White blood cell (leukocyte) count - all weights
wesleycrouse
2022-02-28
Last updated: 2022-04-12
Checks: 7 0
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 107bb6d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
working directory clean
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-d-30000_irnt_allweights.Rmd) and HTML (docs/ukb-d-30000_irnt_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 107bb6d | wesleycrouse | 2022-04-12 | gene set enrichment for supplied gene sets |
| html | 95e0f8e | wesleycrouse | 2022-04-07 | scroll bar |
| Rmd | a4575d7 | wesleycrouse | 2022-04-07 | formating |
| Rmd | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | f7e9822 | wesleycrouse | 2022-04-06 | testing subsections |
| Rmd | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | d0f6e53 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| Rmd | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| html | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| html | b999e70 | wesleycrouse | 2022-04-04 | fixing display of results |
| html | dd02af5 | wesleycrouse | 2022-04-04 | kegg |
| html | 34ca036 | wesleycrouse | 2022-04-04 | kegg for individual tissues |
| Rmd | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | 364b716 | wesleycrouse | 2022-04-02 | TWAS FP based on confidence sets |
| Rmd | c9809d4 | wesleycrouse | 2022-04-01 | additional TWAS FP analyses |
| html | a9dcb4d | wesleycrouse | 2022-04-01 | alternative TWAS FP figures based on confidence sets |
| Rmd | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| html | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| Rmd | 1c45bb5 | wesleycrouse | 2022-03-30 | fixing formatting |
| html | 1c45bb5 | wesleycrouse | 2022-03-30 | fixing formatting |
| Rmd | ad4604a | wesleycrouse | 2022-03-30 | Adding numbers to results |
| html | ad4604a | wesleycrouse | 2022-03-30 | Adding numbers to results |
| html | 18f0b45 | wesleycrouse | 2022-03-24 | format |
| Rmd | 16289f6 | wesleycrouse | 2022-03-24 | improving layout |
| Rmd | 8397beb | wesleycrouse | 2022-03-24 | format |
| html | 8397beb | wesleycrouse | 2022-03-24 | format |
| html | 27e1022 | wesleycrouse | 2022-03-24 | layout |
| html | 85641ef | wesleycrouse | 2022-03-24 | layout |
| Rmd | 717e77e | wesleycrouse | 2022-03-23 | adjusting heatmaps |
| html | 717e77e | wesleycrouse | 2022-03-23 | adjusting heatmaps |
| html | ae26765 | wesleycrouse | 2022-03-23 | plots |
| html | aefd338 | wesleycrouse | 2022-03-23 | adjusting heatmap |
| html | 64ee362 | wesleycrouse | 2022-03-23 | adjusting gene-tissue heatmap |
| html | b5e392d | wesleycrouse | 2022-03-23 | tables |
| Rmd | 1b1fcaf | wesleycrouse | 2022-03-23 | chart for tissue specificity |
| Rmd | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | bbf031d | wesleycrouse | 2022-03-23 | adjusting sections |
| Rmd | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | 60f39e6 | wesleycrouse | 2022-03-22 | additional traits |
| Rmd | 073f2a3 | wesleycrouse | 2022-03-22 | enrichment analysis for all weights |
| Rmd | ba908fe | wesleycrouse | 2022-03-21 | more traits for all weight analysis |
options(width=1000)trait_id <- "ukb-d-30000_irnt"
trait_name <- "White blood cell (leukocyte) count"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions and effects
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
# ctwas_res$region_cs_tag <- paste(ctwas_res$region_tag, ctwas_res$cs_index, sep="_")
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes_index <- ctwas_gene_res$susie_pip>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_genes_index]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#
# twas_genes_index <- abs(ctwas_gene_res$z) > sig_thresh
# twas_genes <- ctwas_gene_res$genename[twas_genes_index]
#
# #gene PIPs and z scores
# gene_pips <- ctwas_gene_res[,c("genename", "region_tag", "susie_pip", "z", "region_cs_tag")]
#
# #total PIPs by region
# regions <- unique(ctwas_gene_res$region_tag)
# region_pips <- data.frame(region=regions, stringsAsFactors=F)
# region_pips$gene_pip <- sapply(regions, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_tag==x])})
# region_pips$snp_pip <- sapply(regions, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_tag==x])})
# region_pips$snp_maxz <- sapply(regions, function(x){max(abs(ctwas_snp_res$z[ctwas_snp_res$region_tag==x]))})
#
# #total PIPs by causal set
# regions_cs <- unique(ctwas_gene_res$region_cs_tag)
# region_cs_pips <- data.frame(region_cs=regions_cs, stringsAsFactors=F)
# region_cs_pips$gene_pip <- sapply(regions_cs, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_cs_tag==x])})
# region_cs_pips$snp_pip <- sapply(regions_cs, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_cs_tag==x])})
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes,
# gene_pips=gene_pips,
# region_pips=region_pips,
# sig_thresh=sig_thresh,
# region_cs_pips=region_cs_pips)
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL,
stringsAsFactors=F)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)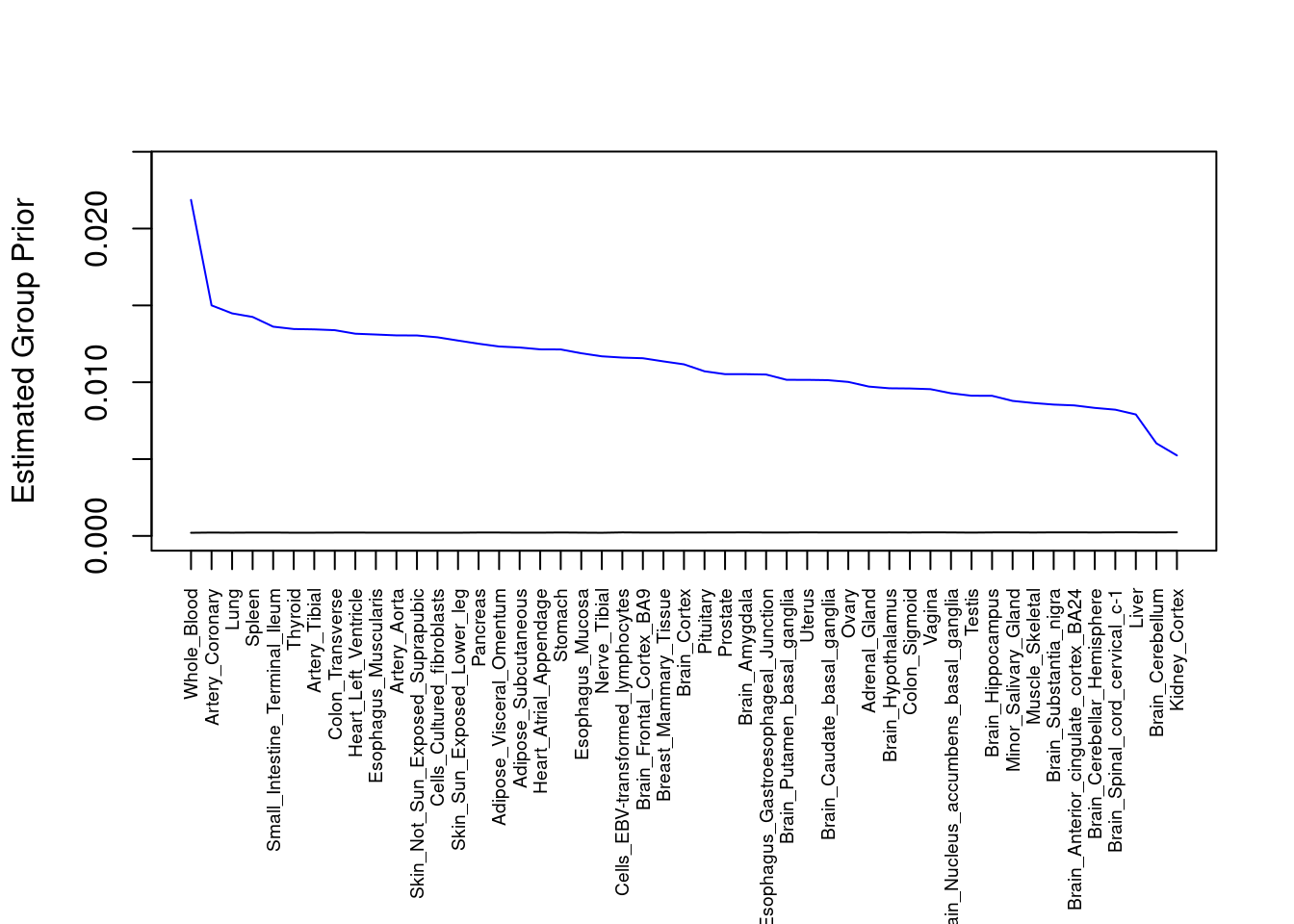
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)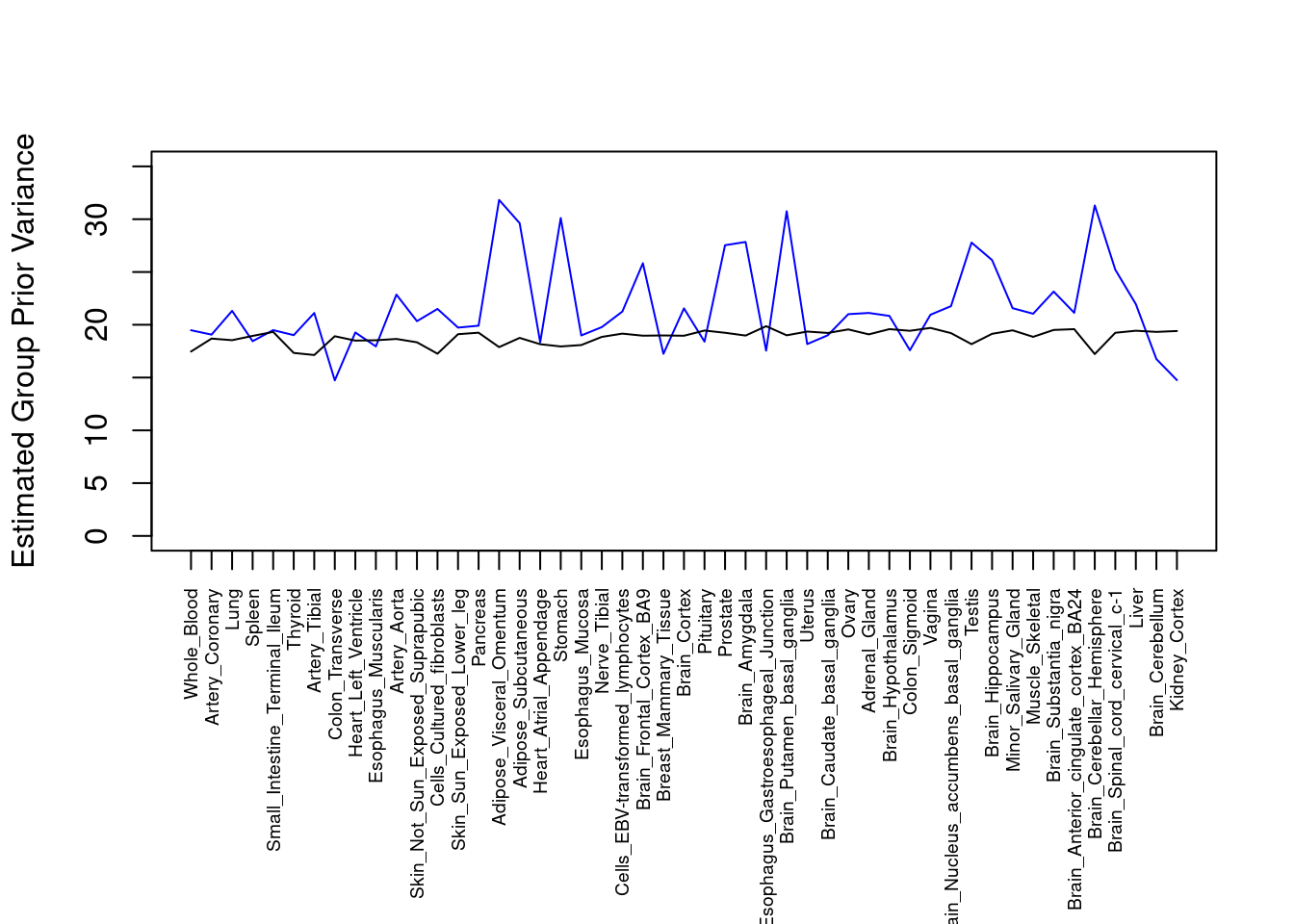
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)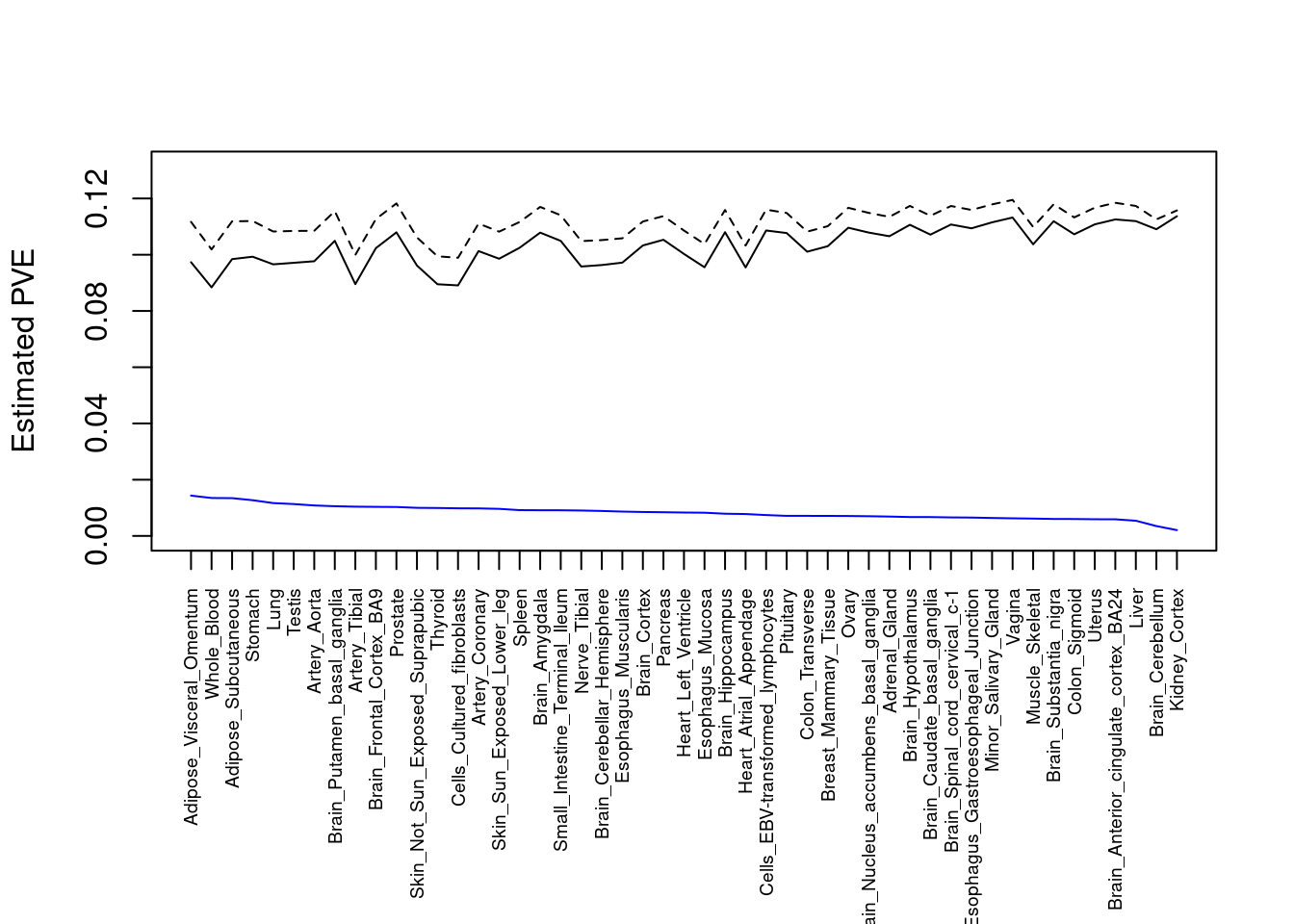
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")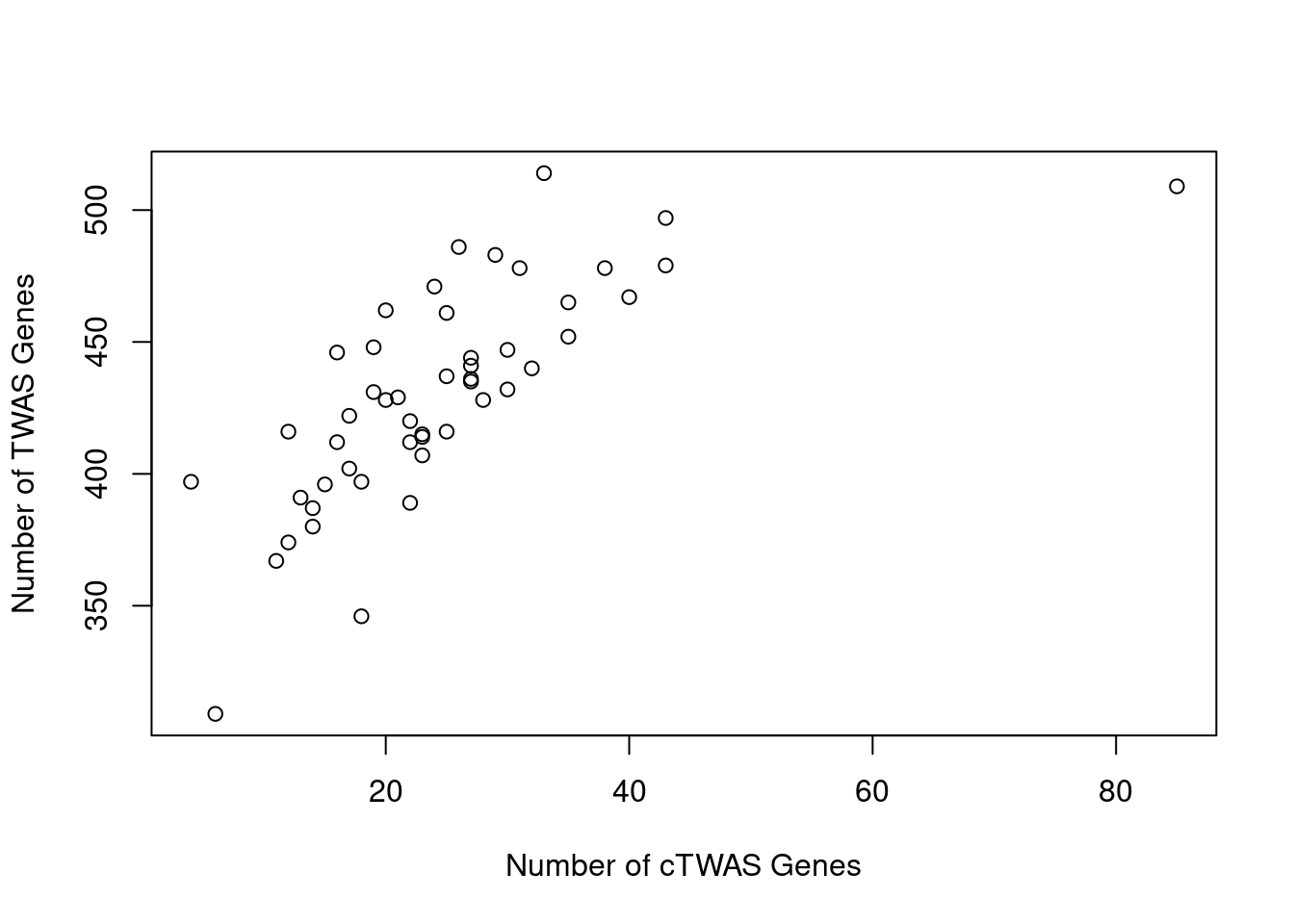
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 397#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 1822Enrichment analysis for cTWAS genes
GO
#enrichment for cTWAS genes using enrichR
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of immune response (GO:0050776) 13/179 0.04385930 LYN;IFITM1;SIGLEC9;ITGB2;HLA-C;ITGAL;TIRAP;FCRLB;JAML;SELL;IRF4;CLEC4G;CD33
2 positive regulation of cytokine-mediated signaling pathway (GO:0001961) 6/36 0.04385930 CSF1;IL7;NLRC5;LAPTM5;CXCR4;AGPAT1
3 lymphocyte mediated immunity (GO:0002449) 4/13 0.04385930 CD70;BTN3A2;CD46;MYO1G
4 neutrophil mediated immunity (GO:0002446) 23/488 0.04385930 KMT2E;SIGLEC9;FCER1G;SLC44A2;ITGB2;HLA-C;ADAM10;PTPRJ;PYGL;ITGAL;ARHGAP45;CNN2;TSPAN14;LAMP1;GM2A;CXCR1;SELL;RAB37;COTL1;BPI;OSTF1;ELANE;CD33
5 amyloid precursor protein catabolic process (GO:0042987) 4/14 0.04385930 ADAM19;PSEN2;ADAM10;ABCA7
6 positive regulation of cytokine production (GO:0001819) 18/335 0.04385930 FCER1G;CRCP;LAPTM5;PIK3CD;BTN3A2;AGPAT1;SCIMP;TIRAP;SCAMP5;IL7;IRF4;PLCG2;HLA-DPB1;POLR2E;CD46;ZFPM1;ELANE;SPTBN1
7 positive regulation of I-kappaB phosphorylation (GO:1903721) 3/6 0.04385930 CX3CR1;PLCG2;AKT1
8 phosphatidylinositol phosphate biosynthetic process (GO:0046854) 6/42 0.04385930 ITPKC;SMG1;TMEM150A;PIK3CD;PIK3R3;IP6K1
