 4
5243 USP44 protein_coding -0.506369197 4
5718 TDG protein_coding -0.288433659 4
12178 C12orf73 protein_coding -1.553859140 4
14626 RP11-575F12.3 lincRNA -0.186786558 5
10866 NOC4L protein_coding 0.594594702 4
10897 DDX51 protein_coding -0.618143356 4
10970 EP400NL protein_coding 0.240202648 5
13585 RP13-977J11.2 lincRNA 0.518461113 4
10413 ZDHHC20 protein_coding -0.772215132 4
1963 SGCG protein_coding 0.109019768 4
10316 LACC1 protein_coding 0.037531417 4
5268 RCBTB2 protein_coding -1.270569149 4
5256 TBC1D4 protein_coding 0.810943103 4
4166 CLYBL protein_coding 1.176324978 4
10909 KLHL33 protein_coding 1.078266890 5
7334 DHRS4 protein_coding 0.259683625 4
7340 DHRS1 protein_coding -2.143355013 5
4318 L3HYPDH protein_coding 0.288050382 4
5780 RAB15 protein_coding 0.006329755 4
3677 DCAF4 protein_coding 1.480577350 4
3688 ACOT2 protein_coding -1.206396425 4
5786 PTGR2 protein_coding -0.916339410 5
10796 ACOT1 protein_coding 0.658176457 4
180 POMT2 protein_coding 0.463089864 6
639 ADCK1 protein_coding -0.780906896 4
546 GALC protein_coding 0.821531163 4
265 CCDC88C protein_coding 0.123727401 4
13650 RP11-1070N10.3 lincRNA 1.203939178 4
10872 SIVA1 protein_coding 0.860653750 4
10533 NOP10 protein_coding -0.909657931 4
12482 GOLGA8B protein_coding 0.420913375 4
4466 EIF2AK4 protein_coding 0.010835172 5
12768 PATL2 protein_coding -0.380215578 4
5612 SPPL2A protein_coding 0.081266251 4
8490 LIPC protein_coding 2.985257644 4
5503 PAQR5 protein_coding 0.904598585 4
2094 CTSH protein_coding 3.428984562 5
10526 IDH2 protein_coding 0.773158504 4
5833 ADAMTS17 protein_coding -2.063478773 4
2021 NPRL3 protein_coding 1.293802591 4
1798 TELO2 protein_coding 0.184463116 5
2061 CPPED1 protein_coding 0.487653845 4
10689 NPIPA1 protein_coding -1.566346740 4
10749 NPIPA5 protein_coding 1.869812585 4
10704 ACSM5 protein_coding -0.525574950 4
9557 FAM192A protein_coding 0.810277775 4
13817 RP11-252A24.3 lincRNA 0.202015770 4
691 ADAT1 protein_coding -2.271438630 5
10827 ZFP1 protein_coding 2.045831428 4
2018 MON1B protein_coding 0.330838688 4
5882 MBTPS1 protein_coding -0.197883832 5
13826 LINC02188 lincRNA -0.211254377 4
7450 RNF166 protein_coding -0.042074860 4
25 DBNDD1 protein_coding -1.819290996 6
4697 CHMP1A protein_coding -1.084918016 4
10449 RPH3AL protein_coding -0.641162542 4
11665 SHPK protein_coding 0.361813715 5
10622 VMO1 protein_coding 3.133140328 4
2621 CAMTA2 protein_coding -0.115870824 4
4499 USP6 protein_coding -0.542041500 5
2690 MYH3 protein_coding -0.937795880 4
4325 RHOT1 protein_coding -0.381424711 4
4768 CCT6B protein_coding 0.169171156 4
8617 SLFN5 protein_coding -0.212653548 4
9543 SLFN11 protein_coding 0.013621326 4
2598 CASC3 protein_coding -3.317317897 4
9689 JUP protein_coding 0.493661739 4
7716 NAGS protein_coding 1.240308747 4
10233 EFCAB13 protein_coding 7.932542707 5
5919 SCRN2 protein_coding 3.003800245 4
8529 COX11 protein_coding -1.714290715 4
13895 RP11-166P13.3 lincRNA -0.327061836 4
13973 MYO15B protein_coding 1.655225290 4
8828 MGAT5B protein_coding 1.631808474 4
10585 MXRA7 protein_coding -0.148266235 4
5952 CCDC40 protein_coding -1.858015176 5
10599 TSPAN10 protein_coding -0.338720883 4
13891 RP11-1055B8.4 lincRNA -0.125751437 4
5970 WDR45B protein_coding 0.650825359 4
1936 LPIN2 protein_coding 0.434124143 5
13913 LINC00667 lincRNA -0.420138960 4
5932 IMPA2 protein_coding -0.859576537 4
977 MOCOS protein_coding 0.630254421 5
8705 KATNAL2 protein_coding -0.822022145 4
4909 CNDP2 protein_coding -0.724962181 4
8598 GALR1 protein_coding 0.276804307 6
2316 MIER2 protein_coding 0.446946951 4
4553 SHC2 protein_coding -0.381734995 5
684 WDR18 protein_coding 1.358541653 5
1593 POLR2E protein_coding -1.958967915 4
4580 ATP8B3 protein_coding -2.743966686 5
11723 CFD protein_coding -0.458322217 5
12702 SCAMP4 protein_coding 0.386606929 4
4220 GPR108 protein_coding 0.153885660 4
10448 TRAPPC5 protein_coding 2.208169538 4
13217 P2RY11 protein_coding -1.099360760 4
13989 ILF3-AS1 lincRNA -1.680045893 4
8736 RAB8A protein_coding -2.225357558 4
2327 JAK3 protein_coding -0.270437783 4
10335 B3GNT3 protein_coding 2.066918160 5
659 SUGP2 protein_coding -4.759323306 4
2345 LSR protein_coding -0.999367319 4
7697 COX7A1 protein_coding -0.337571534 4
3560 CD3EAP protein_coding 0.055655825 4
2308 SIGLEC5 protein_coding -3.600826625 4
13483 SIGLEC14 protein_coding -0.617235528 6
7612 ZNF761 protein_coding -0.807165609 5
8806 ZNF83 protein_coding 0.100355638 8
9296 ZNF160 protein_coding 0.420306051 4
9297 ZNF415 protein_coding 1.020478511 6
11497 ZNF765 protein_coding 0.152206591 4
11996 ZNF525 protein_coding 0.288254365 5
302 NLRP2 protein_coding 0.730400034 4
1189 ZNF586 protein_coding -1.616124312 4
4747 ZNF304 protein_coding -1.089943431 4
11850 ZNF587 protein_coding -0.220575054 4
11510 C20orf96 protein_coding 1.339014986 4
4255 TMEM74B protein_coding -0.407500940 4
1344 CPXM1 protein_coding 0.327553229 4
4254 MCM8 protein_coding 0.276397649 4
11696 ENTPD6 protein_coding -1.936991180 4
6729 DUSP15 protein_coding -1.122158603 4
13751 RP5-1085F17.3 lincRNA -1.259355687 4
4056 SDC4 protein_coding -0.469609700 4
4084 NEURL2 protein_coding -5.148346881 4
6728 JPH2 protein_coding -0.284296504 4
11675 SLC2A10 protein_coding -0.619692208 5
12581 SMIM25 lincRNA -0.314600486 4
6740 CABLES2 protein_coding 1.324157184 4
1869 TCFL5 protein_coding -0.081184724 4
4200 PPDPF protein_coding -0.313736239 4
6012 IFNAR1 protein_coding -3.047194176 4
7479 IFNAR2 protein_coding 0.292538013 4
10932 SH3BGR protein_coding -3.116571305 4
7355 C2CD2 protein_coding -1.584302063 4
10696 MX2 protein_coding -1.092447263 4
7580 CBS protein_coding -1.321471322 6
6015 SIK1 protein_coding 0.243564545 5
10847 UBE2G2 protein_coding -0.224409046 5
14416 LL21NC02-21A1.1 lincRNA 0.422426912 4
10570 YBEY protein_coding 2.388955903 5
14366 AC006946.16 lincRNA -0.784405312 4
1612 SMARCB1 protein_coding 1.728548932 4
1635 SLC35E4 protein_coding 1.402168297 4
10920 TCN2 protein_coding 1.249283592 4
1667 SLC5A4 protein_coding -0.509124597 4
1708 RPL3 protein_coding 0.568767399 4
1669 CYP2D6 protein_coding 0.270522473 5
11989 NAGA protein_coding -1.011002512 4
10705 SERHL2 protein_coding 0.879677859 4
11408 RRP7A protein_coding -0.450122744 4
1497 NUP50 protein_coding -2.090819345 4
1729 FAM118A protein_coding -2.176490239 4
12659 NUP50-AS1 lincRNA -0.791556719 5
13130 ARHGAP8 protein_coding -0.778879342 4
169 MAPK8IP2 protein_coding 0.657549635 4
1701 ARSA protein_coding 0.285684735 5
521 AKR7A2 protein_coding 1.952591686 4
4362 EMC1 protein_coding 1.088366518 4
9138 OTUD3 protein_coding -1.172063931 4
10806 MROH7 protein_coding 3.976137988 4
7879 ACP6 protein_coding 4.613975854 4
9028 HSPBAP1 protein_coding 0.319028943 5
2472 POLD2 protein_coding 1.780745843 4
2417 CCL24 protein_coding 1.961790317 4
11287 ZP3 protein_coding -0.007520099 4
5352 SLC2A8 protein_coding 1.697087549 4
2558 SFXN3 protein_coding 1.830707947 4
5563 BTBD16 protein_coding -0.181379950 6
2871 PARP11 protein_coding -3.090457641 4
4562 CRACR2A protein_coding 1.547976694 4
3647 CCDC92 protein_coding -5.343007339 5
5757 MORN3 protein_coding 0.002443962 4
4278 MCF2L protein_coding -2.343386423 5
14576 RP11-88E10.4 lincRNA -0.242539766 5
7037 ADPRHL1 protein_coding 1.293374116 4
12926 PINLYP protein_coding 2.819929485 4
2302 CCDC114 protein_coding -1.802143687 4
2307 PLA2G4C protein_coding -1.382373754 4
10447 FKRP protein_coding 4.425173545 5
2186 NUCB1 protein_coding -1.455312765 4
4615 TRPM4 protein_coding -2.034168341 4
7712 ALDH16A1 protein_coding -3.466783896 4
1639 DGCR14 protein_coding 2.127548693 4
10823 TXNRD2 protein_coding 2.835183327 4#distribution of number of eQTL for genes with PIP>0.8
table(ctwas_gene_res$num_eqtl[ctwas_gene_res$susie_pip>0.8])/sum(ctwas_gene_res$susie_pip>0.8)
1 2 3 4 5
0.44444444 0.41666667 0.05555556 0.02777778 0.05555556 #genes with 2+ eQTL and PIP>0.8
ctwas_gene_res[ctwas_gene_res$num_eqtl>1 & ctwas_gene_res$susie_pip>0.8,] chrom id pos type region_tag1 region_tag2
7879 1 ENSG00000162836.11 147646379 gene 1 73
5600 2 ENSG00000138439.11 202630335 gene 2 120
8904 3 ENSG00000168291.12 58433123 gene 3 40
13258 5 ENSG00000245937.7 128070567 gene 5 78
7441 7 ENSG00000158604.14 44582687 gene 7 32
4395 7 ENSG00000127948.15 75894921 gene 7 48
1279 7 ENSG00000087087.18 100874758 gene 7 62
1003 8 ENSG00000076554.15 80171625 gene 8 57
1237 8 ENSG00000085719.12 86514210 gene 8 62
7623 9 ENSG00000160447.6 128703005 gene 9 66
1526 10 ENSG00000095485.16 100267650 gene 10 64
3633 12 ENSG00000118971.7 4209750 gene 12 4
3647 12 ENSG00000119242.8 123959862 gene 12 75
7045 16 ENSG00000153786.12 85001918 gene 16 49
5923 17 ENSG00000141338.13 68883786 gene 17 39
3674 18 ENSG00000119537.15 63366845 gene 18 35
161 19 ENSG00000008382.15 4342850 gene 19 5
6023 19 ENSG00000142233.11 48702748 gene 19 33
10447 19 ENSG00000181027.10 46745328 gene 19 33
2199 19 ENSG00000104870.12 49506216 gene 19 34
cs_index susie_pip mu2 region_tag PVE genename
7879 1 0.9932775 21.77810 1_73 6.295220e-05 ACP6
5600 1 0.9695151 38.09714 2_120 1.074898e-04 FAM117B
8904 0 0.9140947 24.02176 3_40 6.390228e-05 PDHB
13258 0 0.8593862 18.23008 5_78 4.559291e-05 LINC01184
7441 2 0.9191981 36.87829 7_32 9.865071e-05 TMED4
4395 1 0.9843314 32.69729 7_48 9.366414e-05 POR
1279 2 0.9450584 25.66844 7_62 7.059571e-05 SRRT
1003 1 0.9370510 18.68606 8_57 5.095669e-05 TPD52
1237 0 0.8147843 18.04139 8_62 4.277922e-05 CPNE3
7623 1 0.9854511 42.73347 9_66 1.225529e-04 PKN3
1526 1 0.9256794 28.62700 10_64 7.711817e-05 CWF19L1
3633 1 0.9318759 19.26549 12_4 5.224666e-05 CCND2
3647 1 0.9604840 26.34034 12_75 7.362610e-05 CCDC92
7045 1 0.9967008 24.23491 16_49 7.029534e-05 ZDHHC7
5923 3 0.9982625 28.49746 17_39 8.278872e-05 ABCA8
3674 1 0.9199769 18.92509 18_35 5.066818e-05 KDSR
161 0 0.9197113 19.83813 19_5 5.309731e-05 MPND
6023 4 0.8492136 47.56594 19_33 1.175529e-04 NTN5
10447 3 0.9482623 20.61788 19_33 5.689744e-05 FKRP
2199 2 0.9967913 10378.34712 19_34 3.010598e-02 FCGRT
gene_type z num_eqtl
7879 protein_coding 4.613976 4
5600 protein_coding 7.877231 2
8904 protein_coding 3.361078 2
13258 lincRNA -3.918127 2
7441 protein_coding 7.688274 2
4395 protein_coding 6.026895 2
1279 protein_coding 4.744831 2
1003 protein_coding -4.121885 2
1237 protein_coding 3.753687 3
7623 protein_coding -6.885356 2
1526 protein_coding 5.813958 2
3633 protein_coding -4.128258 2
3647 protein_coding -5.343007 5
7045 protein_coding -4.891663 3
5923 protein_coding 5.086840 2
3674 protein_coding -4.119957 2
161 protein_coding -4.322018 2
6023 protein_coding 10.033010 2
10447 protein_coding 4.425174 5
2199 protein_coding -4.165895 2cTWAS genes in GO terms enriched for silver standard genes
#reload silver standard genes
known_annotations <- read_xlsx("data/summary_known_genes_annotations.xlsx", sheet="LDL")New names:
* `` -> ...4
* `` -> ...5known_annotations <- unique(known_annotations$`Gene Symbol`)
#GO enrichment analysis for silver standard genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
genes <- known_annotations
GO_enrichment <- enrichr(genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
print(db)
df <- GO_enrichment[[db]]
df <- df[df$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
plotEnrich(GO_enrichment[[db]])
print(df)
}[1] "GO_Biological_Process_2021"
Term
1 cholesterol transport (GO:0030301)
2 cholesterol homeostasis (GO:0042632)
3 sterol homeostasis (GO:0055092)
4 cholesterol efflux (GO:0033344)
5 sterol transport (GO:0015918)
6 cholesterol metabolic process (GO:0008203)
7 sterol metabolic process (GO:0016125)
8 triglyceride-rich lipoprotein particle remodeling (GO:0034370)
9 high-density lipoprotein particle remodeling (GO:0034375)
10 reverse cholesterol transport (GO:0043691)
11 secondary alcohol metabolic process (GO:1902652)
12 regulation of lipoprotein lipase activity (GO:0051004)
13 phospholipid transport (GO:0015914)
14 lipid transport (GO:0006869)
15 acylglycerol homeostasis (GO:0055090)
16 very-low-density lipoprotein particle remodeling (GO:0034372)
17 triglyceride homeostasis (GO:0070328)
18 triglyceride metabolic process (GO:0006641)
19 phospholipid efflux (GO:0033700)
20 chylomicron remodeling (GO:0034371)
21 regulation of cholesterol transport (GO:0032374)
22 chylomicron assembly (GO:0034378)
23 chylomicron remnant clearance (GO:0034382)
24 lipoprotein metabolic process (GO:0042157)
25 diterpenoid metabolic process (GO:0016101)
26 positive regulation of steroid metabolic process (GO:0045940)
27 retinoid metabolic process (GO:0001523)
28 lipid homeostasis (GO:0055088)
29 negative regulation of lipase activity (GO:0060192)
30 positive regulation of cholesterol esterification (GO:0010873)
31 intestinal cholesterol absorption (GO:0030299)
32 negative regulation of cholesterol transport (GO:0032375)
33 intestinal lipid absorption (GO:0098856)
34 acylglycerol metabolic process (GO:0006639)
35 regulation of cholesterol esterification (GO:0010872)
36 phospholipid homeostasis (GO:0055091)
37 very-low-density lipoprotein particle assembly (GO:0034379)
38 positive regulation of lipid metabolic process (GO:0045834)
39 high-density lipoprotein particle assembly (GO:0034380)
40 negative regulation of lipoprotein lipase activity (GO:0051005)
41 negative regulation of lipoprotein particle clearance (GO:0010985)
42 regulation of very-low-density lipoprotein particle remodeling (GO:0010901)
43 intracellular cholesterol transport (GO:0032367)
44 positive regulation of cholesterol transport (GO:0032376)
45 regulation of intestinal cholesterol absorption (GO:0030300)
46 positive regulation of lipoprotein lipase activity (GO:0051006)
47 acylglycerol catabolic process (GO:0046464)
48 positive regulation of lipid biosynthetic process (GO:0046889)
49 positive regulation of triglyceride metabolic process (GO:0090208)
50 positive regulation of triglyceride lipase activity (GO:0061365)
51 regulation of lipid catabolic process (GO:0050994)
52 steroid metabolic process (GO:0008202)
53 positive regulation of lipid catabolic process (GO:0050996)
54 triglyceride catabolic process (GO:0019433)
55 organophosphate ester transport (GO:0015748)
56 phosphatidylcholine metabolic process (GO:0046470)
57 regulation of triglyceride catabolic process (GO:0010896)
58 fatty acid metabolic process (GO:0006631)
59 regulation of fatty acid biosynthetic process (GO:0042304)
60 regulation of sterol transport (GO:0032371)
61 cellular response to low-density lipoprotein particle stimulus (GO:0071404)
62 low-density lipoprotein particle remodeling (GO:0034374)
63 regulation of macrophage derived foam cell differentiation (GO:0010743)
64 organic substance transport (GO:0071702)
65 regulation of cholesterol efflux (GO:0010874)
66 receptor-mediated endocytosis (GO:0006898)
67 secondary alcohol biosynthetic process (GO:1902653)
68 regulation of cholesterol storage (GO:0010885)
69 cholesterol import (GO:0070508)
70 sterol import (GO:0035376)
71 monocarboxylic acid biosynthetic process (GO:0072330)
72 cholesterol biosynthetic process (GO:0006695)
73 positive regulation of cellular metabolic process (GO:0031325)
74 sterol biosynthetic process (GO:0016126)
75 positive regulation of fatty acid metabolic process (GO:0045923)
76 regulation of receptor-mediated endocytosis (GO:0048259)
77 fatty acid biosynthetic process (GO:0006633)
78 regulation of Cdc42 protein signal transduction (GO:0032489)
79 positive regulation of triglyceride catabolic process (GO:0010898)
80 lipid biosynthetic process (GO:0008610)
81 positive regulation of cholesterol efflux (GO:0010875)
82 negative regulation of receptor-mediated endocytosis (GO:0048261)
83 lipoprotein transport (GO:0042953)
84 regulation of lipid metabolic process (GO:0019216)
85 monocarboxylic acid metabolic process (GO:0032787)
86 lipoprotein localization (GO:0044872)
87 lipid catabolic process (GO:0016042)
88 positive regulation of fatty acid biosynthetic process (GO:0045723)
89 intracellular sterol transport (GO:0032366)
90 steroid biosynthetic process (GO:0006694)
91 regulation of lipid biosynthetic process (GO:0046890)
92 regulation of lipase activity (GO:0060191)
93 positive regulation of cellular biosynthetic process (GO:0031328)
94 regulation of amyloid-beta clearance (GO:1900221)
95 phospholipid metabolic process (GO:0006644)
96 regulation of intestinal lipid absorption (GO:1904729)
97 positive regulation of protein catabolic process in the vacuole (GO:1904352)
98 positive regulation of biosynthetic process (GO:0009891)
99 regulation of cholesterol metabolic process (GO:0090181)
100 foam cell differentiation (GO:0090077)
101 positive regulation of cholesterol storage (GO:0010886)
102 macrophage derived foam cell differentiation (GO:0010742)
103 organic hydroxy compound biosynthetic process (GO:1901617)
104 regulation of steroid metabolic process (GO:0019218)
105 organonitrogen compound biosynthetic process (GO:1901566)
106 negative regulation of lipid metabolic process (GO:0045833)
107 regulation of lysosomal protein catabolic process (GO:1905165)
108 positive regulation of amyloid-beta clearance (GO:1900223)
109 cellular lipid catabolic process (GO:0044242)
110 chemical homeostasis (GO:0048878)
111 cholesterol catabolic process (GO:0006707)
112 sterol catabolic process (GO:0016127)
113 steroid hormone biosynthetic process (GO:0120178)
114 regulation of low-density lipoprotein particle clearance (GO:0010988)
115 bile acid metabolic process (GO:0008206)
116 phosphatidylcholine biosynthetic process (GO:0006656)
117 alcohol catabolic process (GO:0046164)
118 organophosphate catabolic process (GO:0046434)
119 regulation of phospholipase activity (GO:0010517)
120 positive regulation of lipid localization (GO:1905954)
121 positive regulation of phospholipid transport (GO:2001140)
122 glycerophospholipid metabolic process (GO:0006650)