9 phosphatidylinositol biosynthetic process (GO:0006661) 10/126 0.04385930 ITPKC;SMG1;MTMR3;TMEM150A;MTMR12;PLEKHA1;PLCG2;PIK3R3;PIK3CD;IP6K1
10 T cell mediated immunity (GO:0002456) 4/16 0.04385930 CD70;BTN3A2;CD46;MYO1G
11 neutrophil activation (GO:0042119) 4/16 0.04385930 KMT2E;CXCL6;PREX1;FCER1G
12 polyol biosynthetic process (GO:0046173) 4/16 0.04385930 CYP27B1;ITPKC;ACER3;IP6K1
13 regulation of I-kappaB phosphorylation (GO:1903719) 3/7 0.04385930 CX3CR1;PLCG2;AKT1
14 epithelial cell differentiation involved in kidney development (GO:0035850) 3/7 0.04385930 SALL1;PAX8;MTSS1
15 neutrophil degranulation (GO:0043312) 22/481 0.04385930 SIGLEC9;FCER1G;SLC44A2;ITGB2;HLA-C;ADAM10;PTPRJ;PYGL;ITGAL;ARHGAP45;CNN2;TSPAN14;LAMP1;GM2A;CXCR1;SELL;RAB37;COTL1;BPI;OSTF1;ELANE;CD33
16 neutrophil activation involved in immune response (GO:0002283) 22/485 0.04385930 SIGLEC9;FCER1G;SLC44A2;ITGB2;HLA-C;ADAM10;PTPRJ;PYGL;ITGAL;ARHGAP45;CNN2;TSPAN14;LAMP1;GM2A;CXCR1;SELL;RAB37;COTL1;BPI;OSTF1;ELANE;CD33
17 regulation of neuron death (GO:1901214) 8/86 0.04385930 DAXX;REST;CSF1;ITGB2;AKT1;CHP1;MAPT;MAP3K5
18 regulation of cell population proliferation (GO:0042127) 30/764 0.04385930 ACVRL1;CSF3;IFITM1;CSF1;FLT3;DOT1L;NPR3;PTPRJ;DLL1;ZFP36L1;DPP4;SLC9A3R1;CYP27B1;PDF;AKT1;TSPO;S1PR2;SH2B3;CD33;LYN;CDKN2D;CADM4;CDKN2A;OSGIN1;ADAM10;SOX11;REST;FES;IL7;ACER3
19 positive regulation of cellular process (GO:0048522) 26/625 0.04385930 CSF3;CSF1;FLT3;DOT1L;PTPRJ;AGPAT1;DLL1;CXCL16;ZFP36L1;DPP4;PDF;RPS6KA1;AKT1;S1PR2;TGM2;LYN;FCER1G;CAV1;ADAM10;SOX11;ABCA7;PTK6;TAL1;IL7;ACER3;MAPT
20 amyloid precursor protein metabolic process (GO:0042982) 4/18 0.04406692 ADAM19;ACHE;PSEN2;ADAM10
21 negative regulation of interleukin-8 production (GO:0032717) 4/18 0.04406692 BPI;CD33;ELANE;MAP2K5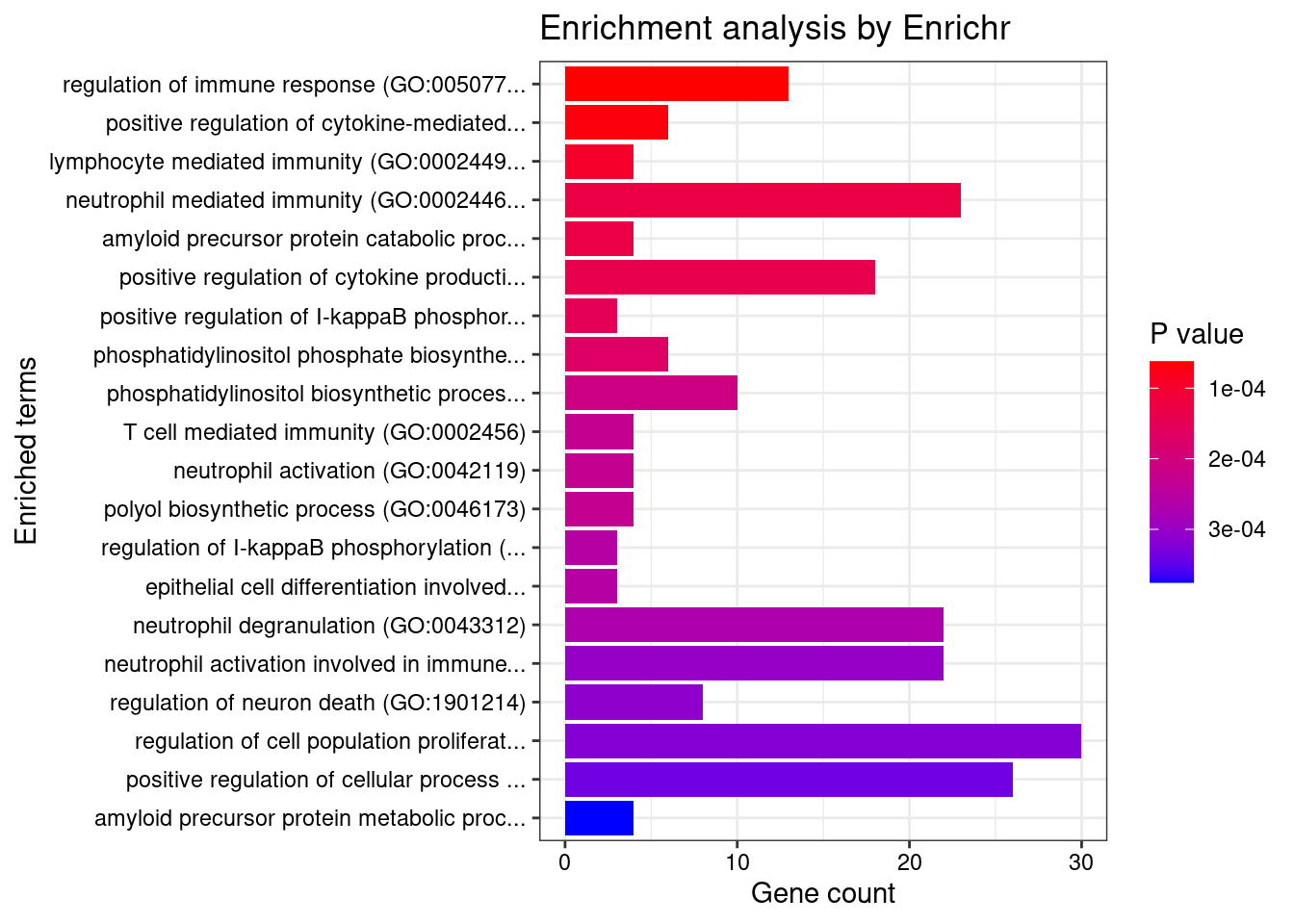
GO_Cellular_Component_2021
Term Overlap Adjusted.P.value Genes
1 secretory granule membrane (GO:0030667) 16/274 0.01391569 SIGLEC9;FCER1G;SLC44A2;ITGB2;HLA-C;ADAM10;PTPRJ;ITPR3;ITGAL;TSPAN14;CXCR1;LAMP1;SELL;RAB37;SCG3;CD33
2 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556) 5/28 0.01391569 SPPL2C;CANX;HLA-DPB1;HLA-C;TAPBP
3 lumenal side of endoplasmic reticulum membrane (GO:0098553) 5/28 0.01391569 SPPL2C;CANX;HLA-DPB1;HLA-C;TAPBP
4 specific granule (GO:0042581) 11/160 0.01870076 CNN2;TSPAN14;RAB37;SLC44A2;ITGB2;ADAM10;PTPRJ;BPI;ITGAL;ELANE;CD33
5 specific granule membrane (GO:0035579) 8/91 0.01871751 TSPAN14;SLC44A2;RAB37;ITGB2;ADAM10;PTPRJ;ITGAL;CD33
6 focal adhesion (GO:0005925) 18/387 0.02696703 MAP2K2;ITGA2;CAV1;ITGB2;ADAM10;ADD1;RAB21;ENAH;CNN2;DPP4;UBOX5;MPRIP;FES;CHP1;TADA1;CD46;MAP4K4;TGM2
7 cell-substrate junction (GO:0030055) 18/394 0.02835393 MAP2K2;ITGA2;CAV1;ITGB2;ADAM10;ADD1;RAB21;ENAH;CNN2;DPP4;UBOX5;MPRIP;FES;CHP1;TADA1;CD46;MAP4K4;TGM2
8 bounding membrane of organelle (GO:0098588) 28/767 0.03917847 SIGLEC9;FLT3;SPPL2C;VPS26A;ITPR3;FURIN;HS6ST1;GLG1;SCAMP5;LAMP1;CXCR1;ST3GAL4;CHP1;ST3GAL6;AP1M2;CSGALNACT1;CAV1;GALNT1;HLA-C;F5;TAPBP;TPST2;TPST1;SELL;HLA-DPB1;SCG3;SHANK3;CD320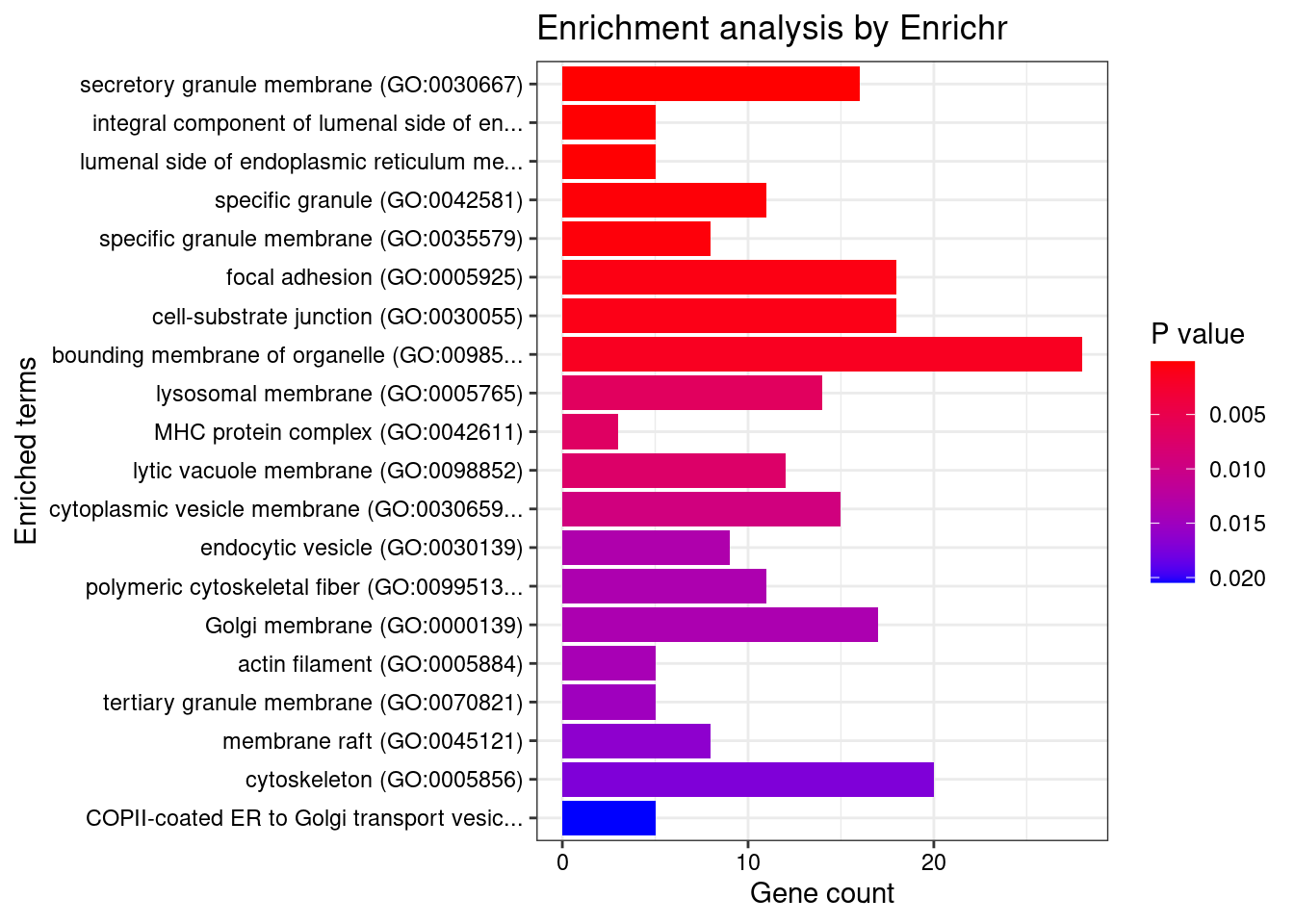
GO_Molecular_Function_2021
Term Overlap Adjusted.P.value Genes
1 C-C chemokine binding (GO:0019957) 6/24 0.002371778 CX3CR1;CXCR1;CXCR4;ACKR3;CCR8;ACKR1
2 chemokine binding (GO:0019956) 6/32 0.005086487 CX3CR1;CXCR1;CXCR4;CCR8;ACKR3;ACKR1
3 C-C chemokine receptor activity (GO:0016493) 5/23 0.005086487 CX3CR1;CXCR1;CXCR4;ACKR3;CCR8
4 C-X-C chemokine receptor activity (GO:0016494) 3/5 0.005086487 CXCR1;ACKR3;CXCR4
5 creatine kinase activity (GO:0004111) 3/5 0.005086487 CKMT1A;CKB;MAP4K4
6 phosphotransferase activity, nitrogenous group as acceptor (GO:0016775) 3/5 0.005086487 CKMT1A;CKB;MAP4K4
7 chemokine receptor activity (GO:0004950) 5/26 0.008107961 CX3CR1;CXCR1;CXCR4;CCR8;ACKR3
8 kinase activity (GO:0016301) 9/112 0.020098510 LYN;ITPKC;CKMT1A;AKT1;PIK3CD;COQ8B;CKB;IP6K1;MAP4K4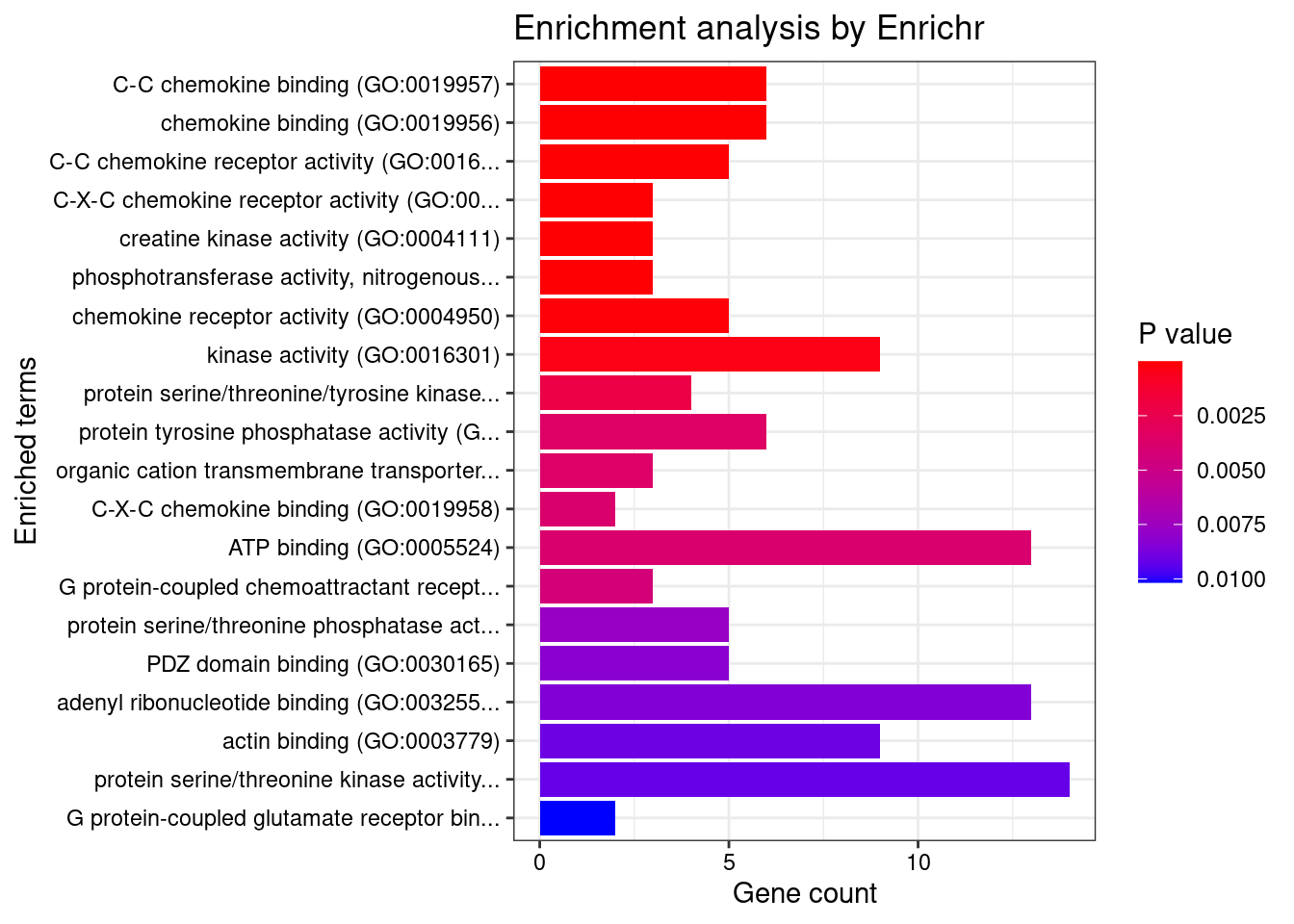
KEGG
#enrichment for cTWAS genes using KEGG
library(WebGestaltR)******************************************* ** Welcome to WebGestaltR ! ** *******************************************background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#listGeneSet()
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...enrichResult[,c("description", "size", "overlap", "FDR", "userId")] description size overlap FDR userId
1 Cytokine-cytokine receptor interaction 253 18 0.04827672 CSF1;IL1RN;CXCL6;CSF3;TNFRSF10A;CXCL16;ACVRL1;CXCR4;TNFSF12;CSF3R;ACKR3;CX3CR1;CXCL2;TNFRSF10C;IL7;CD70;CXCR1;CCR8DisGeNET
#enrichment for cTWAS genes using DisGeNET
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <- disease_enrichment(entities=ctwas_genes, vocabulary = "HGNC", database = "CURATED")RP4-621B10.8 gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.2 gene(s) from the input list not found in DisGeNET CURATEDNUDC gene(s) from the input list not found in DisGeNET CURATEDAPOM gene(s) from the input list not found in DisGeNET CURATEDC18orf25 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDLINC00094 gene(s) from the input list not found in DisGeNET CURATEDFAM102A gene(s) from the input list not found in DisGeNET CURATEDC15orf40 gene(s) from the input list not found in DisGeNET CURATEDTMCC3 gene(s) from the input list not found in DisGeNET CURATEDAC005754.8 gene(s) from the input list not found in DisGeNET CURATEDRP11-45M22.5 gene(s) from the input list not found in DisGeNET CURATEDRP11-387H17.6 gene(s) from the input list not found in DisGeNET CURATEDSUN2 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDBPI gene(s) from the input list not found in DisGeNET CURATEDMEX3A gene(s) from the input list not found in DisGeNET CURATEDIP6K1 gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDSLFN13 gene(s) from the input list not found in DisGeNET CURATEDST3GAL6 gene(s) from the input list not found in DisGeNET CURATEDNECAB2 gene(s) from the input list not found in DisGeNET CURATEDHOXC6 gene(s) from the input list not found in DisGeNET CURATEDKIAA0391 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDCTC-492K19.7 gene(s) from the input list not found in DisGeNET CURATEDNAA38 gene(s) from the input list not found in DisGeNET CURATEDAP1M2 gene(s) from the input list not found in DisGeNET CURATEDZNF311 gene(s) from the input list not found in DisGeNET CURATEDUQCRC1 gene(s) from the input list not found in DisGeNET CURATEDGPR4 gene(s) from the input list not found in DisGeNET CURATEDCSNK1G1 gene(s) from the input list not found in DisGeNET CURATEDPREX1 gene(s) from the input list not found in DisGeNET CURATEDTADA1 gene(s) from the input list not found in DisGeNET CURATEDPAPOLA gene(s) from the input list not found in DisGeNET CURATEDPCDHB3 gene(s) from the input list not found in DisGeNET CURATEDVWA7 gene(s) from the input