123 organic hydroxy compound transport (GO:0015850)
124 negative regulation of macrophage derived foam cell differentiation (GO:0010745)
125 positive regulation of lipid transport (GO:0032370)
126 C21-steroid hormone biosynthetic process (GO:0006700)
127 membrane organization (GO:0061024)
128 positive regulation of endocytosis (GO:0045807)
129 positive regulation of multicellular organismal process (GO:0051240)
130 negative regulation of catabolic process (GO:0009895)
131 negative regulation of lipid catabolic process (GO:0050995)
132 carbohydrate derivative transport (GO:1901264)
133 positive regulation of macrophage derived foam cell differentiation (GO:0010744)
134 protein transport (GO:0015031)
135 fatty acid transport (GO:0015908)
136 positive regulation of lipid storage (GO:0010884)
137 C21-steroid hormone metabolic process (GO:0008207)
138 phospholipid catabolic process (GO:0009395)
139 negative regulation of endocytosis (GO:0045806)
140 regulation of primary metabolic process (GO:0080090)
141 negative regulation of multicellular organismal process (GO:0051241)
142 bile acid biosynthetic process (GO:0006699)
143 regulation of low-density lipoprotein particle receptor catabolic process (GO:0032803)
144 regulation of Rho protein signal transduction (GO:0035023)
145 regulation of small molecule metabolic process (GO:0062012)
146 positive regulation of cellular catabolic process (GO:0031331)
147 artery morphogenesis (GO:0048844)
148 negative regulation of cellular component organization (GO:0051129)
149 glycolipid transport (GO:0046836)
150 positive regulation of lipoprotein particle clearance (GO:0010986)
151 positive regulation of sterol transport (GO:0032373)
152 long-chain fatty acid transport (GO:0015909)
153 response to insulin (GO:0032868)
154 regulation of bile acid metabolic process (GO:1904251)
155 positive regulation of receptor catabolic process (GO:2000646)
156 positive regulation of transport (GO:0051050)
157 negative regulation of endothelial cell proliferation (GO:0001937)
158 negative regulation of endothelial cell migration (GO:0010596)
159 regulation of nitrogen compound metabolic process (GO:0051171)
160 negative regulation of amyloid-beta clearance (GO:1900222)
161 negative regulation of cellular metabolic process (GO:0031324)
162 glycerophospholipid biosynthetic process (GO:0046474)
163 negative regulation of production of molecular mediator of immune response (GO:0002701)
164 unsaturated fatty acid biosynthetic process (GO:0006636)
165 anion transport (GO:0006820)
166 positive regulation of receptor-mediated endocytosis (GO:0048260)
167 negative regulation of cholesterol storage (GO:0010887)
168 regulation of bile acid biosynthetic process (GO:0070857)
169 peptidyl-amino acid modification (GO:0018193)
170 low-density lipoprotein particle receptor catabolic process (GO:0032802)
171 low-density lipoprotein receptor particle metabolic process (GO:0032799)
172 protein oxidation (GO:0018158)
173 positive regulation by host of viral process (GO:0044794)
174 positive regulation of triglyceride biosynthetic process (GO:0010867)
175 receptor internalization (GO:0031623)
176 response to glucose (GO:0009749)
177 positive regulation of cellular component organization (GO:0051130)
178 negative regulation of fatty acid biosynthetic process (GO:0045717)
179 negative regulation of hemostasis (GO:1900047)
180 peptidyl-methionine modification (GO:0018206)
181 ethanol oxidation (GO:0006069)
182 negative regulation of amyloid fibril formation (GO:1905907)
183 negative regulation of protein metabolic process (GO:0051248)
184 unsaturated fatty acid metabolic process (GO:0033559)
185 alpha-linolenic acid metabolic process (GO:0036109)
186 platelet degranulation (GO:0002576)
187 negative regulation of metabolic process (GO:0009892)
188 negative regulation of cell activation (GO:0050866)
189 negative regulation of coagulation (GO:0050819)
190 cGMP-mediated signaling (GO:0019934)
191 intestinal absorption (GO:0050892)
192 receptor metabolic process (GO:0043112)
193 regulation of phagocytosis (GO:0050764)
194 regulation of amyloid fibril formation (GO:1905906)
195 regulation of sequestering of triglyceride (GO:0010889)
196 regulation of triglyceride biosynthetic process (GO:0010866)
197 post-translational protein modification (GO:0043687)
198 regulation of endocytosis (GO:0030100)
199 amyloid fibril formation (GO:1990000)
200 positive regulation of small molecule metabolic process (GO:0062013)
201 cellular response to nutrient levels (GO:0031669)
202 negative regulation of cytokine production involved in immune response (GO:0002719)
203 negative regulation of fatty acid metabolic process (GO:0045922)
204 regulation of cholesterol biosynthetic process (GO:0045540)
205 regulation of steroid biosynthetic process (GO:0050810)
206 positive regulation of catabolic process (GO:0009896)
207 amyloid precursor protein metabolic process (GO:0042982)
208 nitric oxide mediated signal transduction (GO:0007263)
209 positive regulation of nitric-oxide synthase activity (GO:0051000)
210 ethanol metabolic process (GO:0006067)
211 positive regulation of cellular protein catabolic process (GO:1903364)
212 response to fatty acid (GO:0070542)
213 long-term memory (GO:0007616)
214 negative regulation of lipid storage (GO:0010888)
215 linoleic acid metabolic process (GO:0043651)
216 negative regulation of lipid biosynthetic process (GO:0051055)
217 regulation of lipid storage (GO:0010883)
218 regulation of interleukin-1 beta production (GO:0032651)
219 long-chain fatty acid metabolic process (GO:0001676)
220 regulation of cytokine production involved in immune response (GO:0002718)
221 cellular protein metabolic process (GO:0044267)
222 negative regulation of defense response (GO:0031348)
223 transport across blood-brain barrier (GO:0150104)
224 receptor catabolic process (GO:0032801)
225 response to hexose (GO:0009746)
226 regulated exocytosis (GO:0045055)
227 regulation of endothelial cell migration (GO:0010594)
228 negative regulation of protein transport (GO:0051224)
229 positive regulation of binding (GO:0051099)
230 regulation of blood coagulation (GO:0030193)
231 positive regulation of monooxygenase activity (GO:0032770)
232 negative regulation of wound healing (GO:0061045)
233 negative regulation of macromolecule metabolic process (GO:0010605)
234 long-chain fatty acid biosynthetic process (GO:0042759)
235 regulation of developmental growth (GO:0048638)
236 regulation of cell death (GO:0010941)
237 regulation of angiogenesis (GO:0045765)
238 regulation of inflammatory response (GO:0050727)
239 apoptotic cell clearance (GO:0043277)
240 cellular response to peptide hormone stimulus (GO:0071375)
241 negative regulation of blood vessel endothelial cell migration (GO:0043537)
242 phosphate-containing compound metabolic process (GO:0006796)
243 negative regulation of epithelial cell migration (GO:0010633)
244 cellular response to amyloid-beta (GO:1904646)
245 cyclic-nucleotide-mediated signaling (GO:0019935)
246 regulation of receptor internalization (GO:0002090)
247 response to lipid (GO:0033993)
248 regulation of vascular associated smooth muscle cell proliferation (GO:1904705)
249 regulation of protein-containing complex assembly (GO:0043254)
250 ion transport (GO:0006811)
251 negative regulation of response to external stimulus (GO:0032102)
252 regulation of protein binding (GO:0043393)
253 regulation of cellular component biogenesis (GO:0044087)
254 negative regulation of protein secretion (GO:0050709)
255 negative regulation of secretion by cell (GO:1903531)
256 regulation of nitric-oxide synthase activity (GO:0050999)
257 negative regulation of blood coagulation (GO:0030195)
258 cellular response to organic substance (GO:0071310)
259 cellular response to insulin stimulus (GO:0032869)
260 response to amyloid-beta (GO:1904645)
261 establishment of protein localization to extracellular region (GO:0035592)
262 regulation of cellular metabolic process (GO:0031323)
263 regulation of protein metabolic process (GO:0051246)
264 positive regulation of cell differentiation (GO:0045597)
265 negative regulation of cell projection organization (GO:0031345)
266 positive regulation of fat cell differentiation (GO:0045600)
267 negative regulation of BMP signaling pathway (GO:0030514)
268 negative regulation of cellular biosynthetic process (GO:0031327)
269 positive regulation of phagocytosis (GO:0050766)
Overlap Adjusted.P.value
1 28/51 3.291336e-55
2 29/71 1.134840e-52
3 29/72 1.264418e-52
4 16/24 1.003687e-32
5 15/21 2.203564e-31
6 20/77 5.273224e-31
7 19/70 7.882518e-30
8 12/13 1.520527e-27
9 13/18 2.514128e-27
10 12/17 5.731565e-25
11 15/49 2.711706e-24
12 12/21 2.245426e-23
13 15/59 5.665269e-23
14 17/109 2.748742e-22
15 12/25 3.145271e-22
16 9/9 2.283165e-21
17 12/31 7.414146e-21
18 13/55 1.935295e-19
19 9/12 4.196729e-19
20 8/9 5.374944e-18