list not found in DisGeNET CURATEDC6orf47 gene(s) from the input list not found in DisGeNET CURATEDCDK18 gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDMARCH7 gene(s) from the input list not found in DisGeNET CURATEDSLC5A11 gene(s) from the input list not found in DisGeNET CURATEDSPPL2C gene(s) from the input list not found in DisGeNET CURATEDTRMT12 gene(s) from the input list not found in DisGeNET CURATEDSENP3 gene(s) from the input list not found in DisGeNET CURATEDRNF2 gene(s) from the input list not found in DisGeNET CURATEDTTLL12 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDDDX60L gene(s) from the input list not found in DisGeNET CURATEDDHRS12 gene(s) from the input list not found in DisGeNET CURATEDCNOT11 gene(s) from the input list not found in DisGeNET CURATEDCCDC9 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDKANK3 gene(s) from the input list not found in DisGeNET CURATEDMICALL2 gene(s) from the input list not found in DisGeNET CURATEDFAM177A1 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDFRAT1 gene(s) from the input list not found in DisGeNET CURATEDARAP2 gene(s) from the input list not found in DisGeNET CURATEDZFP36L1 gene(s) from the input list not found in DisGeNET CURATEDC5orf64-AS1 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDPIK3R3 gene(s) from the input list not found in DisGeNET CURATEDAC144831.3 gene(s) from the input list not found in DisGeNET CURATEDKIAA1755 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDSLC45A4 gene(s) from the input list not found in DisGeNET CURATEDGLG1 gene(s) from the input list not found in DisGeNET CURATEDGSDMA gene(s) from the input list not found in DisGeNET CURATEDRP4-796I8.1 gene(s) from the input list not found in DisGeNET CURATEDLRRC2 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDCYB561D1 gene(s) from the input list not found in DisGeNET CURATEDRPN1 gene(s) from the input list not found in DisGeNET CURATEDZFPM1 gene(s) from the input list not found in DisGeNET CURATEDEMILIN3 gene(s) from the input list not found in DisGeNET CURATEDHLA-DOB gene(s) from the input list not found in DisGeNET CURATEDDNAH10OS gene(s) from the input list not found in DisGeNET CURATEDGALNT1 gene(s) from the input list not found in DisGeNET CURATEDCADM4 gene(s) from the input list not found in DisGeNET CURATEDSLFN12 gene(s) from the input list not found in DisGeNET CURATEDCDC42BPA gene(s) from the input list not found in DisGeNET CURATEDVKORC1L1 gene(s) from the input list not found in DisGeNET CURATEDRP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDWIPF2 gene(s) from the input list not found in DisGeNET CURATEDBTN2A2 gene(s) from the input list not found in DisGeNET CURATEDFAM212A gene(s) from the input list not found in DisGeNET CURATEDADAM23 gene(s) from the input list not found in DisGeNET CURATEDTBC1D14 gene(s) from the input list not found in DisGeNET CURATEDDUSP14 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDCA11 gene(s) from the input list not found in DisGeNET CURATEDOSCP1 gene(s) from the input list not found in DisGeNET CURATEDTMEM150A gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDGPANK1 gene(s) from the input list not found in DisGeNET CURATEDPDF gene(s) from the input list not found in DisGeNET CURATEDCRCP gene(s) from the input list not found in DisGeNET CURATEDLINC00638 gene(s) from the input list not found in DisGeNET CURATEDRABEP1 gene(s) from the input list not found in DisGeNET CURATEDLINC01184 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATEDCDK11B gene(s) from the input list not found in DisGeNET CURATEDRP11-107M16.2 gene(s) from the input list not found in DisGeNET CURATEDPDCD7 gene(s) from the input list not found in DisGeNET CURATEDTUBG2 gene(s) from the input list not found in DisGeNET CURATEDCDC37 gene(s) from the input list not found in DisGeNET CURATEDRP11-50B3.2 gene(s) from the input list not found in DisGeNET CURATEDGGT5 gene(s) from the input list not found in DisGeNET CURATEDORAOV1 gene(s) from the input list not found in DisGeNET CURATEDPOLR2E gene(s) from the input list not found in DisGeNET CURATEDRP5-894D12.5 gene(s) from the input list not found in DisGeNET CURATEDFGF11 gene(s) from the input list not found in DisGeNET CURATEDRPUSD4 gene(s) from the input list not found in DisGeNET CURATEDBORCS7 gene(s) from the input list not found in DisGeNET CURATEDPRR15L gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDFAM69A gene(s) from the input list not found in DisGeNET CURATEDRP11-738E22.3 gene(s) from the input list not found in DisGeNET CURATEDTSPAN14 gene(s) from the input list not found in DisGeNET CURATEDSIGLEC9 gene(s) from the input list not found in DisGeNET CURATEDSLC2A6 gene(s) from the input list not found in DisGeNET CURATEDRAP1GAP2 gene(s) from the input list not found in DisGeNET CURATEDMYO1G gene(s) from the input list not found in DisGeNET CURATEDNFE2L1 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDSWT1 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDSUMO3 gene(s) from the input list not found in DisGeNET CURATEDCD302 gene(s) from the input list not found in DisGeNET CURATEDTPST2 gene(s) from the input list not found in DisGeNET CURATEDGUCY1B3 gene(s) from the input list not found in DisGeNET CURATEDRP11-442O1.3 gene(s) from the input list not found in DisGeNET CURATEDJAML gene(s) from the input list not found in DisGeNET CURATEDLINC01624 gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDTADA2A gene(s) from the input list not found in DisGeNET CURATEDTPST1 gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDBAZ2B gene(s) from the input list not found in DisGeNET CURATEDZNF180 gene(s) from the input list not found in DisGeNET CURATEDRNF181 gene(s) from the input list not found in DisGeNET CURATEDCATSPER4 gene(s) from the input list not found in DisGeNET CURATEDLINC01623 gene(s) from the input list not found in DisGeNET CURATEDHDHD5 gene(s) from the input list not found in DisGeNET CURATEDZNF114 gene(s) from the input list not found in DisGeNET CURATEDSCIMP gene(s) from the input list not found in DisGeNET CURATEDDTNB gene(s) from the input list not found in DisGeNET CURATEDTTC39C gene(s) from the input list not found in DisGeNET CURATEDUBASH3B gene(s) from the input list not found in DisGeNET CURATEDRP11-556O15.1 gene(s) from the input list not found in DisGeNET CURATEDMTMR12 gene(s) from the input list not found in DisGeNET CURATEDCTC-498M16.4 gene(s) from the input list not found in DisGeNET CURATEDSCG3 gene(s) from the input list not found in DisGeNET CURATEDLRRC25 gene(s) from the input list not found in DisGeNET CURATEDCITED4 gene(s) from the input list not found in DisGeNET CURATEDFCRLB