21 10/25 1.639871e-17
22 8/10 2.436689e-17
23 7/7 1.679428e-16
24 7/9 5.763376e-15
25 11/64 8.362646e-15
26 7/13 2.508790e-13
27 11/92 5.133855e-13
28 10/64 5.133855e-13
29 6/9 3.296018e-12
30 6/9 3.296018e-12
31 6/9 3.296018e-12
32 6/11 1.693836e-11
33 6/11 1.693836e-11
34 8/41 3.013332e-11
35 6/12 3.013332e-11
36 6/12 3.013332e-11
37 6/12 3.013332e-11
38 7/25 4.654994e-11
39 6/13 5.294950e-11
40 5/6 5.459029e-11
41 5/6 5.459029e-11
42 5/6 5.459029e-11
43 6/15 1.393181e-10
44 7/33 3.496187e-10
45 5/8 4.627510e-10
46 5/8 4.627510e-10
47 7/35 5.017364e-10
48 7/35 5.017364e-10
49 6/19 6.556580e-10
50 5/9 9.553575e-10
51 6/21 1.253116e-09
52 9/104 1.493946e-09
53 6/22 1.653549e-09
54 6/23 2.189811e-09
55 6/25 3.733511e-09
56 8/77 3.733511e-09
57 5/12 5.225810e-09
58 9/124 6.552660e-09
59 6/29 9.279729e-09
60 4/5 9.798195e-09
61 5/14 1.207993e-08
62 5/14 1.207993e-08
63 6/31 1.339781e-08
64 9/136 1.355933e-08
65 6/33 1.942867e-08
66 9/143 2.054367e-08
67 6/34 2.282600e-08
68 5/16 2.390304e-08
69 4/6 2.513038e-08
70 4/6 2.513038e-08
71 7/63 2.556641e-08
72 6/35 2.556641e-08
73 8/105 3.517095e-08
74 6/38 4.196695e-08
75 5/18 4.228478e-08
76 6/39 4.816188e-08
77 7/71 5.609765e-08
78 4/8 1.033779e-07
79 4/8 1.033779e-07
80 7/80 1.258618e-07
81 5/23 1.517283e-07
82 5/26 2.906610e-07
83 4/10 2.936601e-07
84 7/92 3.199662e-07
85 8/143 3.471498e-07
86 4/11 4.442138e-07
87 5/29 4.906437e-07
88 4/13 9.252034e-07
89 4/13 9.252034e-07
90 6/65 9.598111e-07
91 5/35 1.249797e-06
92 4/14 1.249797e-06
93 8/180 1.884951e-06
94 4/16 2.212485e-06
95 6/76 2.336232e-06
96 3/5 3.664395e-06
97 3/5 3.664395e-06
98 5/44 3.827136e-06
99 4/21 6.819051e-06
100 3/6 6.952354e-06
101 3/6 6.952354e-06
102 3/6 6.952354e-06
103 5/50 6.991423e-06
104 4/23 9.554179e-06
105 7/158 1.040987e-05
106 4/24 1.121951e-05
107 3/7 1.146235e-05
108 3/7 1.146235e-05
109 4/27 1.788032e-05
110 5/65 2.452122e-05
111 3/9 2.639634e-05
112 3/9 2.639634e-05
113 4/31 3.060279e-05
114 3/10 3.695601e-05
115 4/33 3.856854e-05
116 4/33 3.856854e-05
117 3/11 4.775659e-05
118 3/11 4.775659e-05
119 3/11 4.775659e-05
120 3/11 4.775659e-05
121 3/11 4.775659e-05
122 5/80 6.182763e-05
123 4/40 7.973389e-05
124 3/13 7.973389e-05
125 3/13 7.973389e-05
126 3/15 1.252218e-04
127 7/242 1.420233e-04
128 4/48 1.598563e-04
129 8/345 1.704405e-04
130 4/49 1.709436e-04
131 3/18 2.111817e-04
132 3/18 2.111817e-04
133 3/18 2.111817e-04
134 8/369 2.646441e-04
135 3/20 2.892290e-04
136 3/21 3.341258e-04
137 3/24 4.974036e-04
138 3/24 4.974036e-04
139 3/25 5.597802e-04
140 5/130 5.616142e-04
141 6/214 6.166972e-04
142 3/27 6.934199e-04
143 2/5 7.406583e-04
144 4/73 7.449454e-04
145 3/28 7.586859e-04
146 5/141 7.898802e-04
147 3/30 9.228897e-04
148 4/80 1.034287e-03
149 2/6 1.049782e-03
150 2/6 1.049782e-03
151 2/6 1.049782e-03
152 3/32 1.085012e-03
153 4/84 1.208000e-03
154 2/7 1.428577e-03
155 2/7 1.428577e-03
156 4/91 1.611630e-03
157 3/37 1.625395e-03
158 3/38 1.749224e-03
159 2/8 1.841132e-03
160 2/8 1.841132e-03
161 3/39 1.855101e-03
162 5/177 2.046657e-03
163 2/9 2.304288e-03
164 2/9 2.304288e-03
165 3/43 2.420344e-03
166 3/44 2.575438e-03
167 2/10 2.756295e-03
168 2/10 2.756295e-03
169 2/10 2.756295e-03
170 2/10 2.756295e-03
171 2/10 2.756295e-03
172 2/11 3.303347e-03
173 2/11 3.303347e-03
174 2/11 3.303347e-03
175 3/49 3.337798e-03
176 3/49 3.337798e-03
177 4/114 3.341630e-03
178 2/12 3.781332e-03
179 2/12 3.781332e-03
180 2/12 3.781332e-03
181 2/12 3.781332e-03
182 2/12 3.781332e-03
183 3/52 3.822266e-03
184 3/54 4.245657e-03
185 2/13 4.386588e-03
186 4/125 4.489058e-03
187 3/56 4.645735e-03
188 2/14 4.945884e-03
189 2/14 4.945884e-03
190 2/14 4.945884e-03
191 2/14 4.945884e-03
192 3/58 4.985945e-03
193 3/58 4.985945e-03
194 2/15 5.548828e-03
195 2/15 5.548828e-03
196 2/15 5.548828e-03
197 6/345 5.596173e-03
198 3/61 5.626994e-03
199 3/63 6.147256e-03
200 2/16 6.200855e-03
201 3/66 6.840974e-03
202 2/17 6.840974e-03
203 2/17 6.840974e-03
204 2/17 6.840974e-03
205 2/17 6.840974e-03
206 3/67 7.093595e-03
207 2/18 7.532008e-03
208 2/18 7.532008e-03
209 2/18 7.532008e-03
210 2/19 8.241667e-03
211 2/19 8.241667e-03
212 2/19 8.241667e-03
213 2/19 8.241667e-03
214 2/20 9.094348e-03
215 2/21 9.982653e-03
216 2/22 1.085553e-02
217 2/22 1.085553e-02
218 3/83 1.230478e-02
219 3/83 1.230478e-02
220 2/24 1.273659e-02
221 6/417 1.291939e-02
222 3/85 1.298477e-02
223 3/86 1.336097e-02
224 2/25 1.350640e-02
225 2/25 1.350640e-02
226 4/180 1.399465e-02
227 3/89 1.440735e-02
228 2/26 1.440735e-02
229 3/90 1.479001e-02
230 2/27 1.539039e-02
231 2/28 1.646589e-02
232 2/29 1.757027e-02
233 4/194 1.771224e-02
234 2/30 1.862299e-02
235 2/31 1.977865e-02
236 3/102 2.036757e-02
237 4/203 2.042828e-02
238 4/206 2.141587e-02
239 2/33 2.198466e-02
240 3/106 2.228428e-02
241 2/34 2.311351e-02
242 4/212 2.328380e-02
243 2/35 2.415927e-02
244 2/35 2.415927e-02
245 2/36 2.531623e-02
246 2/36 2.531623e-02
247 3/114 2.646649e-02
248 2/37 2.648825e-02
249 3/116 2.742785e-02
250 3/116 2.742785e-02
251 3/118 2.842391e-02
252 3/118 2.842391e-02
253 2/39 2.842391e-02
254 2/39 2.842391e-02
255 2/39 2.842391e-02
256 2/39 2.842391e-02
257 2/40 2.973753e-02
258 3/123 3.119488e-02
259 3/129 3.533580e-02
260 2/44 3.533580e-02
261 2/46 3.834204e-02
262 2/47 3.980515e-02
263 2/48 4.128649e-02
264 4/258 4.183273e-02
265 2/49 4.262417e-02
266 2/51 4.583569e-02
267 2/52 4.720898e-02
268 2/52 4.720898e-02
269 2/53 4.877011e-02
Genes
1 SCARB1;CETP;LCAT;LIPC;NPC1L1;LIPG;CD36;APOE;LDLRAP1;APOB;LDLR;ABCA1;ABCG8;STARD3;ABCG5;OSBPL5;APOA2;APOA1;APOC3;APOA4;APOA5;NPC1;SOAT1;STAR;NPC2;SOAT2;APOC2;APOC1
2 SCARB1;CETP;MTTP;PCSK9;LPL;LCAT;ABCB11;CYP7A1;LIPC;LIPG;APOE;LDLRAP1;APOB;LDLR;ABCA1;ABCG8;ABCG5;EPHX2;APOA2;APOA1;APOC3;APOA4;APOA5;SOAT1;NPC1;NPC2;SOAT2;APOC2;ANGPTL3
3 SCARB1;CETP;MTTP;PCSK9;LPL;LCAT;ABCB11;CYP7A1;LIPC;LIPG;APOE;LDLRAP1;APOB;LDLR;ABCA1;ABCG8;ABCG5;EPHX2;APOA2;APOA1;APOC3;APOA4;APOA5;SOAT1;NPC1;NPC2;SOAT2;APOC2;ANGPTL3
4 ABCA1;ABCG8;SCARB1;ABCG5;APOA2;APOA1;APOC3;APOA4;APOA5;NPC1;SOAT1;NPC2;SOAT2;APOC2;APOC1;APOE
5 ABCG8;CETP;STARD3;ABCG5;OSBPL5;APOA2;APOA1;LCAT;NPC1;NPC1L1;NPC2;CD36;APOB;LDLRAP1;LDLR
6 ABCA1;STARD3;CETP;OSBPL5;APOA2;APOA1;LCAT;APOA4;HMGCR;APOA5;CYP7A1;CYP27A1;SOAT1;SOAT2;NPC1L1;ANGPTL3;APOE;DHCR7;LDLRAP1;APOB
7 ABCA1;STARD3;CETP;OSBPL5;APOA2;APOA1;LCAT;APOA4;HMGCR;LIPA;CYP7A1;CYP27A1;SOAT1;SOAT2;ANGPTL3;APOE;DHCR7;LDLRAP1;APOB
8 CETP;LIPC;APOC2;APOA2;APOA1;APOC3;LCAT;LPL;APOA4;APOE;APOB;APOA5
9 CETP;SCARB1;APOA2;APOA1;APOC3;LCAT;APOA4;LIPC;APOC2;APOC1;LIPG;APOE;PLTP
10 ABCA1;CETP;SCARB1;LIPC;APOC2;LIPG;APOA2;APOA1;APOC3;LCAT;APOA4;APOE
11 ABCA1;STARD3;CETP;OSBPL5;APOA2;APOA1;LCAT;APOA4;CYP27A1;SOAT1;SOAT2;ANGPTL3;APOE;LDLRAP1;APOB
12 LIPC;SORT1;APOC2;APOH;APOC1;ANGPTL3;APOA1;APOC3;LPL;APOA4;ANGPTL4;APOA5
13 ABCA1;SCARB1;OSBPL5;MTTP;APOA2;APOA1;APOC3;APOA4;APOA5;NPC2;APOC2;APOC1;APOE;LDLR;PLTP
14 ABCA1;SCARB1;ABCG8;CETP;ABCG5;OSBPL5;MTTP;APOA1;APOA4;ABCB11;APOA5;NPC2;NPC1L1;CD36;APOE;LDLR;PLTP
15 CETP;SCARB1;LIPC;APOC2;ANGPTL3;LPL;APOA1;APOC3;APOA4;APOE;ANGPTL4;APOA5
16 CETP;LIPC;APOC2;APOA1;LCAT;LPL;APOA4;APOE;APOA5
17 CETP;SCARB1;LIPC;APOC2;ANGPTL3;LPL;APOA1;APOC3;APOA4;APOE;ANGPTL4;APOA5
18 CETP;APOA2;LPL;APOC3;APOA5;LIPC;LIPI;APOH;LIPG;APOC1;APOE;APOB;LPIN3
19 ABCA1;APOC2;APOC1;APOA2;APOA1;APOC3;APOA4;APOE;APOA5
20 APOC2;APOA2;APOA1;APOC3;LPL;APOA4;APOE;APOB
21 CETP;LRP1;APOC2;LIPG;APOC1;APOA2;TSPO;APOA1;APOA4;APOA5
22 APOC2;MTTP;APOA2;APOA1;APOC3;APOA4;APOE;APOB
23 LIPC;APOC2;APOC1;APOC3;APOE;APOB;LDLR
24 NPC1L1;MTTP;APOA2;APOA1;APOA4;APOE;APOA5
25 LRP1;ADH1B;APOC2;APOA2;APOA1;LPL;APOC3;APOA4;LRP2;APOE;APOB
26 APOC1;APOA2;APOA1;APOA4;APOE;LDLRAP1;APOA5
27 LRP1;ADH1B;APOC2;APOA2;APOA1;LPL;APOC3;APOA4;LRP2;APOE;APOB
28 ABCA1;CETP;LIPG;ANGPTL3;APOA1;APOA4;PPARG;APOE;ABCB11;APOA5
29 SORT1;APOC1;ANGPTL3;APOA2;APOC3;ANGPTL4
30 APOC1;APOA2;APOA1;APOA4;APOE;APOA5
31 ABCG8;ABCG5;NPC1L1;SOAT2;CD36;LDLR
32 ABCG8;ABCG5;APOC2;APOC1;APOA2;APOC3
33 ABCG8;ABCG5;NPC1L1;SOAT2;CD36;LDLR
34 CETP;APOH;APOC1;APOA2;LPL;APOC3;APOE;APOA5
35 APOC1;APOA2;APOA1;APOA4;APOE;APOA5
36 ABCA1;CETP;LIPG;ANGPTL3;APOA1;ABCB11
37 SOAT1;SOAT2;APOC1;MTTP;APOC3;APOB
38 APOA2;ANGPTL3;APOA1;APOA4;PPARG;APOE;APOA5
39 ABCA1;APOA2;APOA1;APOA4;APOE;APOA5
40 SORT1;APOC1;ANGPTL3;APOC3;ANGPTL4
41 LRPAP1;APOC2;APOC1;APOC3;PCSK9
42 APOC2;APOA2;APOA1;APOC3;APOA5
43 ABCA1;NPC1;STAR;NPC2;LDLRAP1;LDLR
44 CETP;LRP1;LIPG;APOA1;PPARG;APOE;PLTP
45 ABCG8;ABCG5;APOA1;APOA4;APOA5
46 APOC2;APOH;APOA1;APOA4;APOA5
47 LIPC;LIPI;LIPG;APOA2;LPL;APOC3;APOA5
48 SCARB1;APOC2;APOA1;APOA4;APOE;LDLR;APOA5
49 SCARB1;APOC2;APOA1;APOA4;APOA5;LDLR
50 APOC2;APOH;APOA1;APOA4;APOA5
51 APOC1;APOA2;ANGPTL3;APOC3;ABCB11;APOA5
52 CYP27A1;STARD3;NPC1;STAR;TSPO;LRP2;ABCB11;LIPA;CYP7A1
53 APOC2;APOA2;ANGPTL3;APOA1;APOA4;APOA5
54 LIPC;LIPI;LIPG;APOC3;LPL;APOA5
55 SCARB1;OSBPL5;NPC2;MTTP;LDLR;PLTP
56 CETP;LIPC;APOA2;APOA1;LCAT;APOA4;APOA5;LPIN3
57 APOC2;APOA1;APOC3;APOA4;APOA5
58 LIPC;LIPI;LIPG;ANGPTL3;LPL;PPARG;CD36;ABCB11;LPIN3