gene(s) from the input list not found in DisGeNET CURATEDTMEM99 gene(s) from the input list not found in DisGeNET CURATEDRP11-219G17.9 gene(s) from the input list not found in DisGeNET CURATEDWBP1L gene(s) from the input list not found in DisGeNET CURATEDCHP1 gene(s) from the input list not found in DisGeNET CURATEDCYB5D1 gene(s) from the input list not found in DisGeNET CURATEDRAB37 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDPDLIM2 gene(s) from the input list not found in DisGeNET CURATEDMED24 gene(s) from the input list not found in DisGeNET CURATEDTMED6 gene(s) from the input list not found in DisGeNET CURATEDOSTF1 gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDRP11-20J15.3 gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDUBALD2 gene(s) from the input list not found in DisGeNET CURATEDADGRF5 gene(s) from the input list not found in DisGeNET CURATEDRP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDRPAIN gene(s) from the input list not found in DisGeNET CURATEDif (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes=ctwas_genes)
#background is union of genes analyzed in all tissue
background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background]})
####################
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
hyp_score_padj gene_set nset ngenes percent padj
1 gwascatalog 5998 217 0.03617873 3.836319e-30
2 mgi_essential 2320 68 0.02931034 1.149336e-04
3 fda_approved_drug_targets 352 13 0.03693182 2.088890e-02
4 clinvar_path_likelypath 2788 61 0.02187948 8.520387e-02
5 core_essentials_hart 266 4 0.01503759 7.135378e-01Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 T cell receptor signaling pathway (GO:0050852) 37/158 0.0001103247 PIK3CD;PTPRJ;SPPL3;PSMB7;PSMD6;PSMD3;PLCG2;PDE4B;EIF2B1;HLA-DQA2;PAK2;HLA-DQA1;HLA-DPA1;NCK1;EIF2B4;HLA-DRB5;BTN2A2;BTN3A1;MOG;BTN2A1;BTN1A1;BTN3A3;HLA-A;BTNL2;BTN3A2;PSMB8;PSMB9;PSMA6;PSMC5;PTPRC;PSMC3;THEMIS2;HLA-DPB1;HLA-DRA;HLA-DRB1;HLA-DQB2;HLA-DQB1
2 antigen receptor-mediated signaling pathway (GO:0050851) 41/185 0.0001103247 BLK;PIK3CD;PTPRJ;SPPL3;CD79B;PSMB7;PSMD6;PSMD3;PLCG2;PDE4B;EIF2B1;HLA-DQA2;PAK2;HLA-DQA1;HLA-DPA1;NCK1;EIF2B4;HLA-DRB5;BTN2A2;BTN3A1;MOG;PLEKHA1;BTN2A1;BTN1A1;BTN3A3;HLA-A;BTNL2;BTN3A2;LIME1;PSMB8;PSMB9;PSMA6;PSMC5;PTPRC;PSMC3;THEMIS2;HLA-DPB1;HLA-DRA;HLA-DRB1;HLA-DQB2;HLA-DQB1
3 interferon-gamma-mediated signaling pathway (GO:0060333) 22/68 0.0001103247 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;TRIM8;IRF4;IRF1;HLA-DPB1;IRF8;HLA-DRA;TRIM26;TRIM38;TRIM31;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
4 positive regulation of cytokine production (GO:0001819) 59/335 0.0006945713 FCN1;CRCP;PIK3CD;BRCA1;TNF;SCAMP5;C1QTNF4;FLOT1;PDE4B;TRIM27;HAVCR2;HLA-DPA1;FCER1G;BTN3A1;SPHK2;IL13;HLA-A;MIF;BTN3A2;HLA-G;TIRAP;EREG;HLA-E;IL17RA;IL1A;IRF4;IL1B;IRF1;RARA;TRIM15;LTB;CD46;PLA2R1;ZFPM1;TMEM106A;ATF4;CEBPG;AGPAT1;AIF1;AGER;PLCG2;POLR2E;NLRP3;LURAP1;ELANE;SPTBN1;CREBBP;CARD9;GLMN;LAPTM5;PTPN11;SCIMP;CCDC88B;IL6;PTPRC;IL7;HLA-DPB1;HSPA1B;HSPA1A
5 cytokine-mediated signaling pathway (GO:0019221) 93/621 0.0009897813 IFITM3;CXCL6;CSF3;IFITM1;IL1RN;SPI1;CSF3R;IFITM2;TNFRSF6B;CXCL8;CSF1;FLT3;PIK3CD;PTPRJ;CXCL1;CXCL3;TNF;CXCL2;TRIM8;PSMD6;PSMD3;CFL1;LEPR;AKT1;TRIM26;CCR5;HLA-DPA1;FCER1G;IL13;HLA-B;HLA-C;TRAF1;HLA-A;MIF;TYK2;HLA-F;SIRT1;HLA-G;TNFRSF1A;EREG;HLA-E;IL17RA;PSMA6;IL1A;IRF4;IL1B;IRF1;BOLA2;CSNK2B;CANX;KIT;LTA;IRF8;LTB;CRK;HLA-DQB2;HLA-DQB1;ALOX15;CDC42;CNN2;PSMB7;UGCG;CXCR2;S1PR1;PTK2B;IL36RN;CD300LF;HLA-DQA2;SH2B3;PAK2;HLA-DQA1;MAPK3;MAP3K3;HLA-DRB5;IL37;LIF;IL36G;PTPN11;PSMB8;PSMB9;IL1F10;POMC;PSMC5;FER;IL6;PSMC3;IL7;HLA-DPB1;HLA-DRA;ACKR3;TRIM38;TRIM31;HLA-DRB1
6 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474) 13/33 0.0020521144 FCER1G;HFE;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-G;HLA-E;TAPBP;CANX;SEC31A
7 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway (GO:0002484) 6/7 0.0020521144 HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E
8 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent (GO:0002486) 6/7 0.0020521144 HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E
9 regulation of cytokine production (GO:0001817) 32/150 0.0020766693 PPP1R11;C5AR2;MAST2;PIK3CD;AGPAT1;TNF;SCAMP5;FLOT1;LURAP1;LYN;BTN2A2;BTN3A1;MOG;BTN2A1;BTN1A1;GLMN;CARD9;HLA-B;BTN3A3;MIF;BTNL2;BTN3A2;EREG;CCDC88B;IL1A;IL6;PTPRC;IRF8;ACKR1;PLA2R1;FCGR2B;HLA-DRB1
10 antigen processing and presentation of endogenous peptide antigen (GO:0002483) 8/14 0.0038076051 TAP2;TAP1;HLA-DRA;HLA-A;HLA-F;HLA-G;HLA-DRB1;HLA-E
11 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent (GO:0002480) 6/8 0.0049908668 HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E
12 antigen processing and presentation of exogenous peptide antigen (GO:0002478) 24/103 0.0049908668 HLA-DRB5;FCER1G;CLTC;HLA-A;HLA-F;KLC1;HLA-E;ACTR1A;HLA-DMA;HLA-DMB;CANX;HLA-DPB1;HLA-DRA;HLA-DOA;FCGR2B;HLA-DOB;HLA-DQA2;AP1M2;HLA-DQA1;HLA-DRB1;HLA-DQB2;SEC31A;HLA-DPA1;HLA-DQB1
13 regulation of telomere capping (GO:1904353) 10/23 0.0049908668 PKIB;RTEL1;USP7;RAD50;TNKS2;ATM;MAPKAPK5;AURKB;SMG6;MAPK3
14 regulation of gene expression (GO:0010468) 138/1079 0.0075146342 SPI1;CSF1;PRDM6;HFE;JMJD1C;TNF;AKT1;TRIM26;DIP2A;TRIM27;SOX7;FADS1;HOXA5;MED1;DAXX;MAP2K2;ZNF282;BTN1A1;ADAM10;SOX11;MIF;ATG14;HIC1;OLFM1;KAT6B;ERP29;TRIM15;ZSCAN26;ADTRP;EZR;TRIM10;ATF4;ZNF398;DHH;PIK3R3;CREBL2;NKAPL;ZFP36L1;NPAT;SBNO1;ATXN1;TERT;PLCG2;ZKSCAN5;APOE;ZNF423;ZNF664;RREB1;WNT3;WNT4;LYN;CPSF4;CREBBP;NDFIP1;BTN2A2;BTN2A1;GLMN;IL36G;TRIP4;SNF8;BTNL2;LARP4;IL1F10;DRAP1;MLXIPL;IL6;OCLN;PTPRC;HNRNPK;REL;ATM;SLC2A4RG;PHF19;ACVRL1;LIN54;GSK3B;GSK3A;CXCL8;CELF1;PHF1;CHD7;PRKAG1;PIK3CD;BRCA1;LIMD1;ATRAID;VSIR;SPOUT1;HINT1;SUFU;MLX;HMG20A;EMILIN1;HAVCR2;TCF19;KDM6B;BTN3A1;IL13;TFEB;BTN3A3;BTN3A2;MED26;SIRT1;POU5F1;TAPBP;IL1A;NR5A2;PTRH2;IL1B;TRIM58;ZNF117;ZNF513;MAPT;CD46;MYCBP;NR1I2;TULP3;AIF1;PABPN1;RNF39;ASCC2;GPBP1L1;MAPK3;TRIM40;TCF7L1;MOG;IL37;EGF;WWP2;CDK11B;REST;TRIM39;TRIM38;TRIM31;MEGF8;ZNF212;HSPA1B;HSPA1A
15 megakaryocyte differentiation (GO:0030219) 7/12 0.0080729197 MED1;MPIG6B;TAL1;UBA5;KIT;SH2B3;ZFPM1
16 regulation of immune response (GO:0050776) 34/179 0.0081873248 IFITM1;ICAM3;ICAM5;ITGAL;FCRLB;FCGR3A;SLAMF7;CD300LF;CD33;MICA;MICB;LYN;ITGA4;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;TIRAP;HLA-E;TREML1;NCR3;CLEC2B;FCGR2A;JAML;IRF4;IRF1;KLRF1;HLA-DRA;FCGR2B;CLEC2D;CLEC4G;HLA-DRB1;NECTIN2
17 embryonic hemopoiesis (GO:0035162) 7/13 0.0136928523 KITLG;KMT2A;TAL1;KIT;SH2B3;ZFPM1;ATF4
18 antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590) 19/78 0.0136928523 FCER1G;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-G;PSMB8;HLA-E;TAPBP;PSMB9;VAMP8;PSMA6;PSMB7;PSMD6;PSMC5;PSMC3;PSMD3