59 APOC2;APOC1;APOA1;APOC3;APOA4;APOA5
60 LRP1;APOC1;TSPO;APOA4
61 ABCA1;LPL;PPARG;CD36;LDLR
62 CETP;LIPC;APOA2;APOB;LPA
63 ABCA1;CETP;LPL;PPARG;CD36;APOB
64 ABCA1;ABCG8;CETP;ABCG5;APOA1;APOA4;LRP2;APOA5;PLTP
65 CETP;LRP1;APOA1;PPARG;APOE;PLTP
66 SCARB1;LRP1;APOA1;CD36;LRP2;APOE;LDLRAP1;APOB;LDLR
67 NPC1L1;APOA1;APOA4;HMGCR;DHCR7;APOA5
68 ABCA1;SCARB1;LPL;PPARG;APOB
69 SCARB1;APOA1;CD36;LDLR
70 SCARB1;APOA1;CD36;LDLR
71 CYP27A1;LIPC;LIPI;LIPG;LPL;ABCB11;CYP7A1
72 NPC1L1;APOA1;APOA4;HMGCR;DHCR7;APOA5
73 APOC1;APOA2;PCSK9;APOA1;APOA4;PPARG;APOE;APOA5
74 NPC1L1;APOA1;APOA4;HMGCR;DHCR7;APOA5
75 APOC2;APOA1;APOA4;PPARG;APOA5
76 LRPAP1;APOC2;APOC1;APOC3;LDLRAP1;APOA5
77 FADS3;LIPC;LIPI;EPHX2;LIPG;LPL;FADS1
78 ABCA1;APOA1;APOC3;APOE
79 APOC2;APOA1;APOA4;APOA5
80 LIPC;STAR;LIPI;LIPG;LPL;HMGCR;FADS1
81 LRP1;APOA1;PPARG;APOE;PLTP
82 LRPAP1;APOC2;APOC1;PCSK9;APOC3
83 LRP1;PPARG;CD36;APOB
84 NPC2;APOC2;APOC1;APOC3;PPARG;HMGCR;DHCR7
85 NPC1;ADH1B;ANGPTL3;LPL;PPARG;VDAC1;CD36;ABCB11
86 LRP1;PPARG;CD36;APOB
87 LIPC;LIPI;LIPG;LPL;APOA4
88 APOC2;APOA1;APOA4;APOA5
89 ABCA1;NPC1;STAR;NPC2
90 CYP27A1;STAR;HMGCR;DHCR7;ABCB11;CYP7A1
91 STAR;APOA1;APOA4;APOE;APOA5
92 LIPC;APOA2;ANGPTL3;LPL
93 SCARB1;STAR;APOC2;APOA1;APOA4;CD36;APOA5;LDLR
94 LRPAP1;LRP1;HMGCR;APOE
95 LIPG;APOA2;ANGPTL3;LPL;LCAT;FADS1
96 APOA1;APOA4;APOA5
97 LRP1;LRP2;LDLR
98 APOA1;APOA4;APOE;CD36;APOA5
99 EPHX2;APOE;LDLRAP1;KPNB1
100 SOAT1;SOAT2;PPARG
101 SCARB1;LPL;APOB
102 SOAT1;SOAT2;PPARG
103 CYP27A1;HMGCR;DHCR7;ABCB11;CYP7A1
104 STAR;EPHX2;APOE;ABCB11
105 VAPA;VAPB;APOA2;APOA1;LCAT;APOE;LPIN3
106 APOC2;APOC1;APOA2;APOC3
107 LRP1;LRP2;LDLR
108 LRPAP1;LRP1;APOE
109 LIPG;APOA2;ANGPTL3;LPIN3
110 CETP;ANGPTL3;APOA4;PPARG;ABCB11
111 CYP27A1;APOE;CYP7A1
112 CYP27A1;APOE;CYP7A1
113 STARD3;STAR;TSPO;DHCR7
114 APOC3;PCSK9;LDLRAP1
115 CYP27A1;NPC1;ABCB11;CYP7A1
116 APOA2;LCAT;APOA1;LPIN3
117 CYP27A1;APOE;CYP7A1
118 LIPG;ANGPTL3;APOA2
119 LRP1;APOC2;ANGPTL3
120 LRP1;LPL;APOB
121 CETP;APOA1;APOE
122 CETP;APOA1;LCAT;APOA4;APOA5
123 ABCG8;ABCG5;NPC2;ABCB11
124 ABCA1;CETP;PPARG
125 CETP;LRP1;APOE
126 STARD3;STAR;TSPO
127 NPC1;VAPA;VAPB;LRP2;LDLRAP1;APOB;LDLR
128 LRP1;APOE;LDLRAP1;APOA5
129 GHR;ABCA1;LRPAP1;LRP1;APOC2;CD36;APOE;APOA5
130 APOC1;APOA2;APOC3;HMGCR
131 APOC1;APOA2;APOC3
132 SCARB1;NPC2;PLTP
133 LPL;CD36;APOB
134 ABCA1;LRP1;MTTP;PPARG;CD36;LRP2;APOE;APOB
135 PPARG;APOE;CD36
136 SCARB1;LPL;APOB
137 STARD3;STAR;TSPO
138 LIPG;APOA2;ANGPTL3
139 APOC2;APOC1;APOC3
140 PPARG;HMGCR;APOE;DHCR7;LDLR
141 LRPAP1;APOA2;APOA1;APOC3;APOA4;HMGCR
142 CYP27A1;ABCB11;CYP7A1
143 PCSK9;APOE
144 ABCA1;APOA1;APOC3;APOE
145 EPHX2;APOE;ABCB11
146 APOC2;APOA1;APOA4;APOE;APOA5
147 LRP1;ANGPTL3;LRP2
148 APOA2;APOA1;APOC3;APOA4
149 NPC2;PLTP
150 LIPG;LDLRAP1
151 CETP;LIPG
152 PPARG;APOE;CD36
153 SORT1;PCSK9;PPARG;LPIN3
154 ABCB11;CYP7A1
155 PCSK9;APOE
156 LRP1;APOA2;APOA1;APOE
157 APOH;PPARG;APOE
158 APOH;PPARG;APOE
159 APOE;LDLR
160 LRPAP1;HMGCR
161 LRPAP1;PCSK9;APOE
162 LIPI;APOA2;APOA1;LCAT;LPIN3
163 APOA2;APOA1
164 FADS3;FADS1
165 TSPO;VDAC2;VDAC1
166 PCSK9;LDLRAP1;APOA5
167 ABCA1;PPARG
168 STAR;CYP7A1
169 APOA2;APOA1
170 MYLIP;PCSK9
171 MYLIP;PCSK9
172 APOA2;APOA1
173 VAPA;APOE
174 SCARB1;LDLR
175 LRP1;CD36;LDLRAP1
176 APOA2;LPL;CYP7A1
177 LRP1;APOC2;APOE;APOA5
178 APOC1;APOC3
179 APOH;APOE
180 APOA2;APOA1
181 ALDH2;ADH1B
182 APOE;LDLR
183 HMGCR;APOE;LDLR
184 FADS3;FADS2;FADS1
185 FADS2;FADS1
186 ITIH4;APOH;APOA1;CD36
187 APOC2;APOC1;APOC3
188 APOE;LDLR
189 APOH;APOE
190 APOE;CD36
191 NPC1L1;CD36
192 LRP1;CD36;LDLRAP1
193 SCARB1;APOA2;APOA1
194 APOE;LDLR
195 LPL;PPARG
196 SCARB1;LDLR
197 APOA2;PCSK9;APOA1;APOE;APOB;APOA5
198 LRPAP1;LRP1;APOE
199 APOA1;APOA4;CD36
200 PPARG;LDLRAP1
201 PCSK9;LPL;FADS1
202 APOA2;APOA1
203 APOC1;APOC3
204 APOE;KPNB1
205 STAR;CYP7A1
206 APOA2;ANGPTL3;APOA5
207 APOE;LDLRAP1
208 APOE;CD36
209 SCARB1;APOE
210 ALDH2;ADH1B
211 PCSK9;APOE
212 LPL;CD36
213 APOE;LDLR
214 ABCA1;PPARG
215 FADS2;FADS1
216 APOC1;APOC3
217 LPL;APOB
218 APOA1;LPL;CD36
219 FADS2;EPHX2;FADS1
220 APOA2;APOA1
221 APOA2;PCSK9;APOA1;APOE;APOB;APOA5
222 APOA1;PPARG;APOE
223 LRP1;CD36;LRP2
224 MYLIP;PCSK9
225 APOA2;LPL
226 ITIH4;APOH;APOA1;CD36
227 SCARB1;APOH;APOE
228 HMGCR;APOE
229 LRP1;PPARG;APOE
230 APOH;APOE
231 SCARB1;APOE
232 APOH;APOE
233 LRPAP1;PCSK9;APOE;LDLR
234 EPHX2;FADS1
235 GHR;APOE
236 LRPAP1;LRP1;CD36
237 APOH;ANGPTL3;PPARG;ANGPTL4
238 APOA1;LPL;PPARG;APOE
239 SCARB1;LRP1
240 PCSK9;PPARG;LPIN3
241 PPARG;APOE
242 EPHX2;ANGPTL3;LPL;LCAT
243 APOH;APOE
244 LRP1;CD36
245 APOE;CD36
246 LRPAP1;PCSK9
247 APOA4;PPARG;CD36
248 PPARG;LDLRAP1
249 ABCA1;CD36;APOE
250 TSPO;VDAC2;VDAC1
251 APOA1;PPARG;APOE
252 LRPAP1;LRP1;LDLRAP1
253 APOE;CD36
254 HMGCR;APOE
255 HMGCR;APOE
256 SCARB1;APOE
257 APOH;APOE
258 GHR;LRP2;LDLRAP1
259 PCSK9;PPARG;LPIN3
260 LRP1;CD36
261 ABCA1;MTTP
262 NPC2;ABCB11
263 APOE;LDLR
264 LPL;PPARG;CD36;APOB
265 MYLIP;APOE
266 LPL;PPARG
267 PPARG;LRP2
268 APOC1;APOC3
269 APOA2;APOA1
[1] "GO_Cellular_Component_2021"
Term Overlap
1 high-density lipoprotein particle (GO:0034364) 12/19
2 chylomicron (GO:0042627) 10/10
3 triglyceride-rich plasma lipoprotein particle (GO:0034385) 10/15
4 very-low-density lipoprotein particle (GO:0034361) 10/15
5 early endosome (GO:0005769) 13/266
6 low-density lipoprotein particle (GO:0034362) 4/7
7 spherical high-density lipoprotein particle (GO:0034366) 4/8
8 endoplasmic reticulum lumen (GO:0005788) 10/285
9 endocytic vesicle membrane (GO:0030666) 8/158
10 endoplasmic reticulum membrane (GO:0005789) 14/712
11 lysosome (GO:0005764) 11/477
12 lytic vacuole (GO:0000323) 8/219
13 endocytic vesicle (GO:0030139) 7/189
14 clathrin-coated endocytic vesicle membrane (GO:0030669) 5/69
15 clathrin-coated endocytic vesicle (GO:0045334) 5/85
16 clathrin-coated vesicle membrane (GO:0030665) 5/90
17 lysosomal membrane (GO:0005765) 8/330
18 intracellular organelle lumen (GO:0070013) 12/848
19 collagen-containing extracellular matrix (GO:0062023) 8/380
20 endocytic vesicle lumen (GO:0071682) 3/21
21 organelle outer membrane (GO:0031968) 5/142
22 ATP-binding cassette (ABC) transporter complex (GO:0043190) 2/6
23 lytic vacuole membrane (GO:0098852) 6/267
24 endosome membrane (GO:0010008) 6/325
25 mitochondrial outer membrane (GO:0005741) 4/126
26 platelet dense granule lumen (GO:0031089) 2/14
27 vesicle (GO:0031982) 5/226
28 endolysosome membrane (GO:0036020) 2/17
29 basolateral plasma membrane (GO:0016323) 4/151
30 cytoplasmic vesicle membrane (GO:0030659) 6/380
31 platelet dense granule (GO:0042827) 2/21
32 lysosomal lumen (GO:0043202) 3/86
33 endolysosome (GO:0036019) 2/25
34 secretory granule lumen (GO:0034774) 5/316
35 brush border membrane (GO:0031526) 2/37
36 mitochondrial envelope (GO:0005740) 3/127
37 extracellular membrane-bounded organelle (GO:0065010) 2/56
38 extracellular vesicle (GO:1903561) 2/59
39 vacuolar lumen (GO:0005775) 3/161
40 caveola (GO:0005901) 2/60
Adjusted.P.value
1 5.261200e-24
2 6.209923e-24
3 9.203512e-21
4 9.203512e-21
5 1.246888e-10
6 7.731648e-08
7 1.321996e-07
8 6.153361e-07
9 8.075627e-07
10 1.200431e-06
11 6.211359e-06
12 7.353950e-06
13 2.938912e-05
14 2.938912e-05
15 7.671871e-05
16 9.509306e-05
17 1.065748e-04
18 1.698308e-04
19 2.574496e-04
20 2.574496e-04
21 6.432532e-04
22 8.164449e-04
23 1.420908e-03
24 3.827987e-03
25 3.898444e-03
26 4.116966e-03
27 4.199032e-03
28 5.675277e-03
29 6.551600e-03
30 6.795770e-03
31 7.845057e-03
32 1.043474e-02
33 1.043474e-02
34 1.423039e-02
35 2.126720e-02
36 2.763580e-02
37 4.460396e-02
38 4.700422e-02
39 4.700422e-02
40 4.700422e-02
Genes
1 CETP;APOC2;APOH;APOC1;APOA2;APOA1;APOC3;LCAT;APOA4;APOE;APOA5;PLTP
2 APOC2;APOH;APOC1;APOA2;APOA1;APOC3;APOA4;APOE;APOB;APOA5
3 APOC2;APOH;APOC1;APOA2;APOA1;APOC3;APOA4;APOE;APOB;APOA5
4 APOC2;APOH;APOC1;APOA2;APOA1;APOC3;APOA4;APOE;APOB;APOA5
5 LRP1;SORT1;APOA2;PCSK9;APOA1;APOC3;APOA4;APOC2;LIPG;APOE;LDLRAP1;APOB;LDLR
6 APOC2;APOE;APOB;APOA5
7 APOC2;APOA2;APOA1;APOC3
8 LRPAP1;LIPC;MTTP;APOA2;PCSK9;APOA1;APOA4;APOE;APOB;APOA5
9 SCARB1;LRP1;CD36;LRP2;APOE;LDLRAP1;APOB;LDLR
10 ABCA1;STARD3;HMGCR;CYP7A1;FADS2;NCEH1;SOAT1;VAPA;SOAT2;VAPB;DHCR7;APOB;FADS1;LPIN3
11 SCARB1;STARD3;NPC1;LRP1;NPC2;SORT1;PCSK9;LRP2;APOB;LIPA;LDLR
12 SCARB1;NPC1;NPC2;SORT1;PCSK9;LRP2;LIPA;LDLR
13 ABCA1;SCARB1;LRP1;APOA1;CD36;APOE;APOB
14 LRP2;APOE;LDLRAP1;APOB;LDLR
15 LRP2;APOE;LDLRAP1;APOB;LDLR
16 LRP2;APOE;LDLRAP1;APOB;LDLR
17 SCARB1;STARD3;NPC1;LRP1;VAPA;PCSK9;LRP2;LDLR
18 CYP27A1;LIPC;ALDH2;MTTP;APOA2;PCSK9;APOA1;APOA4;APOE;APOB;APOA5;KPNB1
19 ITIH4;APOH;ANGPTL3;APOA1;APOC3;APOA4;ANGPTL4;APOE
20 APOA1;APOE;APOB
21 VDAC3;TSPO;VDAC2;VDAC1;DHCR7
22 ABCG8;ABCG5
23 SCARB1;STARD3;NPC1;LRP1;PCSK9;LRP2
24 STARD3;SORT1;PCSK9;ABCB11;APOB;LDLR
25 VDAC3;TSPO;VDAC2;VDAC1
26 ITIH4;APOH
27 ABCA1;CETP;VAPA;APOA1;APOE
28 PCSK9;LDLR
29 LRP1;MTTP;ABCB11;LDLR
30 SCARB1;LRP1;SORT1;CD36;APOB;LDLR
31 ITIH4;APOH
32 NPC2;APOB;LIPA
33 PCSK9;LDLR
34 ITIH4;NPC2;APOH;APOA1;KPNB1
35 LRP2;CD36
36 STAR;VDAC2;VDAC1
37 APOA1;APOE
38 APOA1;APOE
39 NPC2;APOB;LIPA
40 SCARB1;CD36
[1] "GO_Molecular_Function_2021"
Term
1 cholesterol binding (GO:0015485)
2 sterol binding (GO:0032934)
3 cholesterol transfer activity (GO:0120020)
4 sterol transfer activity (GO:0120015)
5 lipoprotein particle receptor binding (GO:0070325)
6 phosphatidylcholine-sterol O-acyltransferase activator activity (GO:0060228)
7 lipoprotein particle binding (GO:0071813)
8 low-density lipoprotein particle binding (GO:0030169)
9 lipase inhibitor activity (GO:0055102)
10 low-density lipoprotein particle receptor binding (GO:0050750)
11 lipoprotein lipase activity (GO:0004465)
12 amyloid-beta binding (GO:0001540)
13 triglyceride lipase activity (GO:0004806)
14 lipase activity (GO:0016298)
15 lipase binding (GO:0035473)
16 apolipoprotein A-I binding (GO:0034186)
17 apolipoprotein receptor binding (GO:0034190)
18 phospholipase A1 activity (GO:0008970)
19 lipase activator activity (GO:0060229)
20 carboxylic ester hydrolase activity (GO:0052689)
21 voltage-gated anion channel activity (GO:0008308)
22 voltage-gated ion channel activity (GO:0005244)
23 phosphatidylcholine transporter activity (GO:0008525)
24 protein heterodimerization activity (GO:0046982)
25 phosphatidylcholine transfer activity (GO:0120019)
26 phospholipase activity (GO:0004620)
27 O-acyltransferase activity (GO:0008374)
28 high-density lipoprotein particle binding (GO:0008035)
29 ceramide transfer activity (GO:0120017)
30 clathrin heavy chain binding (GO:0032050)
31 phospholipase inhibitor activity (GO:0004859)
32 protein homodimerization activity (GO:0042803)
33 anion channel activity (GO:0005253)
34 phospholipid transfer activity (GO:0120014)
35 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen (GO:0016709)
36 steroid hydroxylase activity (GO:0008395)
37 NADP binding (GO:0050661)
38 peptidase inhibitor activity (GO:0030414)
39 endopeptidase regulator activity (GO:0061135)
Overlap Adjusted.P.value
1 17/50 2.288174e-28
2 17/60 4.390322e-27
3 11/18 5.744987e-22
4 11/19 1.020647e-21
5 10/28 4.677379e-17
6 6/6 3.484605e-14
7 8/24 2.055320e-13
8 6/17 3.140441e-10
9 5/10 1.805253e-09
10 6/23 2.016360e-09
11 4/5 9.113232e-09
12 7/80 1.430772e-07
13 5/23 1.612041e-07
14 6/49 1.859889e-07
15 3/5 3.788104e-06
16 3/5 3.788104e-06
17 3/6 7.112969e-06
18 3/10 3.991032e-05
19 3/12 6.897596e-05
20 5/96 1.501407e-04
21 3/16 1.501407e-04
22 3/16 1.501407e-04
23 3/18 2.082322e-04
24 6/188 3.016202e-04
25 2/5 7.035280e-04
26 4/73 7.035280e-04
27 3/30 8.567836e-04
28 2/6 9.653527e-04
29 2/8 1.732107e-03
30 2/9 2.147965e-03
31 2/10 2.592555e-03
32 8/636 7.345938e-03
33 3/68 7.879809e-03
34 2/22 1.181407e-02
35 2/36 2.870121e-02
36 2/36 2.870121e-02
37 2/36 2.870121e-02
38 2/40 3.429432e-02
39 2/46 4.375411e-02
Genes
1 ABCA1;STARD3;CETP;OSBPL5;APOA2;APOA1;APOC3;APOA4;APOA5;NPC1;SOAT1;STAR;NPC2;SOAT2;TSPO;VDAC2;VDAC1
2 ABCA1;STARD3;CETP;OSBPL5;APOA2;APOA1;APOC3;APOA4;APOA5;NPC1;SOAT1;STAR;NPC2;SOAT2;TSPO;VDAC2;VDAC1
3 ABCA1;ABCG8;CETP;ABCG5;NPC2;MTTP;APOA2;APOA1;APOA4;APOB;PLTP
4 ABCA1;ABCG8;CETP;ABCG5;NPC2;MTTP;APOA2;APOA1;APOA4;APOB;PLTP
5 LRPAP1;LRP1;APOA2;PCSK9;APOA1;APOC3;APOE;APOB;LDLRAP1;APOA5
6 APOC1;APOA2;APOA1;APOA4;APOE;APOA5
7 SCARB1;LIPC;APOA2;LPL;PCSK9;CD36;LDLR;PLTP
8 SCARB1;LIPC;PCSK9;CD36;LDLR;PLTP
9 APOC2;APOC1;ANGPTL3;APOA2;APOC3
10 LRPAP1;PCSK9;APOE;APOB;LDLRAP1;APOA5
11 LIPC;LIPI;LIPG;LPL
12 LRPAP1;LRP1;APOA1;CD36;APOE;LDLRAP1;LDLR
13 LIPC;LIPI;LIPG;LCAT;LPL
14 LIPC;LIPI;LIPG;LCAT;LPL;LIPA
15 LRPAP1;APOB;APOA5
16 ABCA1;SCARB1;LCAT
17 APOA2;APOA1;PCSK9
18 LIPC;LIPG;LPL
19 APOC2;APOH;APOA5
20 LIPC;LIPG;LPL;LCAT;LIPA
21 VDAC3;VDAC2;VDAC1
22 VDAC3;VDAC2;VDAC1
23 ABCA1;MTTP;PLTP
24 ABCG8;ABCG5;VAPA;VAPB;MTTP;APOA2
25 MTTP;PLTP
26 LIPC;LIPI;LIPG;LPL
27 SOAT1;SOAT2;LCAT
28 APOA2;PLTP
29 MTTP;PLTP
30 LRP1;LDLR
31 APOC1;ANGPTL3
32 GHR;STARD3;VAPB;EPHX2;APOA2;LPL;APOA4;APOE
33 VDAC3;VDAC2;VDAC1
34 MTTP;PLTP
35 CYP27A1;CYP7A1
36 CYP27A1;CYP7A1
37 HMGCR;DHCR7
38 ITIH4;LPA
39 ITIH4;LPAGO_known_annotations <- do.call(rbind, GO_enrichment)
GO_known_annotations <- GO_known_annotations[GO_known_annotations$Adjusted.P.value<0.05,]
#GO enrichment analysis for cTWAS genes
genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
GO_enrichment <- enrichr(genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.GO_ctwas_genes <- do.call(rbind, GO_enrichment)
#optionally subset to only significant GO terms
#GO_ctwas_genes <- GO_ctwas_genes[GO_ctwas_genes$Adjusted.P.value<0.05,]
#identify cTWAS genes in silver standard enriched GO terms
GO_ctwas_genes <- GO_ctwas_genes[GO_ctwas_genes$Term %in% GO_known_annotations$Term,]
overlap_genes <- lapply(GO_ctwas_genes$Genes, function(x){unlist(strsplit(x, ";"))})
overlap_genes <- -sort(-table(unlist(overlap_genes)))
#ctwas genes in silver standard enriched GO terms, not already in silver standard
overlap_genes[!(names(overlap_genes) %in% known_annotations)]
GPAM ABCA8 SCD SPHK2 ACP6 CPNE3 POR ZDHHC7 ACVR1C
12 10 10 6 5 4 3 3 2
KDSR PARP9 PDHB PLPPR2 NTN5 SPTY2D1 TMED4 TPD52
2 2 2 2 1 1 1 1 save(overlap_genes, file=paste0(results_dir, "/overlap_genes.Rd"))
load(paste0(results_dir, "/overlap_genes.Rd"))
overlap_genes <- overlap_genes[!(names(overlap_genes) %in% known_annotations)]
overlap_genes
GPAM ABCA8 SCD SPHK2 ACP6 CPNE3 POR ZDHHC7 ACVR1C
12 10 10 6 5 4 3 3 2
KDSR PARP9 PDHB PLPPR2 NTN5 SPTY2D1 TMED4 TPD52
2 2 2 2 1 1 1 1 overlap_genes <- names(overlap_genes)
#ctwas_gene_res[ctwas_gene_res$genename %in% overlap_genes, report_cols,]Results for Paper
out_table <- ctwas_gene_res
report_cols <- report_cols[!(report_cols %in% c("mu2", "PVE"))]
report_cols <- c(report_cols,"silver","GO_overlap_silver", "bystander")
#reload silver standard genes
known_annotations <- read_xlsx("data/summary_known_genes_annotations.xlsx", sheet="LDL")New names:
* `` -> ...4
* `` -> ...5known_annotations <- unique(known_annotations$`Gene Symbol`)
out_table$silver <- F
out_table$silver[out_table$genename %in% known_annotations] <- T
#create extended bystanders list (all silver standard, not just imputed silver standard)
library(biomaRt)
library(GenomicRanges)
ensembl <- useEnsembl(biomart="ENSEMBL_MART_ENSEMBL", dataset="hsapiens_gene_ensembl")
G_list <- getBM(filters= "chromosome_name", attributes= c("hgnc_symbol","chromosome_name","start_position","end_position","gene_biotype"), values=1:22, mart=ensembl)
G_list <- G_list[G_list$hgnc_symbol!="",]
G_list <- G_list[G_list$gene_biotype %in% c("protein_coding","lncRNA"),]
G_list$start <- G_list$start_position
G_list$end <- G_list$end_position
G_list_granges <- makeGRangesFromDataFrame(G_list, keep.extra.columns=T)
known_annotations_positions <- G_list[G_list$hgnc_symbol %in% known_annotations,]
half_window <- 1000000
known_annotations_positions$start <- known_annotations_positions$start_position - half_window
known_annotations_positions$end <- known_annotations_positions$end_position + half_window
known_annotations_positions$start[known_annotations_positions$start<1] <- 1
known_annotations_granges <- makeGRangesFromDataFrame(known_annotations_positions, keep.extra.columns=T)
bystanders_extended <- findOverlaps(known_annotations_granges,G_list_granges)
bystanders_extended <- unique(subjectHits(bystanders_extended))
bystanders_extended <- G_list$hgnc_symbol[bystanders_extended]
bystanders_extended <- unique(bystanders_extended[!(bystanders_extended %in% known_annotations)])
save(bystanders_extended, file=paste0(results_dir, "/bystanders_extended.Rd"))
load(paste0(results_dir, "/bystanders_extended.Rd"))
#add extended bystanders list to output
out_table$bystander <- F
out_table$bystander[out_table$genename %in% bystanders_extended] <- T
#reload GO overlaps with silver standard
load(paste0(results_dir, "/overlap_genes.Rd"))
out_table$GO_overlap_silver <- NA
out_table$GO_overlap_silver[out_table$susie_pip>0.8] <- 0
for (i in names(overlap_genes)){
out_table$GO_overlap_silver[out_table$genename==i] <- overlap_genes[i]
}cTWAS identifies high-confidence liver genes associated with LDL cholesterol
full.gene.pip.summary <- data.frame(gene_name = ctwas_gene_res$genename,
gene_pip = ctwas_gene_res$susie_pip,
gene_id = ctwas_gene_res$id,
chr = as.integer(ctwas_gene_res$chrom),
start = ctwas_gene_res$pos / 1e3,
is_highlight = F, stringsAsFactors = F) %>% as_tibble()
full.gene.pip.summary$is_highlight <- full.gene.pip.summary$gene_pip > 0.80
don <- full.gene.pip.summary %>%
# Compute chromosome size
group_by(chr) %>%
summarise(chr_len=max(start)) %>%
# Calculate cumulative position of each chromosome
mutate(tot=cumsum(chr_len)-chr_len) %>%
dplyr::select(-chr_len) %>%
# Add this info to the initial dataset
left_join(full.gene.pip.summary, ., by=c("chr"="chr")) %>%
# Add a cumulative position of each SNP
arrange(chr, start) %>%
mutate( BPcum=start+tot)
axisdf <- don %>% group_by(chr) %>% summarize(center=( max(BPcum) + min(BPcum) ) / 2 )
x_axis_labels <- axisdf$chr