19 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (GO:0002479) 18/73 0.0170252114 HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-G;PSMB8;HLA-E;TAPBP;PSMB9;VAMP8;PSMA6;PSMB7;PSMD6;PSMC5;PSMC3;PSMD3
20 cellular response to interferon-gamma (GO:0071346) 25/121 0.0176416941 AIF1;TRIM8;SYNCRIP;TRIM26;HLA-DQA2;HLA-DQA1;HLA-DPA1;HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;IRF4;IRF1;HLA-DPB1;HLA-DRA;IRF8;TRIM38;TRIM31;TDGF1;HLA-DRB1;HLA-DQB2;HLA-DQB1
21 negative regulation of extrinsic apoptotic signaling pathway in absence of ligand (GO:2001240) 9/23 0.0216159067 IL1A;TERT;IL1B;EYA4;AKT1;TNF;HSPA1B;MAP2K5;HSPA1A
22 negative regulation of signal transduction in absence of ligand (GO:1901099) 9/23 0.0216159067 IL1A;TERT;IL1B;EYA4;AKT1;TNF;HSPA1B;MAP2K5;HSPA1A
23 transcription by RNA polymerase II (GO:0006366) 50/320 0.0220333588 SETD2;GMEB2;ELL;CCNT1;THRA;MAML1;ZC3H8;ARRB2;LSM10;SNAPC4;FAM170A;NELFE;ABT1;MED1;MED26;NRBP1;NCOR1;NR5A2;TFDP1;RARA;ATF4;PPARD;NOTCH4;NR1I2;C5AR1;TAF9;RTF1;RPAP2;HDAC7;MNT;PABPN1;POLR2B;POLR2E;E2F2;NRBF2;RREB1;CPSF4;CREBBP;NR1D1;GTF2H2;GTF2H3;SUPT5H;GTF2H4;DRAP1;CDK7;TFCP2;NFIC;TAF8;NABP1;TAF7
24 regulation of tumor necrosis factor production (GO:0032680) 25/124 0.0222530612 C5AR2;PTPRJ;NLRC3;AGER;VSIR;C1QTNF4;DDT;PLCG2;TRIM27;CD33;HAVCR2;SPHK2;IL37;NFKBIL1;PTPN11;MIF;TIRAP;HLA-E;POMC;IL1A;IL6;PTPRC;RARA;BPI;TMEM106A
25 antigen processing and presentation of exogenous peptide antigen via MHC class II (GO:0019886) 21/98 0.0295851235 HLA-DRB5;FCER1G;CLTC;KLC1;ACTR1A;HLA-DMA;HLA-DMB;CANX;HLA-DPB1;HLA-DRA;HLA-DOA;FCGR2B;HLA-DOB;HLA-DQA2;AP1M2;HLA-DQA1;HLA-DRB1;HLA-DQB2;SEC31A;HLA-DPA1;HLA-DQB1
26 negative regulation of leukocyte mediated cytotoxicity (GO:0001911) 7/15 0.0295851235 PTPRC;ARRB2;HLA-F;FCGR2B;HLA-G;MICA;HLA-E
27 cellular response to lipopolysaccharide (GO:0071222) 24/120 0.0305047677 LYN;CXCL6;CXCL8;IL37;GFI1;NFKBIL1;IL36G;CXCL1;NR1D1;CXCL3;TNF;CXCL2;IL1F10;IL1A;IL6;PABPN1;IL1B;PDE4B;ADAM9;NLRP3;IL36RN;BPI;CCR5;GIT1
28 positive regulation of phagocytosis (GO:0050766) 14/53 0.0329088793 FCER1G;PTPRJ;ABCA7;TULP1;MERTK;TNF;SIRPB1;C2;C4B;C4A;PTPRC;IL1B;CD300LF;FCGR2B
29 antigen processing and presentation of peptide antigen via MHC class II (GO:0002495) 21/100 0.0345332873 HLA-DRB5;FCER1G;CLTC;KLC1;ACTR1A;HLA-DMA;HLA-DMB;CANX;HLA-DPB1;HLA-DRA;HLA-DOA;FCGR2B;HLA-DOB;HLA-DQA2;AP1M2;HLA-DQA1;HLA-DRB1;HLA-DQB2;SEC31A;HLA-DPA1;HLA-DQB1
30 cellular response to molecule of bacterial origin (GO:0071219) 23/115 0.0345332873 CXCL6;CXCL8;IL37;GFI1;NFKBIL1;IL36G;CXCL1;NR1D1;CXCL3;CXCL2;TIRAP;IL1F10;IL1A;IL6;PABPN1;IL1B;PDE4B;ADAM9;NLRP3;IL36RN;CCR5;FCGR2B;GIT1
31 regulation of primary metabolic process (GO:0080090) 25/130 0.0345332873 ACACA;OAZ2;PSMB7;PSMD6;SIN3A;OPA3;PSMD3;TNFAIP8L3;APOE;MED1;SREBF1;NQO1;CREBBP;ABCA7;NR1D1;LSS;AZIN1;PSMB8;PSMB9;PSMA6;PSMC5;NCOR1;GPAM;PSMC3;PPARD
32 regulation of interleukin-2 production (GO:0032663) 13/48 0.0345332873 GLMN;LAPTM5;HDAC7;IL1A;PTPRC;IRF4;IL1B;PDE4B;PLCG2;TRIM27;EZR;SPTBN1;HAVCR2
33 positive regulation of apoptotic cell clearance (GO:2000427) 5/8 0.0345332873 C4B;C4A;ABCA7;CD300LF;C2
34 regulation of apoptotic cell clearance (GO:2000425) 5/8 0.0345332873 C4B;C4A;CD300LF;C2;TGM2
35 negative regulation of natural killer cell mediated cytotoxicity (GO:0045953) 7/16 0.0345332873 HLA-B;ARRB2;HLA-A;HLA-F;HLA-G;MICA;HLA-E
36 negative regulation of telomere maintenance (GO:0032205) 7/16 0.0345332873 RTEL1;RAD50;SLX4;TNKS2;PINX1;ATM;SMG6
37 Fc receptor mediated stimulatory signaling pathway (GO:0002431) 17/74 0.0345332873 BLK;LYN;FCER1G;ARPC1B;WIPF2;ARPC1A;CDC42;FCGR3A;FCGR2A;ARPC2;ELMO1;PLCG2;FCGR2B;CRK;NCK1;MAPK3;MYO1G
38 response to endoplasmic reticulum stress (GO:0034976) 22/110 0.0394871632 GSK3B;CXCL8;ATF6B;UBA5;SEC16A;CCDC47;ALOX15;TMTC3;TNFRSF10B;RNF5;BBC3;SCAMP5;BAG6;TMEM259;FLOT1;UFC1;RNF183;BAK1;CDK5RAP3;MAP3K5;ATF4;NCK1
39 protein phosphorylation (GO:0006468) 68/496 0.0493393067 ACVRL1;DDR1;GSK3B;GSK3A;CCNT1;PRAG1;MAML1;FLT3;MAST2;PRKAG1;TESK2;PIK3CD;AATK;STK19;IGF1R;SLK;CAMKV;RPS6KA1;AKT1;MAP3K5;BRD2;DAPK2;MINK1;DYRK1A;ADAM10;TYK2;NRBP1;PEAK1;CSNK2B;KIT;RARA;MAPKAPK5;TRIB2;CSNK1G1;BLK;PKN3;AURKB;GRK1;STK36;GRK4;ERBB2;PTK2B;MARK4;PAK2;MARK3;IP6K3;MAP4K4;MAPK3;LYN;PRRT1;CDK18;MAP3K3;NEK6;GTF2H3;HIPK1;CDC42BPA;MERTK;GTF2H4;CDK11B;CDK7;FER;RPS6KB1;FES;CDK10;COQ8B;ATM;TDGF1;CDK15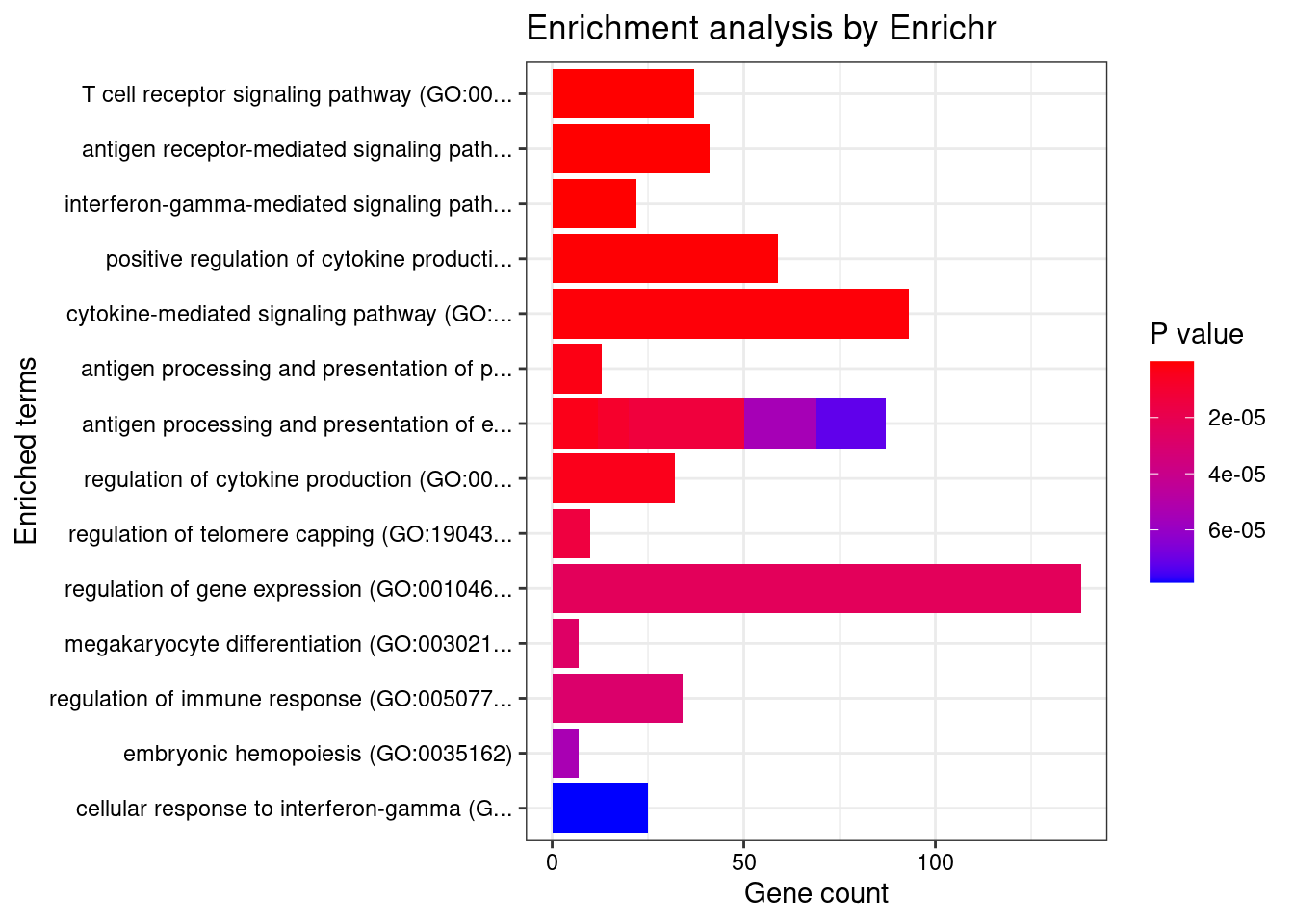
GO_Cellular_Component_2021
Term Overlap Adjusted.P.value Genes
1 MHC protein complex (GO:0042611) 18/20 1.019156e-14 HLA-DRB5;HFE;HLA-B;HLA-C;HLA-A;HLA-F;HLA-E;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
2 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556) 18/28 1.148924e-10 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-G;HLA-E;TAPBP;CANX;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
3 lumenal side of endoplasmic reticulum membrane (GO:0098553) 18/28 1.148924e-10 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-G;HLA-E;TAPBP;CANX;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
4 MHC class II protein complex (GO:0042613) 12/13 3.492150e-10 HLA-DRB5;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
5 ER to Golgi transport vesicle membrane (GO:0012507) 18/54 4.905787e-05 HLA-DRB5;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;HLA-DQA2;CNIH3;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1;SEC31A
6 transport vesicle membrane (GO:0030658) 19/60 4.905787e-05 HLA-DRB5;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;HLA-DQA2;CNIH3;HLA-DQB2;VAMP2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1;SEC31A
7 coated vesicle membrane (GO:0030662) 18/55 4.905787e-05 HLA-DRB5;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;HLA-DQA2;CNIH3;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1;SEC31A
8 COPII-coated ER to Golgi transport vesicle (GO:0030134) 20/79 8.949178e-04 HLA-DRB5;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;TMED6;HLA-DQA2;CNIH3;HLA-DQB2;HLA-DQA1;CNIH4;HLA-DRB1;HLA-DPA1;HLA-DQB1;SEC31A
9 MHC class I protein complex (GO:0042612) 5/6 1.427197e-03 HFE;HLA-B;HLA-C;HLA-A;HLA-E
10 endocytic vesicle membrane (GO:0030666) 30/158 3.133267e-03 CLTC;ADRB2;TGOLN2;APOE;HLA-DQA2;HLA-DQA1;WNT3;ATP6V0A1;WNT4;HLA-DPA1;HLA-DRB5;EGF;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-G;EREG;HLA-E;TAPBP;VAMP8;NOSTRIN;HLA-DPB1;HLA-DRA;HLA-DRB1;HLA-DQB2;VAMP2;HLA-DQB1
11 focal adhesion (GO:0005925) 58/387 3.547843e-03 MDC1;PRAG1;RPL31;ARPC1B;CLTC;RPL10A;LIMD1;SLC6A4;RPL7A;RPS16;MPRIP;RPS18;CFL1;FLOT1;CHP1;ITGB6;RPS10;TNS3;GIT1;TGM2;CAP1;MAP2K2;ITGA4;ITGA2;ADAM10;ENAH;PEAK1;ADAM9;RPL27;EZR;CD46;FBLN7;GRB7;ASAP3;IQGAP1;ADD1;CDC42;RAB21;CNN2;DPP4;LIMA1;INPP5E;GNA12;PTK2B;FLNB;RPL18;MAP4K4;MAPK3;EHD3;UBOX5;HNRNPK;PTPRC;ARPC2;FES;TADA1;HSPA1B;NECTIN2;HSPA1A