x_axis_labels[seq(1,21,2)] <- ""
ggplot(don, aes(x=BPcum, y=gene_pip)) +
# Show all points
ggrastr::geom_point_rast(aes(color=as.factor(chr)), size=2) +
scale_color_manual(values = rep(c("grey", "skyblue"), 22 )) +
# custom X axis:
# scale_x_continuous(label = axisdf$chr,
# breaks= axisdf$center,
# guide = guide_axis(n.dodge = 2)) +
scale_x_continuous(label = x_axis_labels,
breaks = axisdf$center) +
scale_y_continuous(expand = c(0, 0), limits = c(0,1.25), breaks=(1:5)*0.2, minor_breaks=(1:10)*0.1) + # remove space between plot area and x axis
# Add highlighted points
ggrastr::geom_point_rast(data=subset(don, is_highlight==T), color="orange", size=2) +
# Add label using ggrepel to avoid overlapping
ggrepel::geom_label_repel(data=subset(don, is_highlight==T),
aes(label=gene_name),
size=4,
min.segment.length = 0,
label.size = NA,
fill = alpha(c("white"),0)) +
# Custom the theme:
theme_bw() +
theme(
text = element_text(size = 14),
legend.position="none",
panel.border = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
) +
xlab("Chromosome") +
ylab("cTWAS PIP")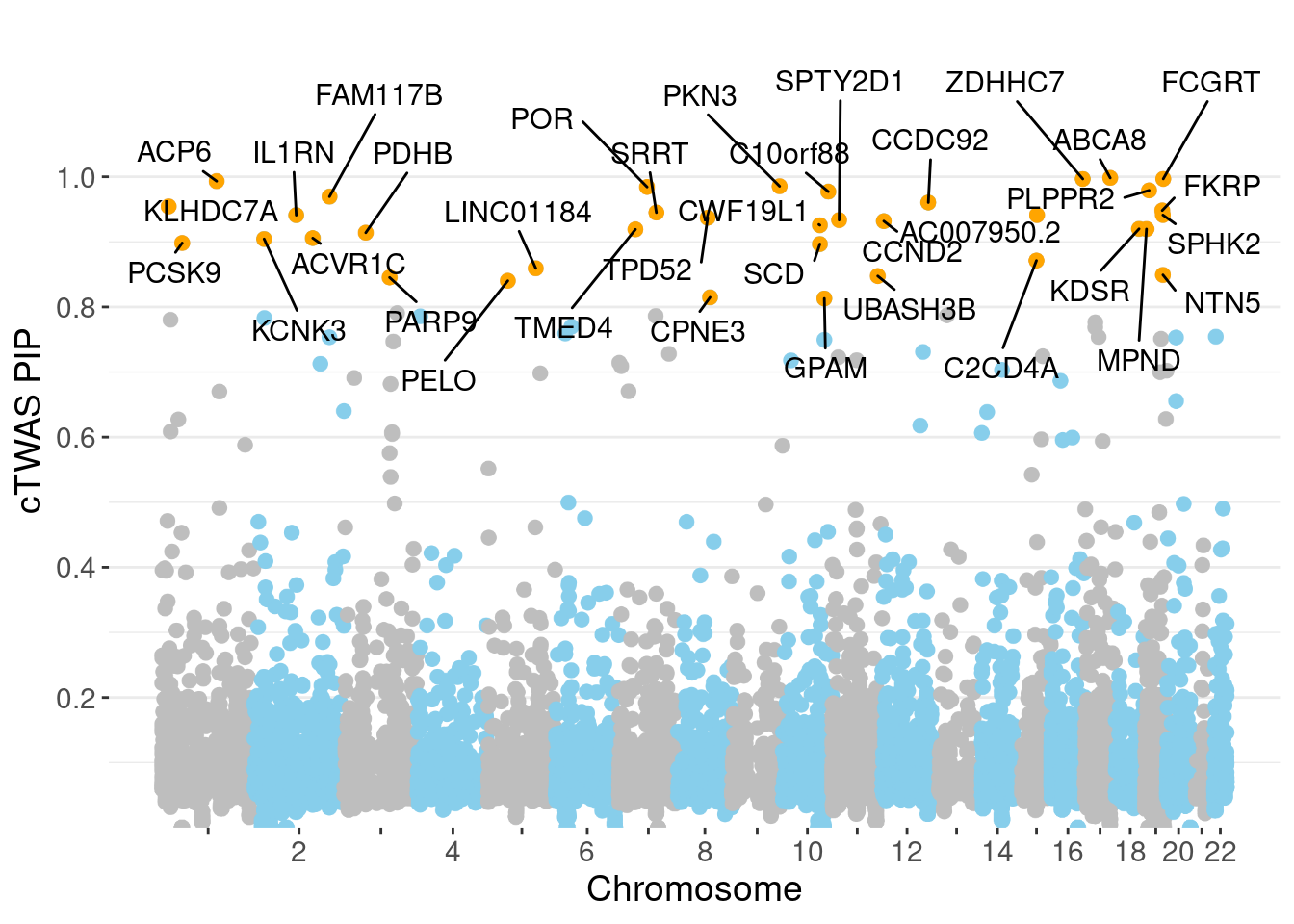
#number of SNPs at PIP>0.8 threshold
sum(out_table$susie_pip>0.8)[1] 36#number of SNPs at PIP>0.5 threshold
sum(out_table$susie_pip>0.5)[1] 91#genes with PIP>0.8
head(out_table[order(-out_table$susie_pip),report_cols], sum(out_table$susie_pip>0.8)) genename region_tag susie_pip z num_eqtl silver
5923 ABCA8 17_39 0.9982625 5.086840 2 FALSE
2199 FCGRT 19_34 0.9967913 -4.165895 2 FALSE
7045 ZDHHC7 16_49 0.9967008 -4.891663 3 FALSE
7879 ACP6 1_73 0.9932775 4.613976 4 FALSE
7623 PKN3 9_66 0.9854511 -6.885356 2 FALSE
4395 POR 7_48 0.9843314 6.026895 2 FALSE
2313 PLPPR2 19_10 0.9790947 3.965665 1 FALSE
3730 C10orf88 10_77 0.9771524 -6.762952 1 FALSE
5600 FAM117B 2_120 0.9695151 7.877231 2 FALSE
3647 CCDC92 12_75 0.9604840 -5.343007 5 FALSE
10255 KLHDC7A 1_13 0.9544827 4.124187 1 FALSE
10447 FKRP 19_33 0.9482623 4.425174 5 FALSE
1279 SRRT 7_62 0.9450584 4.744831 2 FALSE
632 SPHK2 19_33 0.9412590 -8.721460 1 FALSE
14551 AC007950.2 15_29 0.9412296 5.555780 1 FALSE
5327 IL1RN 2_67 0.9410797 4.455379 1 FALSE
1003 TPD52 8_57 0.9370510 -4.121885 2 FALSE
10272 SPTY2D1 11_13 0.9335133 -5.557123 1 FALSE
3633 CCND2 12_4 0.9318759 -4.128258 2 FALSE
1526 CWF19L1 10_64 0.9256794 5.813958 2 FALSE
3674 KDSR 18_35 0.9199769 -4.119957 2 FALSE
161 MPND 19_5 0.9197113 -4.322018 2 FALSE
7441 TMED4 7_32 0.9191981 7.688274 2 FALSE
8904 PDHB 3_40 0.9140947 3.361078 2 FALSE
4022 ACVR1C 2_94 0.9057641 -4.185879 1 FALSE
9348 KCNK3 2_16 0.9044393 -4.772296 1 FALSE
9036 PCSK9 1_34 0.8982431 17.210869 1 TRUE
1559 SCD 10_64 0.8966611 -4.541468 1 FALSE
11865 C2CD4A 15_28 0.8713009 4.535165 1 FALSE
13258 LINC01184 5_78 0.8593862 -3.918127 2 FALSE
6023 NTN5 19_33 0.8492136 10.033010 2 FALSE
7079 UBASH3B 11_74 0.8475160 4.906621 1 FALSE
5607 PARP9 3_76 0.8452748 3.744644 1 FALSE
6967 PELO 5_30 0.8402073 8.522224 1 FALSE
1237 CPNE3 8_62 0.8147843 3.753687 3 FALSE
3724 GPAM 10_70 0.8129956 4.000718 1 FALSE
GO_overlap_silver bystander
5923 10 FALSE
2199 0 FALSE
7045 3 FALSE
7879 5 FALSE
7623 0 FALSE
4395 3 FALSE
2313 2 TRUE
3730 0 FALSE
5600 0 FALSE
3647 0 TRUE
10255 0 FALSE
10447 0 FALSE
1279 0 FALSE
632 6 FALSE
14551 0 FALSE
5327 0 FALSE
1003 1 FALSE
10272 1 FALSE
3633 0 FALSE
1526 0 FALSE
3674 2 FALSE
161 0 FALSE
7441 1 TRUE
8904 2 FALSE
4022 2 FALSE
9348 0 FALSE
9036 37 FALSE
1559 10 FALSE
11865 0 FALSE
13258 0 FALSE
6023 1 FALSE
7079 0 FALSE
5607 2 FALSE
6967 0 FALSE
1237 4 FALSE
3724 12 FALSEhead(out_table[order(-out_table$susie_pip),report_cols[-(7:8)]], sum(out_table$susie_pip>0.8)) genename region_tag susie_pip z num_eqtl silver
5923 ABCA8 17_39 0.9982625 5.086840 2 FALSE
2199 FCGRT 19_34 0.9967913 -4.165895 2 FALSE
7045 ZDHHC7 16_49 0.9967008 -4.891663 3 FALSE
7879 ACP6 1_73 0.9932775 4.613976 4 FALSE
7623 PKN3 9_66 0.9854511 -6.885356 2 FALSE
4395 POR 7_48 0.9843314 6.026895 2 FALSE
2313 PLPPR2 19_10 0.9790947 3.965665 1 FALSE
3730 C10orf88 10_77 0.9771524 -6.762952 1 FALSE
5600 FAM117B 2_120 0.9695151 7.877231 2 FALSE
3647 CCDC92 12_75 0.9604840 -5.343007 5 FALSE
10255 KLHDC7A 1_13 0.9544827 4.124187 1 FALSE
10447 FKRP 19_33 0.9482623 4.425174 5 FALSE
1279 SRRT 7_62 0.9450584 4.744831 2 FALSE
632 SPHK2 19_33 0.9412590 -8.721460 1 FALSE
14551 AC007950.2 15_29 0.9412296 5.555780 1 FALSE
5327 IL1RN 2_67 0.9410797 4.455379 1 FALSE
1003 TPD52 8_57 0.9370510 -4.121885 2 FALSE
10272 SPTY2D1 11_13 0.9335133 -5.557123 1 FALSE
3633 CCND2 12_4 0.9318759 -4.128258 2 FALSE
1526 CWF19L1 10_64 0.9256794 5.813958 2 FALSE
3674 KDSR 18_35 0.9199769 -4.119957 2 FALSE
161 MPND 19_5 0.9197113 -4.322018 2 FALSE
7441 TMED4 7_32 0.9191981 7.688274 2 FALSE
8904 PDHB 3_40 0.9140947 3.361078 2 FALSE
4022 ACVR1C 2_94 0.9057641 -4.185879 1 FALSE
9348 KCNK3 2_16 0.9044393 -4.772296 1 FALSE
9036 PCSK9 1_34 0.8982431 17.210869 1 TRUE
1559 SCD 10_64 0.8966611 -4.541468 1 FALSE
11865 C2CD4A 15_28 0.8713009 4.535165 1 FALSE
13258 LINC01184 5_78 0.8593862 -3.918127 2 FALSE
6023 NTN5 19_33 0.8492136 10.033010 2 FALSE
7079 UBASH3B 11_74 0.8475160 4.906621 1 FALSE
5607 PARP9 3_76 0.8452748 3.744644 1 FALSE
6967 PELO 5_30 0.8402073 8.522224 1 FALSE
1237 CPNE3 8_62 0.8147843 3.753687 3 FALSE
3724 GPAM 10_70 0.8129956 4.000718 1 FALSEhead(out_table[order(-out_table$susie_pip),report_cols[c(1,7:8)]], sum(out_table$susie_pip>0.8)) genename GO_overlap_silver bystander
5923 ABCA8 10 FALSE
2199 FCGRT 0 FALSE
7045 ZDHHC7 3 FALSE
7879 ACP6 5 FALSE
7623 PKN3 0 FALSE
4395 POR 3 FALSE
2313 PLPPR2 2 TRUE
3730 C10orf88 0 FALSE
5600 FAM117B 0 FALSE
3647 CCDC92 0 TRUE
10255 KLHDC7A 0 FALSE
10447 FKRP 0 FALSE
1279 SRRT 0 FALSE
632 SPHK2 6 FALSE
14551 AC007950.2 0 FALSE
5327 IL1RN 0 FALSE
1003 TPD52 1 FALSE
10272 SPTY2D1 1 FALSE
3633 CCND2 0 FALSE
1526 CWF19L1 0 FALSE
3674 KDSR 2 FALSE
161 MPND 0 FALSE
7441 TMED4 1 TRUE
8904 PDHB 2 FALSE
4022 ACVR1C 2 FALSE
9348 KCNK3 0 FALSE
9036 PCSK9 37 FALSE
1559 SCD 10 FALSE
11865 C2CD4A 0 FALSE
13258 LINC01184 0 FALSE
6023 NTN5 1 FALSE
7079 UBASH3B 0 FALSE
5607 PARP9 2 FALSE
6967 PELO 0 FALSE
1237 CPNE3 4 FALSE
3724 GPAM 12 FALSEcTWAS avoids false positives when multiple genes are in a region
TNKS is a silver standard (assumed true positive gene) that is correctly detected. The bystander gene RP11-115J16.2 is significant using TWAS but has low PIP using cTWAS.
# #TNKS gene
# locus_plot4("8_12", label="cTWAS")
#
# out_table[out_table$region_tag=="8_12",report_cols[-(7:8)]]
# out_table[out_table$region_tag=="8_12",report_cols[c(1,7:8)]]FADS1 is a silver standard gene (assumed true positive gene) that is correctly detected. There are 5 significant TWAS genes at this locus, including FADS2, another silver standard gene. FADS2 is not detected due to its high LD with FADS1. The remaining 3 bystander genes at this locus have low PIP using cTWAS.
# #FADS1 gene
# locus_plot3("11_34", focus="FADS1")
#
# out_table[out_table$region_tag=="11_34",report_cols[-(7:8)]]
# out_table[out_table$region_tag=="11_34",report_cols[c(1,7:8)]]
#
# #number of significant TWAS genes at this locus
# sum(abs(out_table$z[out_table$region_tag=="11_34"])>sig_thresh)cTWAS avoids false positives when SNPs in a region are (considerably) more significant
POLK is a gene that is significant using TWAS but not detected using TWAS. cTWAS places a high posterior probability on SNPs are this locus. OpenTargets suggets that the causal gene at this locus is HMGCR (note: different GWAS, similar population), which is not imputed in our dataset. cTWAS selected the variants at this locus because the causal gene is not imputed. Note that MR-JTI claims POLK is causal using their method, and their paper includes a discussion of its potential relevance to LDL.
locus_plot("5_45", label="TWAS")
#locus_plot("5_45", label="TWAS", rerun_ctwas = T)
out_table[out_table$region_tag=="5_45",report_cols[-(7:8)]] genename region_tag susie_pip z num_eqtl silver
3095 PDE8B 5_45 0.06221375 0.2387889 1 FALSE
4860 AP3B1 5_45 0.11964878 1.3922880 1 FALSE
4861 ZBED3 5_45 0.06166792 -0.1260374 1 FALSE
6413 CRHBP 5_45 0.06784777 -0.6328738 1 FALSE
8186 F2RL2 5_45 0.08093380 -0.9027896 2 FALSE
8191 F2RL1 5_45 0.23897109 2.2141016 3 FALSE
8192 AGGF1 5_45 0.08974170 -1.0343064 2 FALSE
8193 WDR41 5_45 0.06701744 0.4828811 3 FALSE
9379 TBCA 5_45 0.06373028 0.3199763 1 FALSE
10456 F2R 5_45 0.06798428 -0.6619550 1 FALSEout_table[out_table$region_tag=="5_45",report_cols[c(1,7:8)]] genename GO_overlap_silver bystander
3095 PDE8B NA FALSE
4860 AP3B1 NA FALSE
4861 ZBED3 NA FALSE
6413 CRHBP NA FALSE
8186 F2RL2 NA FALSE
8191 F2RL1 NA FALSE
8192 AGGF1 NA FALSE
8193 WDR41 NA FALSE
9379 TBCA NA FALSE
10456 F2R NA FALSEcTWAS is more precise than TWAS in distinguishing silver standard and bystander genes
load(paste0(results_dir, "/known_annotations.Rd"))
load(paste0(results_dir, "/bystanders.Rd"))
#remove genes without imputed expression from bystander list