12 clathrin-coated endocytic vesicle membrane (GO:0030669) 17/69 3.547843e-03 HLA-DRB5;EGF;CLTC;ADRB2;EREG;VAMP8;TGOLN2;HLA-DPB1;HLA-DRA;APOE;HLA-DQA2;HLA-DQB2;VAMP2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
13 cell-substrate junction (GO:0030055) 58/394 5.091093e-03 MDC1;PRAG1;RPL31;ARPC1B;CLTC;RPL10A;LIMD1;SLC6A4;RPL7A;RPS16;MPRIP;RPS18;CFL1;FLOT1;CHP1;ITGB6;RPS10;TNS3;GIT1;TGM2;CAP1;MAP2K2;ITGA4;ITGA2;ADAM10;ENAH;PEAK1;ADAM9;RPL27;EZR;CD46;FBLN7;GRB7;ASAP3;IQGAP1;ADD1;CDC42;RAB21;CNN2;DPP4;LIMA1;INPP5E;GNA12;PTK2B;FLNB;RPL18;MAP4K4;MAPK3;EHD3;UBOX5;HNRNPK;PTPRC;ARPC2;FES;TADA1;HSPA1B;NECTIN2;HSPA1A
14 transcription factor TFIIH holo complex (GO:0005675) 6/12 8.561067e-03 CDK7;GTF2H2C;MMS19;GTF2H2;GTF2H3;GTF2H4
15 lysosome (GO:0005764) 66/477 9.940301e-03 IFITM3;SLC35F6;IFITM1;IFITM2;SLC44A2;CLTC;VLDLR;LIPA;SLC2A6;BBC3;LAMP1;NAGLU;GM2A;FLOT1;ACP2;HLA-DOA;GUSB;HLA-DOB;HLA-DPA1;RAB2A;STARD3;USP6;SPHK2;SLC11A2;TFEB;PRSS16;HLA-F;TMEM150C;VAMP8;RRAGC;TMBIM1;DAGLB;HLA-DQB2;HLA-DQB1;CALCRL;PSEN2;ADRB2;DPP4;HLA-DMA;SNX1;HLA-DMB;SNX2;NEU1;CXCR2;GC;SLC17A1;HLA-DQA2;HLA-DQA1;SLC17A3;AP1M2;SLC17A4;ATP6V0A1;CD164;HLA-DRB5;EGF;FUCA1;LAPTM5;AP3D1;OCLN;SNX14;HLA-DPB1;PLEKHM1;HLA-DRA;PPT2;RNF183;HLA-DRB1
16 lytic vacuole membrane (GO:0098852) 41/267 1.483728e-02 IFITM3;SLC35F6;IFITM1;IFITM2;SLC44A2;PSEN2;VLDLR;SLC2A6;DPP4;HLA-DMA;HLA-DMB;LAMP1;FLOT1;HLA-DOA;HLA-DQA2;HLA-DOB;HLA-DQA1;AP1M2;ATP6V0A1;HLA-DPA1;RAB2A;STARD3;HLA-DRB5;SPHK2;EGF;SLC11A2;TFEB;AP3D1;LAPTM5;HLA-F;TMEM150C;VAMP8;OCLN;TMBIM1;HLA-DPB1;HLA-DRA;RNF183;DAGLB;HLA-DRB1;HLA-DQB2;HLA-DQB1
17 clathrin-coated endocytic vesicle (GO:0045334) 17/85 3.317813e-02 HLA-DRB5;EGF;CLTC;ADRB2;EREG;VAMP8;TGOLN2;HLA-DPB1;HLA-DRA;APOE;HLA-DQA2;HLA-DQB2;VAMP2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
18 phagocytic vesicle membrane (GO:0030670) 11/45 3.963181e-02 VAMP8;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-G;ATP6V0A1;HLA-E;TAPBP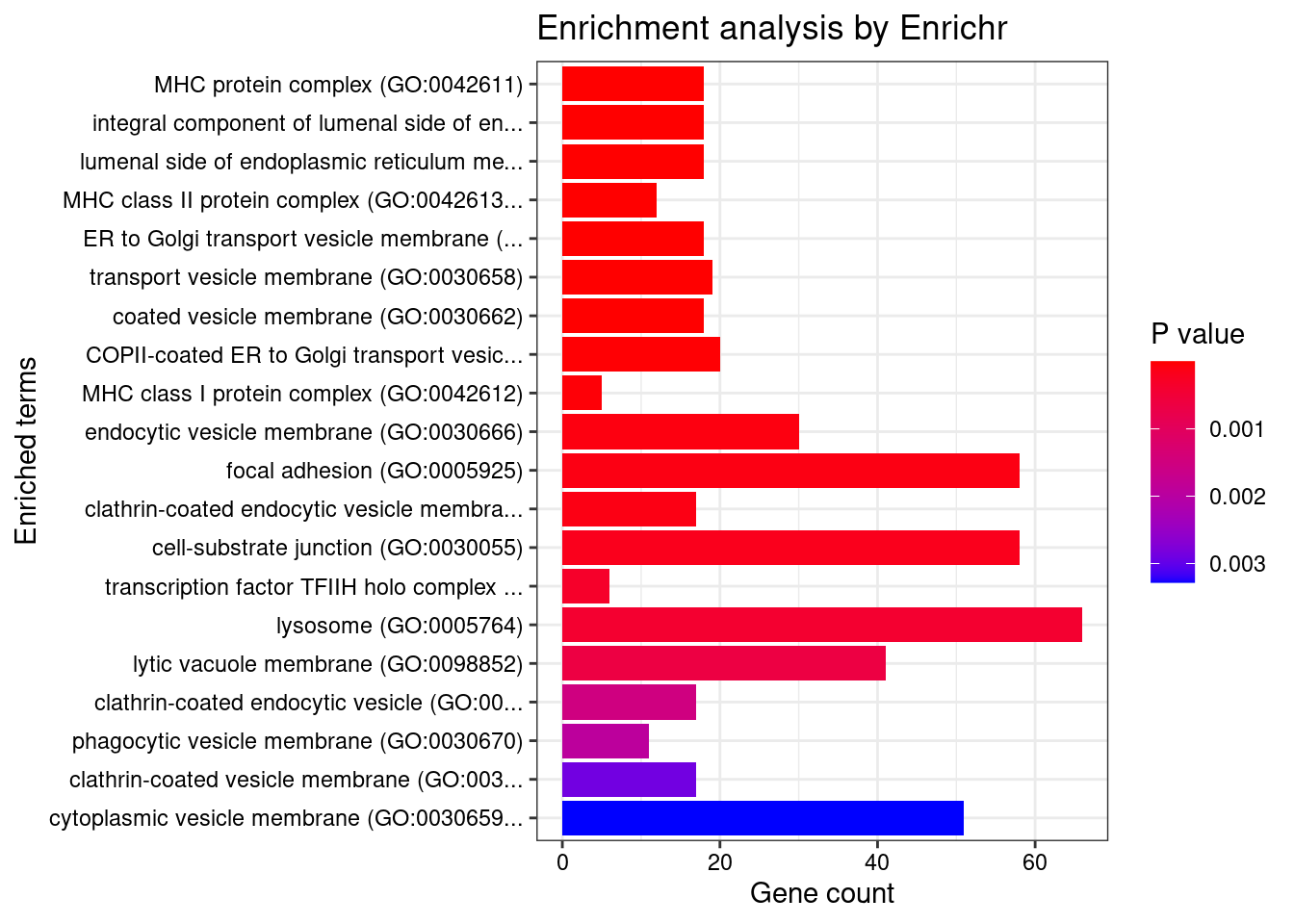
GO_Molecular_Function_2021
Term Overlap Adjusted.P.value Genes
1 MHC class II receptor activity (GO:0032395) 9/10 3.215513e-06 HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
2 myosin heavy chain binding (GO:0032036) 5/7 4.620947e-02 MYBPC3;SPTBN5;MYL2;MYL3;CORO1A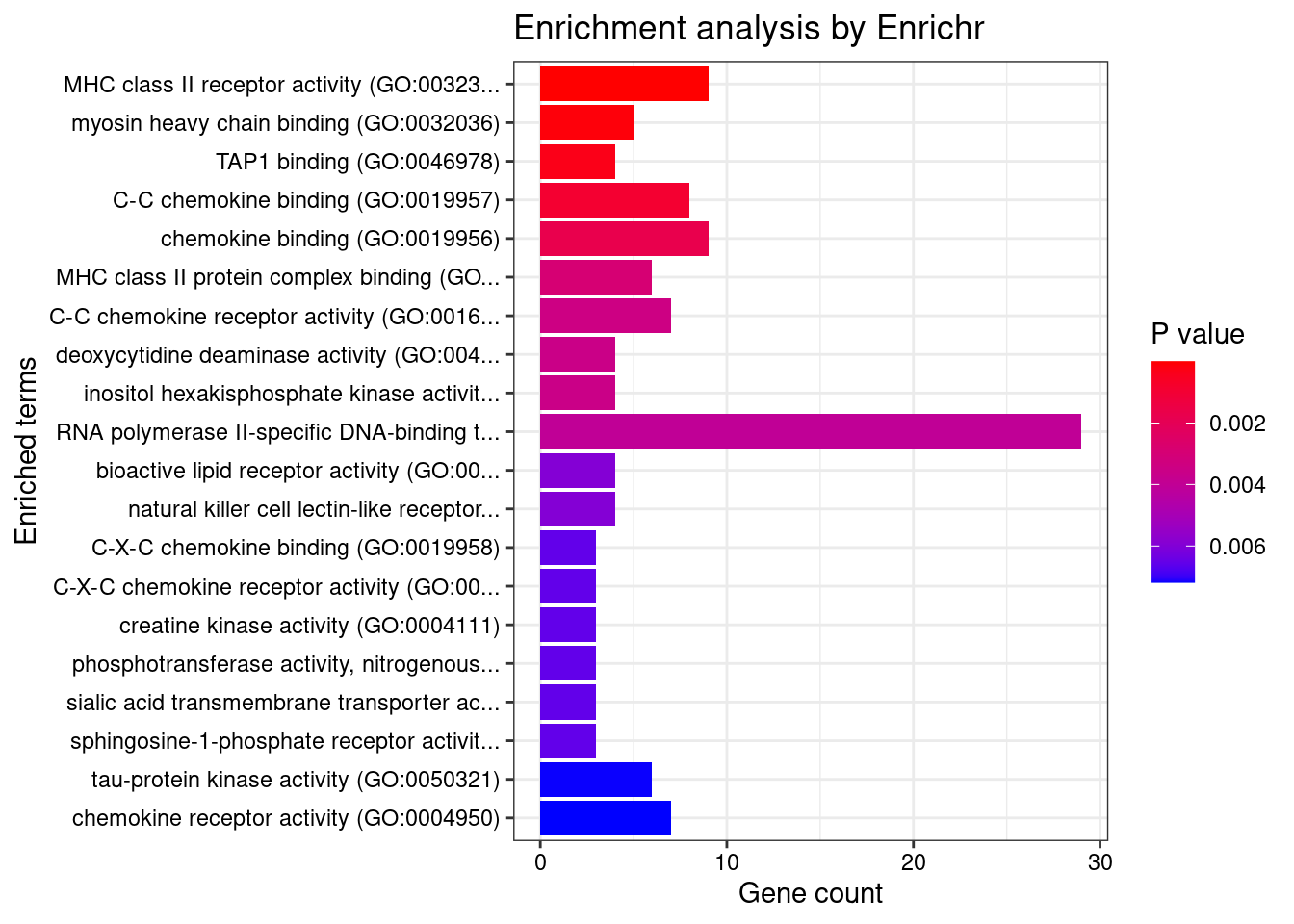
Enrichment analysis for cTWAS genes in top tissues separately
GO
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Adipose_Visceral_Omentum
Number of cTWAS Genes in Tissue: 43
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Whole_Blood
Number of cTWAS Genes in Tissue: 85
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 amyloid precursor protein metabolic process (GO:0042982) 3/18 0.04849071 ADAM19;ACHE;PSEN2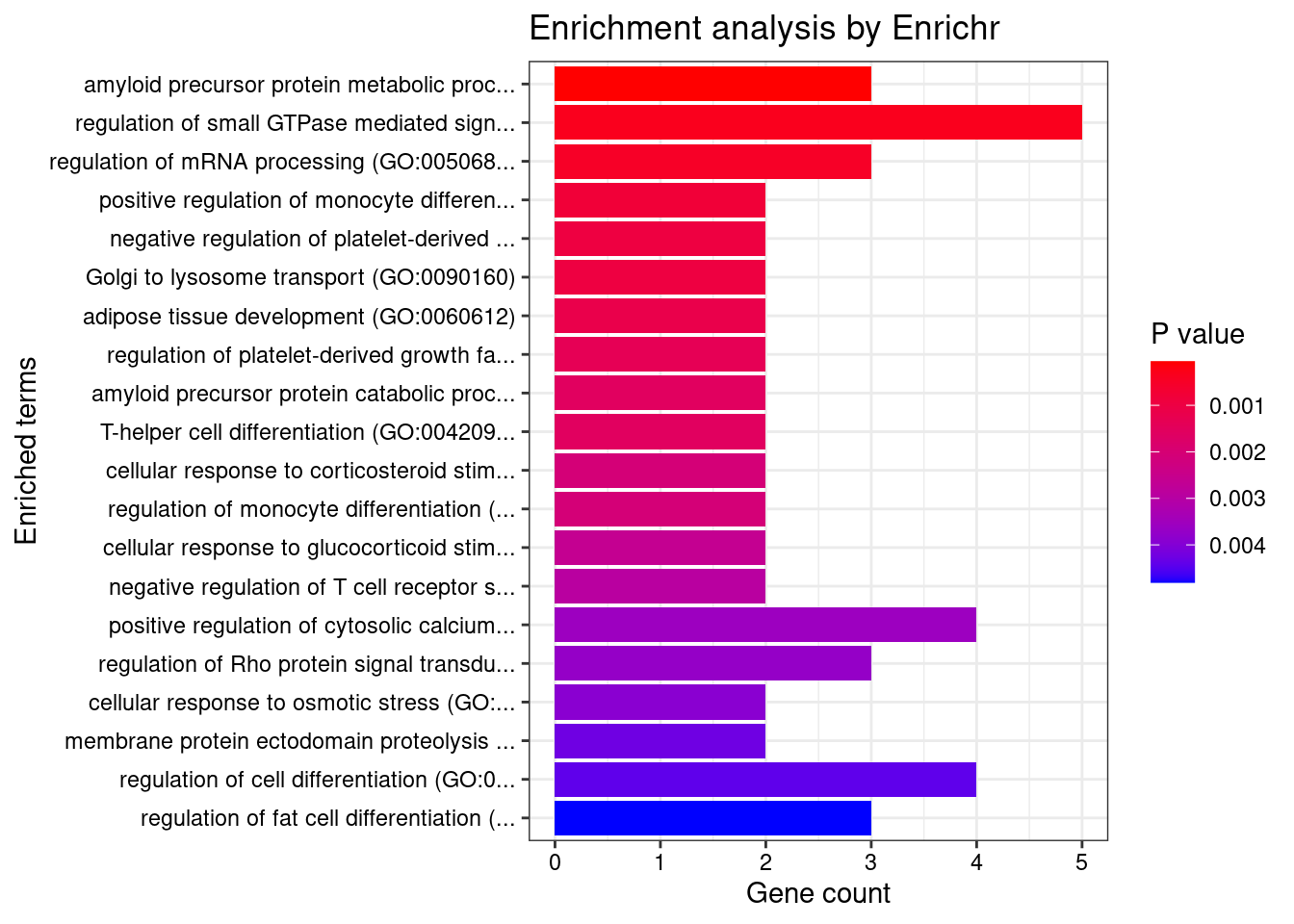
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 38
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)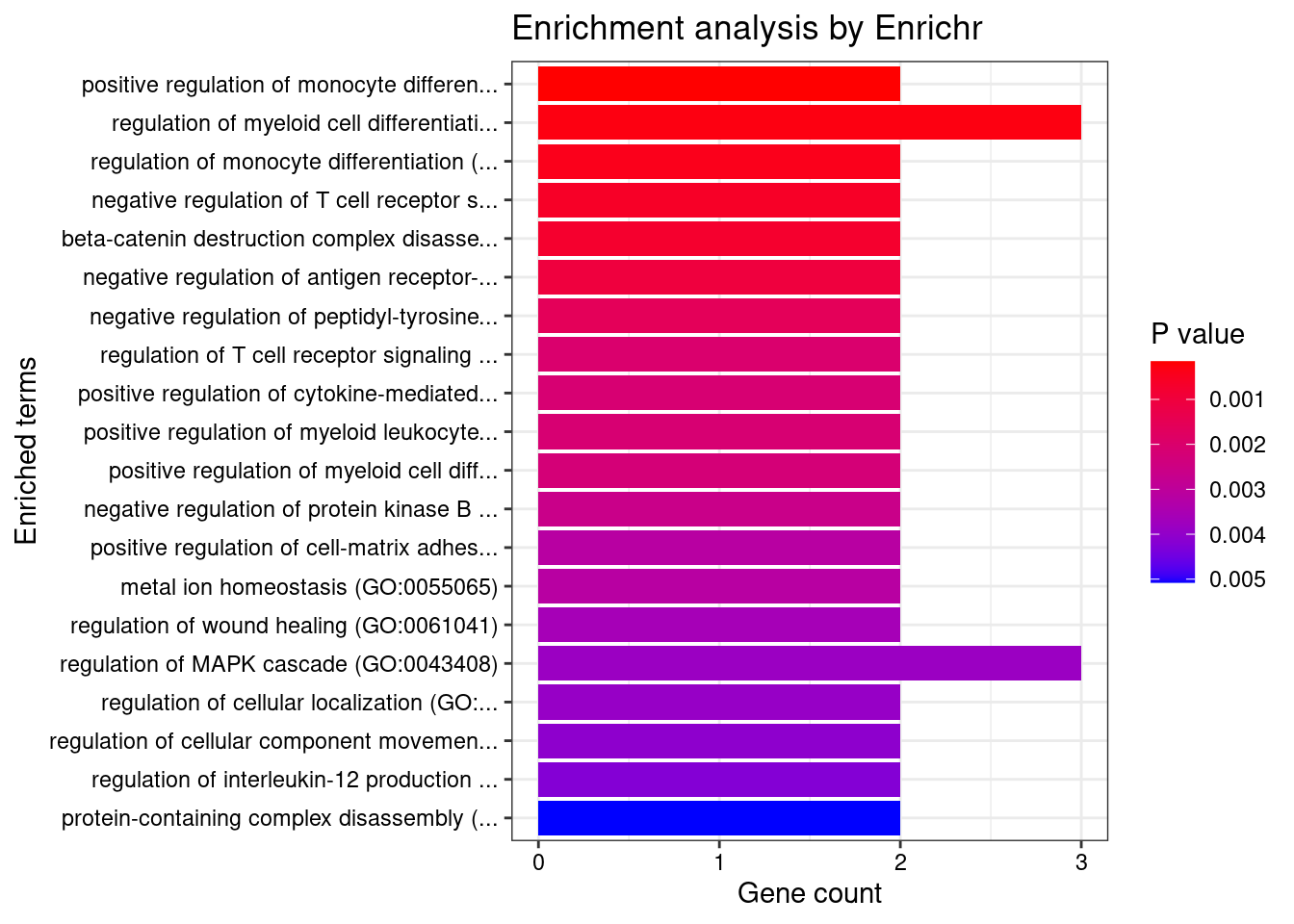
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Stomach
Number of cTWAS Genes in Tissue: 32
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)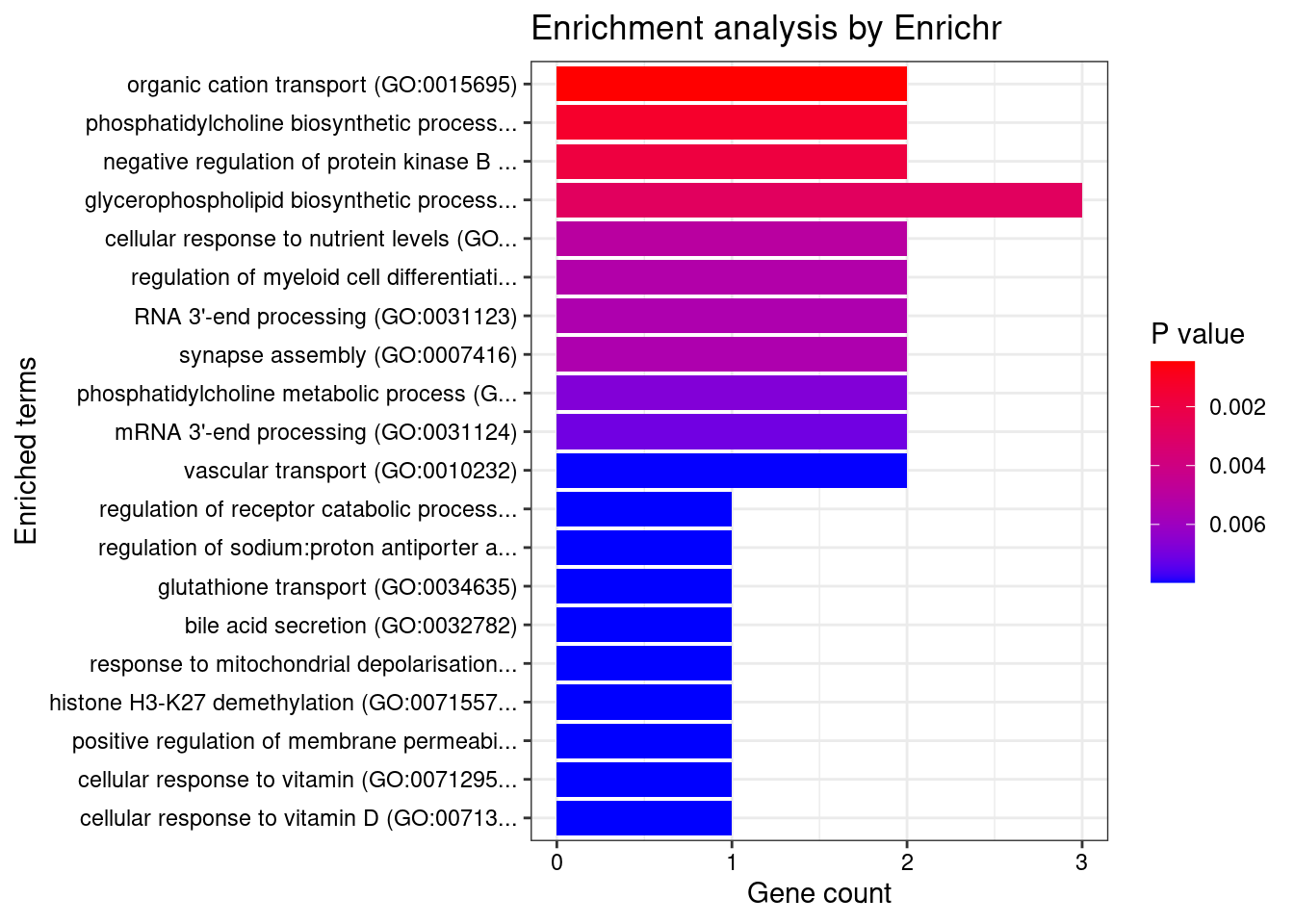
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Lung
Number of cTWAS Genes in Tissue: 33
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)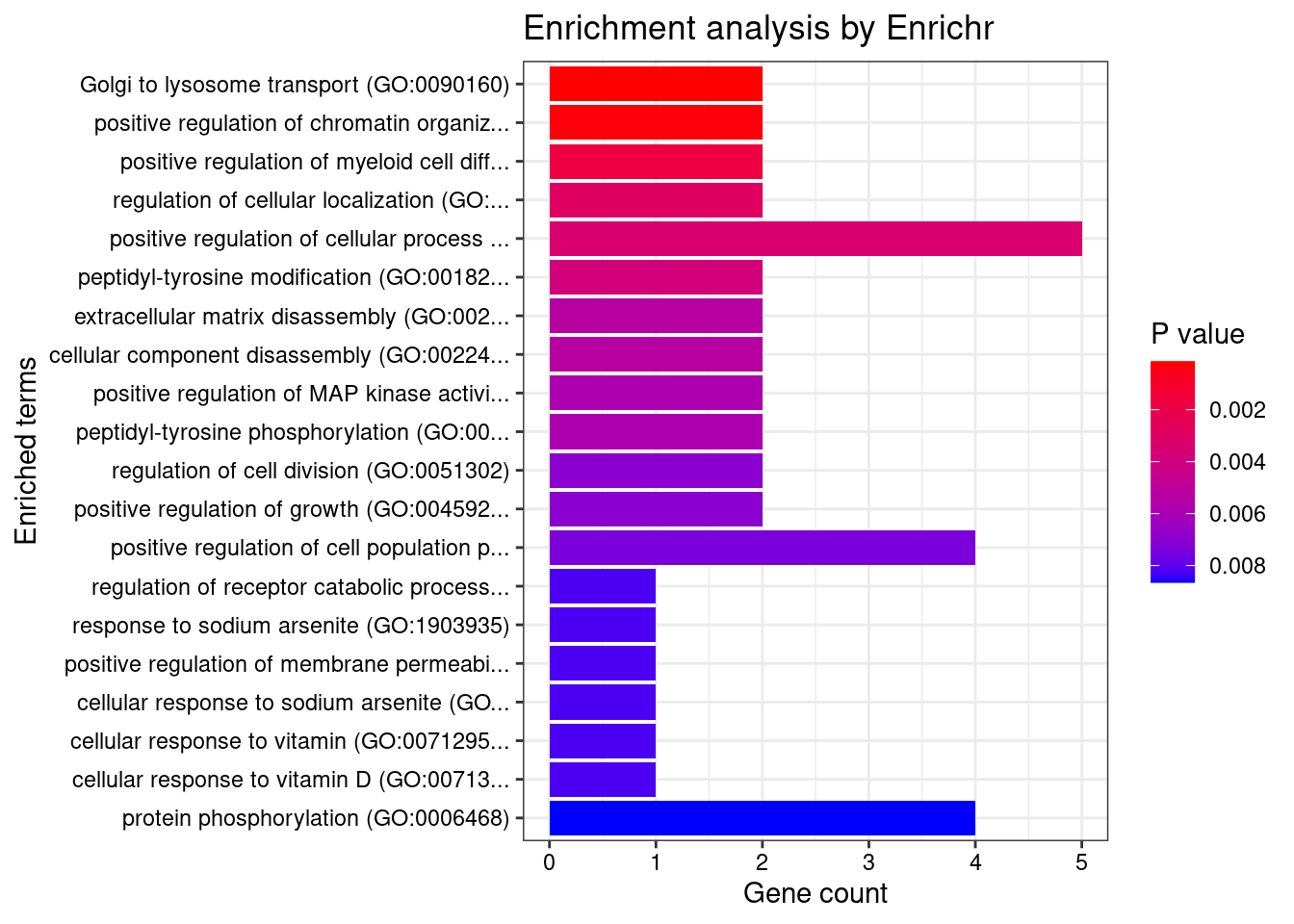
KEGG
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- NULL
try(enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_tissue, referenceGene=background_tissue,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F))
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
} Adipose_Visceral_Omentum
Number of cTWAS Genes in Tissue: 43
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Whole_Blood
Number of cTWAS Genes in Tissue: 85
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 38
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Stomach
Number of cTWAS Genes in Tissue: 32
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Lung
Number of cTWAS Genes in Tissue: 33
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_tissue, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
} Adipose_Visceral_Omentum
Number of cTWAS Genes in Tissue: 43LRRC25 gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDLINC01624 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDWBP1L gene(s) from the input list not found in DisGeNET CURATEDCHP1 gene(s) from the input list not found in DisGeNET CURATEDGALNT1 gene(s) from the input list not found in DisGeNET CURATEDMARCH7 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDMED24 gene(s) from the input list not found in DisGeNET CURATEDRP5-894D12.5 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDFAM212A gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDFAM177A1 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDFRAT1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
7 Arthralgia 0.03630088 1/18 2/9703
44 Heart Diseases 0.03630088 2/18 46/9703
49 Hepatic Veno-Occlusive Disease 0.03630088 1/18 2/9703
55 Infarction 0.03630088 1/18 2/9703
71 Measles 0.03630088 1/18 1/9703
83 Neutropenia 0.03630088 2/18 21/9703
101 Thrombocytopenia 0.03630088 2/18 29/9703
112 Lymphoma, AIDS-Related 0.03630088 1/18 2/9703
124 Polyarthralgia 0.03630088 1/18 2/9703
136 Cyclic neutropenia 0.03630088 1/18 1/9703
147 Complement deficiency disease 0.03630088 2/18 33/9703
151 Bone Marrow Neoplasms 0.03630088 1/18 1/9703
175 Impairment, Light Touch Sensation 0.03630088 1/18 2/9703
176 Pain Sensation Diminished 0.03630088 1/18 2/9703
177 Pinprick Sensation Diminished 0.03630088 1/18 2/9703
178 Position Sense Disorders 0.03630088 1/18 2/9703
179 Proprioceptive Disorders 0.03630088 1/18 2/9703
180 Thermal Sensation Disorders 0.03630088 1/18 2/9703
181 Somatosensory Disorders 0.03630088 1/18 2/9703
209 DEAFNESS, AUTOSOMAL RECESSIVE 68 0.03630088 1/18 1/9703
210 EPILEPSY, BENIGN NEONATAL, 2 0.03630088 1/18 1/9703
215 HEMOLYTIC UREMIC SYNDROME, ATYPICAL, SUSCEPTIBILITY TO, 2 0.03630088 1/18 1/9703
218 Decreased serum complement C4b 0.03630088 1/18 1/9703
224 Cyclic Hematopoesis 0.03630088 1/18 1/9703
3 Agranulocytosis 0.04259481 1/18 4/9703
30 Scleroderma 0.04259481 1/18 4/9703
43 Headache 0.04259481 1/18 5/9703
52 Hyperbilirubinemia 0.04259481 1/18 4/9703
58 leiomyosarcoma 0.04259481 1/18 5/9703
84 Ocular Headache 0.04259481 1/18 5/9703
93 Localized scleroderma 0.04259481 1/18 3/9703
96 Soft Tissue Neoplasms 0.04259481 1/18 3/9703
97 Staphylococcal Infections 0.04259481 1/18 5/9703
99 Stomatitis 0.04259481 1/18 4/9703
126 HELLP Syndrome 0.04259481 1/18 4/9703
132 Leiomyosarcoma, Epithelioid 0.04259481 1/18 5/9703
133 Leiomyosarcoma, Myxoid 0.04259481 1/18 5/9703
143 Linear Scleroderma 0.04259481 1/18 3/9703
156 Throbbing Headache 0.04259481 1/18 5/9703
157 Bilateral Headache 0.04259481 1/18 5/9703
160 Generalized Headache 0.04259481 1/18 5/9703
168 Orthostatic Headache 0.04259481 1/18 5/9703
169 Periorbital Headache 0.04259481 1/18 5/9703
170 Retro-Ocular Headache 0.04259481 1/18 5/9703
171 Sharp Headache 0.04259481 1/18 5/9703
172 Vertex Headache 0.04259481 1/18 5/9703
184 Hemicrania 0.04259481 1/18 5/9703
198 Staphylococcus aureus infection 0.04259481 1/18 5/9703
204 Morphea 0.04259481 1/18 3/9703
206 Oral Mucositis 0.04259481 1/18 4/9703
211 Neutropenia, Severe Congenital, Autosomal Dominant 1 0.04259481 1/18 4/9703
105 Varicosity 0.04650929 1/18 6/9703
113 Neoplasms, Second Primary 0.04650929 1/18 6/9703
116 Neoplasms, Therapy-Associated 0.04650929 1/18 6/9703
135 Familial benign neonatal epilepsy 0.04650929 1/18 6/9703
183 Treatment related secondary malignancy 0.04650929 1/18 6/9703
Whole_Blood
Number of cTWAS Genes in Tissue: 85ZFPM1 gene(s) from the input list not found in DisGeNET CURATEDTPST1 gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDCDC42BPA gene(s) from the input list not found in DisGeNET CURATEDBAZ2B gene(s) from the input list not found in DisGeNET CURATEDRNF181 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDDDX60L gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDHDHD5 gene(s) from the input list not found in DisGeNET CURATEDTBC1D14 gene(s) from the input list not found in DisGeNET CURATEDTTC39C gene(s) from the input list not found in DisGeNET CURATEDCCDC9 gene(s) from the input list not found in DisGeNET CURATEDCA11 gene(s) from the input list not found in DisGeNET CURATEDMTMR12 gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDARAP2 gene(s) from the input list not found in DisGeNET CURATEDZFP36L1 gene(s) from the input list not found in DisGeNET CURATEDLRRC25 gene(s) from the input list not found in DisGeNET CURATEDRABEP1 gene(s) from the input list not found in DisGeNET CURATEDCITED4 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDKIAA0391 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDOSTF1 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDUQCRC1 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDGPR4 gene(s) from the input list not found in DisGeNET CURATEDGLG1 gene(s) from the input list not found in DisGeNET CURATED
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 38RP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDC18orf25 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDC15orf40 gene(s) from the input list not found in DisGeNET CURATEDARAP2 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDSWT1 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDFRAT1 gene(s) from the input list not found in DisGeNET CURATEDCADM4 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
8 Anorexia 0.03232892 2/24 18/9703
52 Freckles 0.03232892 1/24 1/9703
67 Learning Disorders 0.03232892 2/24 29/9703
74 Melanosis 0.03232892 1/24 1/9703
75 Chloasma 0.03232892 1/24 1/9703
78 Movement Disorders 0.03232892 2/24 29/9703
101 Relapsing Fever 0.03232892 1/24 1/9703
103 Sclerosis 0.03232892 1/24 1/9703
138 Cerebellar Gait Ataxia 0.03232892 1/24 1/9703
171 Alcohol-Induced Disorders 0.03232892 1/24 1/9703
176 Etat Marbre 0.03232892 2/24 23/9703
224 Schnitzler Syndrome 0.03232892 1/24 1/9703
251 Adult Learning Disorders 0.03232892 2/24 29/9703
252 Learning Disturbance 0.03232892 2/24 29/9703
253 Learning Disabilities 0.03232892 2/24 29/9703
285 Gait Ataxia, Sensory 0.03232892 1/24 1/9703
286 Gait Ataxia 0.03232892 1/24 1/9703
315 Developmental Academic Disorder 0.03232892 2/24 29/9703
318 Mucinous carcinoma of breast 0.03232892 1/24 1/9703
338 ALCOHOL SENSITIVITY, ACUTE 0.03232892 1/24 1/9703
339 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 4 (finding) 0.03232892 1/24 1/9703
340 Lipodystrophy, Congenital Generalized, Type 3 0.03232892 1/24 1/9703
341 INTERLEUKIN 1 RECEPTOR ANTAGONIST DEFICIENCY 0.03232892 1/24 1/9703
346 METHYLMALONIC ACIDURIA, TRANSIENT, DUE TO TRANSCOBALAMIN RECEPTOR DEFECT 0.03232892 1/24 1/9703
360 Recurrent fevers 0.03232892 1/24 1/9703
362 PARTIAL LIPODYSTROPHY, CONGENITAL CATARACTS, AND NEURODEGENERATION SYNDROME 0.03232892 1/24 1/9703
363 PULMONARY HYPERTENSION, PRIMARY, 3 0.03232892 1/24 1/9703
366 Abnormality of skull size 0.03232892 1/24 1/9703
379 Methylmalonic aciduria due to transcobalamin receptor defect 0.03232892 1/24 1/9703
Stomach
Number of cTWAS Genes in Tissue: 32CRCP gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDTADA1 gene(s) from the input list not found in DisGeNET CURATEDNAA38 gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDLINC01184 gene(s) from the input list not found in DisGeNET CURATEDNECAB2 gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDDUSP14 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDVWA7 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATED
Lung
Number of cTWAS Genes in Tissue: 33RP11-219G17.9 gene(s) from the input list not found in DisGeNET CURATEDRP4-621B10.8 gene(s) from the input list not found in DisGeNET CURATEDST3GAL6 gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDZNF180 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDMYO1G gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDADGRF5 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDGPANK1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
3 Anovulation 0.02628324 1/17 1/9703
47 Cyclic neutropenia 0.02628324 1/17 1/9703
49 Conduction disorder of the heart 0.02628324 1/17 1/9703
61 Acute myeloid leukemia, minimal differentiation 0.02628324 1/17 1/9703
80 Increased risk of sudden cardiac death 0.02628324 1/17 1/9703
96 Cyclic Hematopoesis 0.02628324 1/17 1/9703
99 ALZHEIMER DISEASE 18 0.02628324 1/17 1/9703
23 Childhood Acute Lymphoblastic Leukemia 0.03306891 2/17 52/9703
24 L2 Acute Lymphoblastic Leukemia 0.03306891 2/17 50/9703
55 Renal carnitine transport defect 0.03306891 1/17 2/9703
82 Precursor T-Cell Lymphoblastic Leukemia-Lymphoma 0.03306891 2/17 53/9703
85 Mixed phenotype acute leukemia 0.03306891 1/17 2/9703
50 Propionic acidemia 0.03673588 1/17 3/9703
76 Pancreatic Endocrine Carcinoma 0.03673588 1/17 3/9703
83 Precursor Cell Lymphoblastic Leukemia Lymphoma 0.03673588 2/17 61/9703
11 Coxsackievirus Infections 0.03926264 1/17 6/9703
26 Acute myelomonocytic leukemia 0.03926264 1/17 5/9703
28 Leukocytosis 0.03926264 1/17 6/9703
31 Myocarditis 0.03926264 1/17 6/9703
43 Pleocytosis 0.03926264 1/17 6/9703
59 Reticulate acropigmentation of Kitamura 0.03926264 1/17 4/9703
67 Alcohol Related Neurodevelopmental Disorder 0.03926264 1/17 5/9703
69 Carditis 0.03926264 1/17 6/9703
79 Neutropenia, Severe Congenital, Autosomal Dominant 1 0.03926264 1/17 4/9703