unrelated_genes <- unrelated_genes[unrelated_genes %in% ctwas_gene_res$genename]
#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#sensitivity / recall
sensitivity <- rep(NA,2)
names(sensitivity) <- c("ctwas", "TWAS")
sensitivity["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
sensitivity["TWAS"] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
sensitivityctwas TWAS
0.025 0.325 #specificity / (1 - False Positive Rate)
specificity <- rep(NA,2)
names(specificity) <- c("ctwas", "TWAS")
specificity["ctwas"] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
specificity["TWAS"] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
specificity ctwas TWAS
0.9970414 0.9304734 #precision / PPV / (1 - False Discovery Rate)
precision <- rep(NA,2)
names(precision) <- c("ctwas", "TWAS")
precision["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
precision["TWAS"] <- sum(twas_genes %in% known_annotations)/length(twas_genes)
precision ctwas TWAS
0.3333333 0.2166667 #store sensitivity and specificity calculations for plots
sensitivity_plot <- sensitivity
specificity_plot <- specificity
#precision / PPV by PIP threshold
pip_range <- c(0.5, 0.8, 1)
precision_range <- rep(NA, length(pip_range))
number_detected <- rep(NA, length(pip_range))
for (i in 1:length(pip_range)){
pip_upper <- pip_range[i]
if (i==1){
pip_lower <- 0
} else {
pip_lower <- pip_range[i-1]
}
#assign ctwas genes using PIP threshold
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip_lower]
number_detected[i] <- length(ctwas_genes)
precision_range[i] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
}
names(precision_range) <- paste0(">= ", c(0, pip_range[-length(pip_range)]))
precision_range <- precision_range*100
precision_range <- c(precision_range, precision["TWAS"]*100)
names(precision_range)[4] <- "TWAS Bonferroni"
number_detected <- c(number_detected, length(twas_genes))
barplot(precision_range, ylim=c(0,100), main="Precision for Distinguishing Silver Standard and Bystander Genes", xlab="PIP Threshold for Detection", ylab="% of Detected Genes in Silver Standard")
abline(h=20, lty=2)
abline(h=40, lty=2)
abline(h=60, lty=2)
abline(h=80, lty=2)
xx <- barplot(precision_range, add=T, col=c(rep("darkgrey",3), "white"))
text(x = xx, y = rep(0, length(number_detected)), label = paste0(number_detected, " detected"), pos = 3, cex=0.8)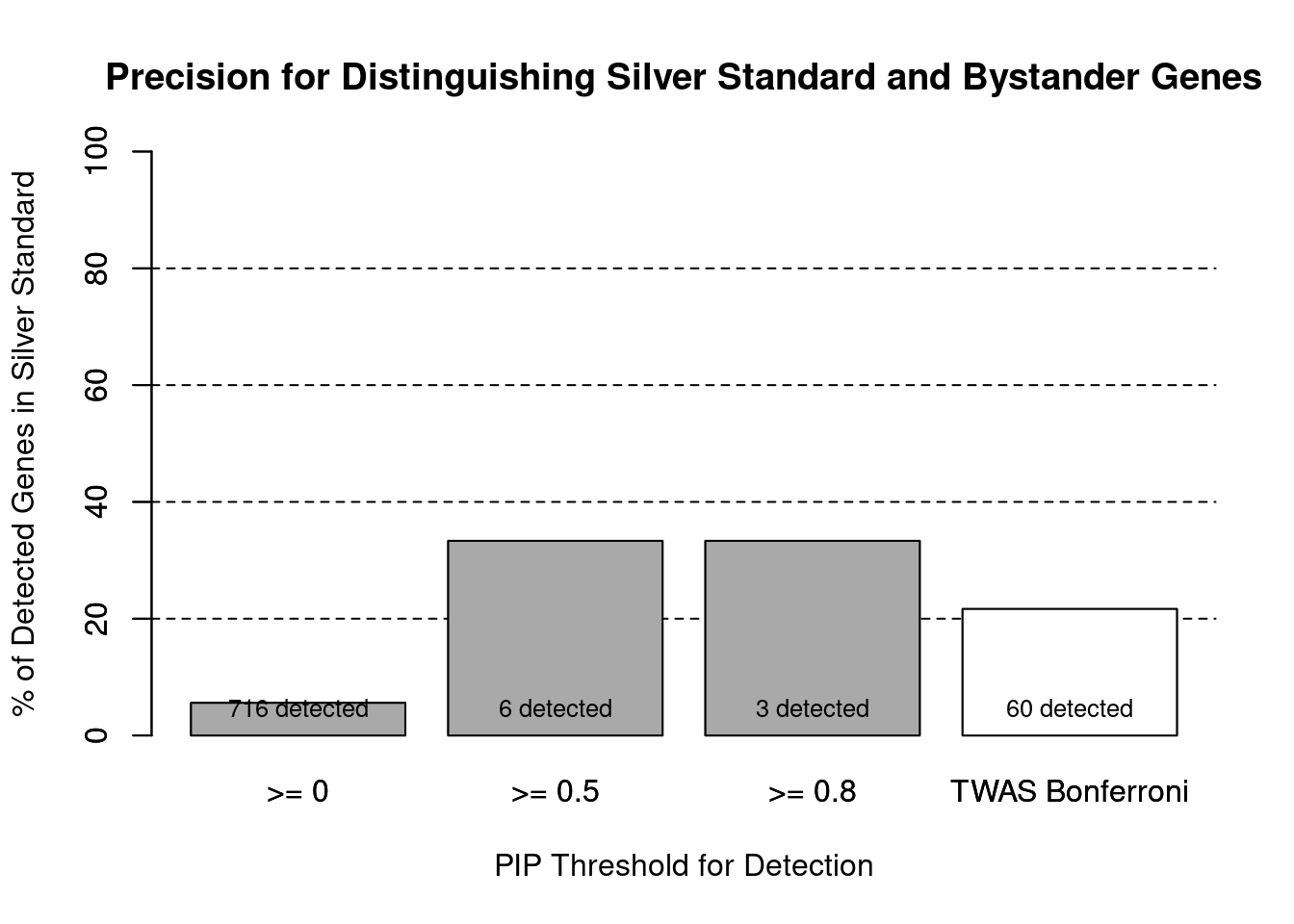
#text(x = xx, y = precision_range, label = paste0(round(precision_range,1), "%"), pos = 3, cex=0.8, offset = 1.5)
#false discovery rate by PIP threshold
barplot(100-precision_range, ylim=c(0,100), main="False Discovery Rate for Distinguishing Silver Standard and Bystander Genes", xlab="PIP Threshold for Detection", ylab="% Bystanders in Detected Genes")
abline(h=20, lty=2)
abline(h=40, lty=2)
abline(h=60, lty=2)
abline(h=80, lty=2)
xx <- barplot(100-precision_range, add=T, col=c(rep("darkgrey",3), "white"))
text(x = xx, y = rep(0, length(number_detected)), label = paste0(number_detected, " detected"), pos = 3, cex=0.8)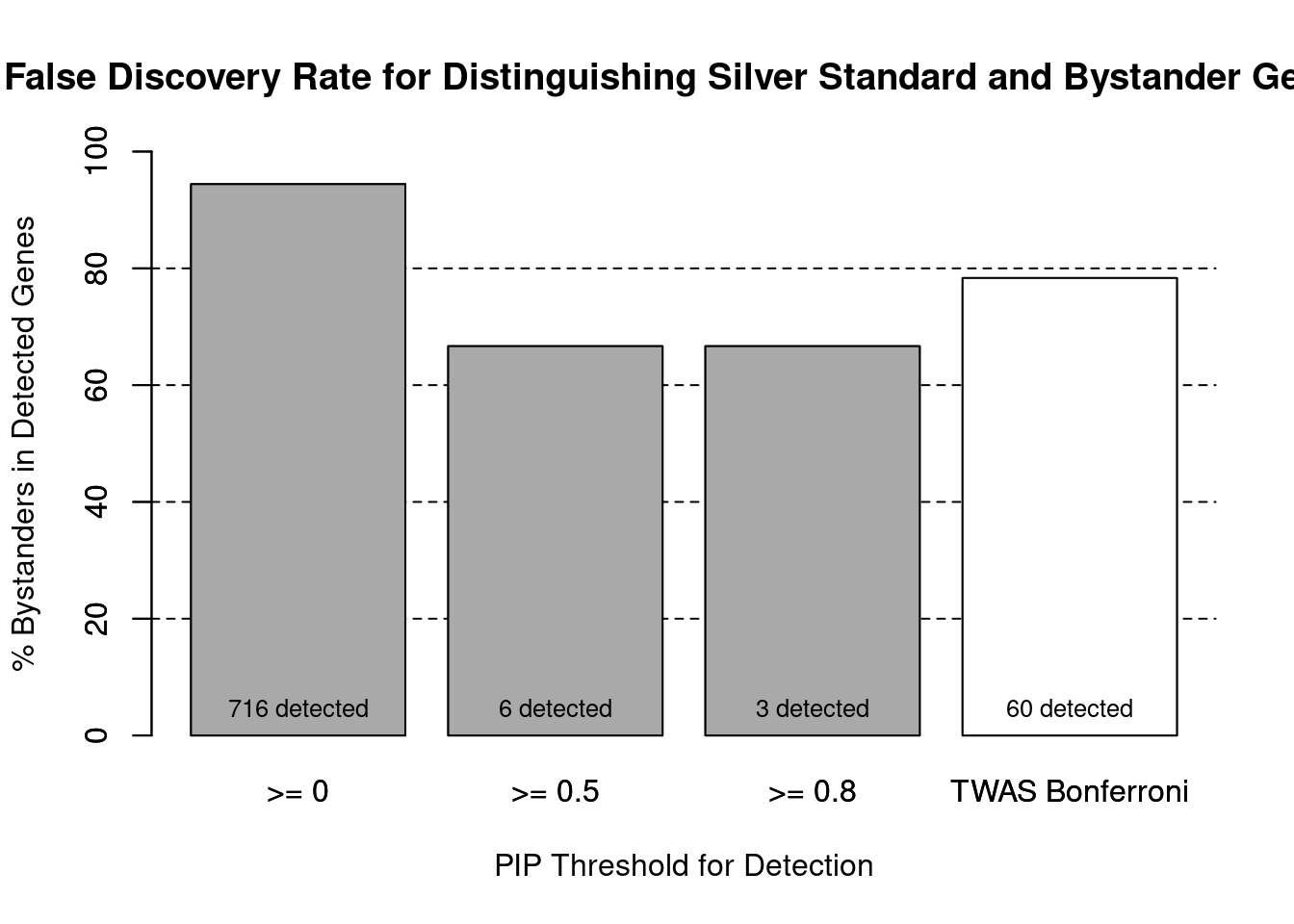
#text(x = xx, y = precision_range, label = paste0(round(precision_range,1), "%"), pos = 3, cex=0.8, offset = 1.5)Undetected silver standard genes have low TWAS z-scores or stronger signal from nearby variants
For all 69 silver standard genes, sequentially bin each gene using the following criteria: 1) gene not imputed; 2) gene detected by cTWAS at PIP>0.8; 3) gene insignificant by TWAS; 4) gene nearby a detected silver standard gene; 5) gene nearby a detected bystander gene; 6) gene nearby a detected SNP; 7) inconclusive.
#reload silver standard genes
known_annotations <- read_xlsx("data/summary_known_genes_annotations.xlsx", sheet="LDL")New names:
* `` -> ...4
* `` -> ...5known_annotations <- unique(known_annotations$`Gene Symbol`)
#categorize silver standard genes by case
silver_standard_case <- c()
uncertain_regions <- matrix(NA, 0, 2)
for (i in 1:length(known_annotations)){
current_gene <- known_annotations[i]
if (current_gene %in% ctwas_gene_res$genename) {
if (ctwas_gene_res$susie_pip[ctwas_gene_res$genename == current_gene] > 0.8){
silver_standard_case <- c(silver_standard_case, "Detected (PIP > 0.8)")
} else {
if (abs(ctwas_gene_res$z[ctwas_gene_res$genename == current_gene]) < sig_thresh){
silver_standard_case <- c(silver_standard_case, "Insignificant z-score")
} else {
current_region <- ctwas_gene_res$region_tag[ctwas_gene_res$genename == current_gene]
current_gene_res <- ctwas_gene_res[ctwas_gene_res$region_tag==current_region,]
current_snp_res <- ctwas_snp_res[ctwas_snp_res$region_tag==current_region,]
if (any(current_gene_res$susie_pip>0.8)){
if (any(current_gene_res$genename[current_gene_res$susie_pip>0.8] %in% known_annotations)){
silver_standard_case <- c(silver_standard_case, "Nearby Silver Standard Gene")
} else {
silver_standard_case <- c(silver_standard_case, "Nearby Bystander Gene")
}
} else {
#if (any(current_snp_res$susie_pip>0.8)){
if (sum(current_snp_res$susie_pip)>0.8){
silver_standard_case <- c(silver_standard_case, "Nearby SNP(s)")
} else {
silver_standard_case <- c(silver_standard_case, "Inconclusive")
uncertain_regions <- rbind(uncertain_regions, c(current_gene, ctwas_gene_res$region_tag[ctwas_gene_res$genename == current_gene]))
print(c(current_gene, ctwas_gene_res$region_tag[ctwas_gene_res$genename == current_gene]))
}
}
}
}
} else {
silver_standard_case <- c(silver_standard_case, "Not Imputed")
}
}
names(silver_standard_case) <- known_annotations
#table of outcomes for silver standard genes
-sort(-table(silver_standard_case))silver_standard_case
Not Imputed Insignificant z-score Nearby SNP(s)
29 27 11
Detected (PIP > 0.8) Nearby Bystander Gene
1 1 #show inconclusive genes
silver_standard_case[silver_standard_case=="Inconclusive"]named character(0)# for (i in 1:nrow(uncertain_regions)){
# locus_plot3(uncertain_regions[i,2], focus=uncertain_regions[i,1])
# }
#pie chart of outcomes for silver standard genes
df <- data.frame(-sort(-table(silver_standard_case)))
names(df) <- c("Outcome", "Frequency")
#df <- df[df$Outcome!="Not Imputed",] #exclude genes not imputed
df$Outcome <- droplevels(df$Outcome) #exclude genes not imputed
bp<- ggplot(df, aes(x=Outcome, y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity", position=position_dodge()) +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + theme(legend.position = "none")
bp
pie <- ggplot(df, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
# locus_plot3(focus="KPNB1", region_tag="17_27")
# locus_plot3(focus="LPIN3", region_tag="20_25")
# locus_plot3(focus="LIPC", region_tag="15_26")cTWAS detects some genes that are not significant using a stringent TWAS threshold
TTC39B is a member of the Dyslipidaemia term in the disease_GLAD4U. This gene was not included in our silver standard. This gene is not significant using TWAS but is detected by cTWAS.
#locus_plot3(focus="TTC39B", region_tag="9_13")
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] ctwas_0.1.31 forcats_0.4.0 stringr_1.4.0
[4] dplyr_1.0.7 purrr_0.3.4 readr_1.4.0
[7] tidyr_1.1.0 tidyverse_1.3.0 tibble_3.1.2
[10] GenomicRanges_1.36.0 GenomeInfoDb_1.20.0 IRanges_2.18.1
[13] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[16] readxl_1.3.1 WebGestaltR_0.4.4 disgenet2r_0.99.2
[19] enrichR_3.0 cowplot_1.0.0 ggplot2_3.3.3
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_1.4-1 rjson_0.2.20
[4] ellipsis_0.3.2 rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.3.1 rstudioapi_0.10 farver_2.1.0
[10] ggrepel_0.8.1 bit64_4.0.5 AnnotationDbi_1.46.0
[13] fansi_0.5.0 lubridate_1.7.4 xml2_1.3.2
[16] logging_0.10-108 codetools_0.2-16 doParallel_1.0.16
[19] cachem_1.0.5 knitr_1.23 jsonlite_1.6
[22] workflowr_1.6.2 apcluster_1.4.8 Cairo_1.5-12.2
[25] broom_0.7.9 dbplyr_1.4.2 compiler_3.6.1
[28] httr_1.4.1 backports_1.1.4 assertthat_0.2.1
[31] Matrix_1.2-18 fastmap_1.1.0 cli_3.0.1
[34] later_0.8.0 htmltools_0.3.6 prettyunits_1.0.2
[37] tools_3.6.1 igraph_1.2.4.1 gtable_0.3.0
[40] glue_1.4.2 GenomeInfoDbData_1.2.1 reshape2_1.4.3
[43] doRNG_1.8.2 Rcpp_1.0.6 Biobase_2.44.0
[46] cellranger_1.1.0 vctrs_0.3.8 svglite_1.2.2
[49] iterators_1.0.13 xfun_0.8 rvest_0.3.5
[52] lifecycle_1.0.0 rngtools_1.5 XML_3.98-1.20
[55] zlibbioc_1.30.0 scales_1.1.0 hms_1.1.0
[58] promises_1.0.1 yaml_2.2.0 curl_3.3
[61] memoise_2.0.0 ggrastr_0.2.3 gdtools_0.1.9
[64] stringi_1.4.3 RSQLite_2.2.7 foreach_1.5.1
[67] rlang_0.4.11 pkgconfig_2.0.3 bitops_1.0-6
[70] evaluate_0.14 lattice_0.20-38 labeling_0.3
[73] bit_4.0.4 tidyselect_1.1.0 plyr_1.8.4
[76] magrittr_2.0.1 R6_2.5.0 generics_0.0.2
[79] DBI_1.1.1 pgenlibr_0.3.1 pillar_1.6.1
[82] haven_2.3.1 whisker_0.3-2 withr_2.4.1
[85] RCurl_1.98-1.1 modelr_0.1.8 crayon_1.4.1
[88] utf8_1.2.1 rmarkdown_1.13 progress_1.2.2
[91] grid_3.6.1 data.table_1.14.0 blob_1.2.1
[94] git2r_0.26.1 reprex_0.3.0 digest_0.6.20
[97] httpuv_1.5.1 munsell_0.5.0 beeswarm_0.2.3
[100] vipor_0.4.5