91 Alcohol Related Birth Defect 0.03926264 1/17 6/9703
95 Partial Fetal Alcohol Syndrome 0.03926264 1/17 5/9703
97 dowling-degos disease 0.03926264 1/17 5/9703
103 Dowling-Degos disease 1 0.03926264 1/17 4/9703
34 Pancreatic Neoplasm 0.04181118 2/17 100/9703
57 Malignant neoplasm of pancreas 0.04181118 2/17 102/9703
75 Stage IV Skin Melanoma 0.04181118 1/17 7/9703
86 Acute erythroleukemia 0.04181118 1/17 8/9703
87 Acute erythroleukemia - M6a subtype 0.04181118 1/17 8/9703
88 Acute myeloid leukemia FAB-M6 0.04181118 1/17 8/9703
89 Acute erythroleukemia - M6b subtype 0.04181118 1/17 8/9703Gene sets curated by Macarthur Lab
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_tissue=ctwas_genes_tissue)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_tissue]})
##########
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_tissue)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
} Adipose_Visceral_Omentum
Number of cTWAS Genes in Tissue: 43
gene_set nset ngenes percent padj
1 gwascatalog 4065 25 0.006150062 0.001540967
2 mgi_essential 1494 9 0.006024096 0.141744428
3 fda_approved_drug_targets 221 1 0.004524887 0.879039698
4 clinvar_path_likelypath 1892 5 0.002642706 0.977102873
5 core_essentials_hart 182 0 0.000000000 1.000000000
Whole_Blood
Number of cTWAS Genes in Tissue: 85
gene_set nset ngenes percent padj
1 gwascatalog 3692 48 0.013001083 4.377469e-05
2 mgi_essential 1344 18 0.013392857 3.020787e-02
3 fda_approved_drug_targets 181 3 0.016574586 2.691302e-01
4 clinvar_path_likelypath 1701 12 0.007054674 8.351675e-01
5 core_essentials_hart 164 0 0.000000000 1.000000e+00
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 38
gene_set nset ngenes percent padj
1 gwascatalog 4064 23 0.005659449 0.0009620228
2 mgi_essential 1489 9 0.006044325 0.0459384928
3 fda_approved_drug_targets 224 3 0.013392857 0.0459384928
4 clinvar_path_likelypath 1892 8 0.004228330 0.2262656280
5 core_essentials_hart 185 0 0.000000000 1.0000000000
Stomach
Number of cTWAS Genes in Tissue: 32
gene_set nset ngenes percent padj
1 gwascatalog 3903 20 0.005124263 0.00188930
2 fda_approved_drug_targets 207 3 0.014492754 0.04174893
3 mgi_essential 1416 8 0.005649718 0.04466689
4 core_essentials_hart 184 0 0.000000000 1.00000000
5 clinvar_path_likelypath 1800 3 0.001666667 1.00000000
Lung
Number of cTWAS Genes in Tissue: 33
gene_set nset ngenes percent padj
1 gwascatalog 4183 18 0.004303132 0.02550935
2 mgi_essential 1491 7 0.004694836 0.17593387
3 fda_approved_drug_targets 230 2 0.008695652 0.18586659
4 clinvar_path_likelypath 1946 4 0.002055498 0.91806415
5 core_essentials_hart 189 0 0.000000000 1.00000000Summary of results across tissues
weight_groups <- as.data.frame(matrix(c("Adipose_Subcutaneous", "Adipose",
"Adipose_Visceral_Omentum", "Adipose",
"Adrenal_Gland", "Endocrine",
"Artery_Aorta", "Cardiovascular",
"Artery_Coronary", "Cardiovascular",
"Artery_Tibial", "Cardiovascular",
"Brain_Amygdala", "CNS",
"Brain_Anterior_cingulate_cortex_BA24", "CNS",
"Brain_Caudate_basal_ganglia", "CNS",
"Brain_Cerebellar_Hemisphere", "CNS",
"Brain_Cerebellum", "CNS",
"Brain_Cortex", "CNS",
"Brain_Frontal_Cortex_BA9", "CNS",
"Brain_Hippocampus", "CNS",
"Brain_Hypothalamus", "CNS",
"Brain_Nucleus_accumbens_basal_ganglia", "CNS",
"Brain_Putamen_basal_ganglia", "CNS",
"Brain_Spinal_cord_cervical_c-1", "CNS",
"Brain_Substantia_nigra", "CNS",
"Breast_Mammary_Tissue", "None",
"Cells_Cultured_fibroblasts", "Skin",
"Cells_EBV-transformed_lymphocytes", "Blood or Immune",
"Colon_Sigmoid", "Digestive",
"Colon_Transverse", "Digestive",
"Esophagus_Gastroesophageal_Junction", "Digestive",
"Esophagus_Mucosa", "Digestive",
"Esophagus_Muscularis", "Digestive",
"Heart_Atrial_Appendage", "Cardiovascular",
"Heart_Left_Ventricle", "Cardiovascular",
"Kidney_Cortex", "None",
"Liver", "None",
"Lung", "None",
"Minor_Salivary_Gland", "None",
"Muscle_Skeletal", "None",
"Nerve_Tibial", "None",
"Ovary", "None",
"Pancreas", "None",
"Pituitary", "Endocrine",
"Prostate", "None",
"Skin_Not_Sun_Exposed_Suprapubic", "Skin",
"Skin_Sun_Exposed_Lower_leg", "Skin",
"Small_Intestine_Terminal_Ileum", "Digestive",
"Spleen", "Blood or Immune",
"Stomach", "Digestive",
"Testis", "Endocrine",
"Thyroid", "Endocrine",
"Uterus", "None",
"Vagina", "None",
"Whole_Blood", "Blood or Immune"),
nrow=49, ncol=2, byrow=T), stringsAsFactors=F)
colnames(weight_groups) <- c("weight", "group")
#display tissue groups
print(weight_groups) weight group
1 Adipose_Subcutaneous Adipose
2 Adipose_Visceral_Omentum Adipose
3 Adrenal_Gland Endocrine
4 Artery_Aorta Cardiovascular
5 Artery_Coronary Cardiovascular
6 Artery_Tibial Cardiovascular
7 Brain_Amygdala CNS
8 Brain_Anterior_cingulate_cortex_BA24 CNS
9 Brain_Caudate_basal_ganglia CNS
10 Brain_Cerebellar_Hemisphere CNS
11 Brain_Cerebellum CNS
12 Brain_Cortex CNS
13 Brain_Frontal_Cortex_BA9 CNS
14 Brain_Hippocampus CNS
15 Brain_Hypothalamus CNS
16 Brain_Nucleus_accumbens_basal_ganglia CNS
17 Brain_Putamen_basal_ganglia CNS
18 Brain_Spinal_cord_cervical_c-1 CNS
19 Brain_Substantia_nigra CNS
20 Breast_Mammary_Tissue None
21 Cells_Cultured_fibroblasts Skin
22 Cells_EBV-transformed_lymphocytes Blood or Immune
23 Colon_Sigmoid Digestive
24 Colon_Transverse Digestive
25 Esophagus_Gastroesophageal_Junction Digestive
26 Esophagus_Mucosa Digestive
27 Esophagus_Muscularis Digestive
28 Heart_Atrial_Appendage Cardiovascular
29 Heart_Left_Ventricle Cardiovascular
30 Kidney_Cortex None
31 Liver None
32 Lung None
33 Minor_Salivary_Gland None
34 Muscle_Skeletal None
35 Nerve_Tibial None
36 Ovary None
37 Pancreas None
38 Pituitary Endocrine
39 Prostate None
40 Skin_Not_Sun_Exposed_Suprapubic Skin
41 Skin_Sun_Exposed_Lower_leg Skin
42 Small_Intestine_Terminal_Ileum Digestive
43 Spleen Blood or Immune
44 Stomach Digestive
45 Testis Endocrine
46 Thyroid Endocrine
47 Uterus None
48 Vagina None
49 Whole_Blood Blood or Immunegroups <- unique(weight_groups$group)
df_group <- list()
for (i in 1:length(groups)){
group <- groups[i]
weights <- weight_groups$weight[weight_groups$group==group]
df_group[[group]] <- list(ctwas=unique(unlist(lapply(df[weights], function(x){x$ctwas}))),
background=unique(unlist(lapply(df[weights], function(x){x$gene_pips$genename}))))
}
output <- output[sapply(weight_groups$weight, match, output$weight),,drop=F]
output$group <- weight_groups$group
output$n_ctwas_group <- sapply(output$group, function(x){length(df_group[[x]][["ctwas"]])})
output$n_ctwas_group[output$group=="None"] <- 0
#barplot of number of cTWAS genes in each tissue
output <- output[order(-output$n_ctwas),,drop=F]
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(output$n_ctwas, names.arg=output$weight, las=2, ylab="Number of cTWAS Genes", cex.names=0.6, main="Number of cTWAS Genes by Tissue")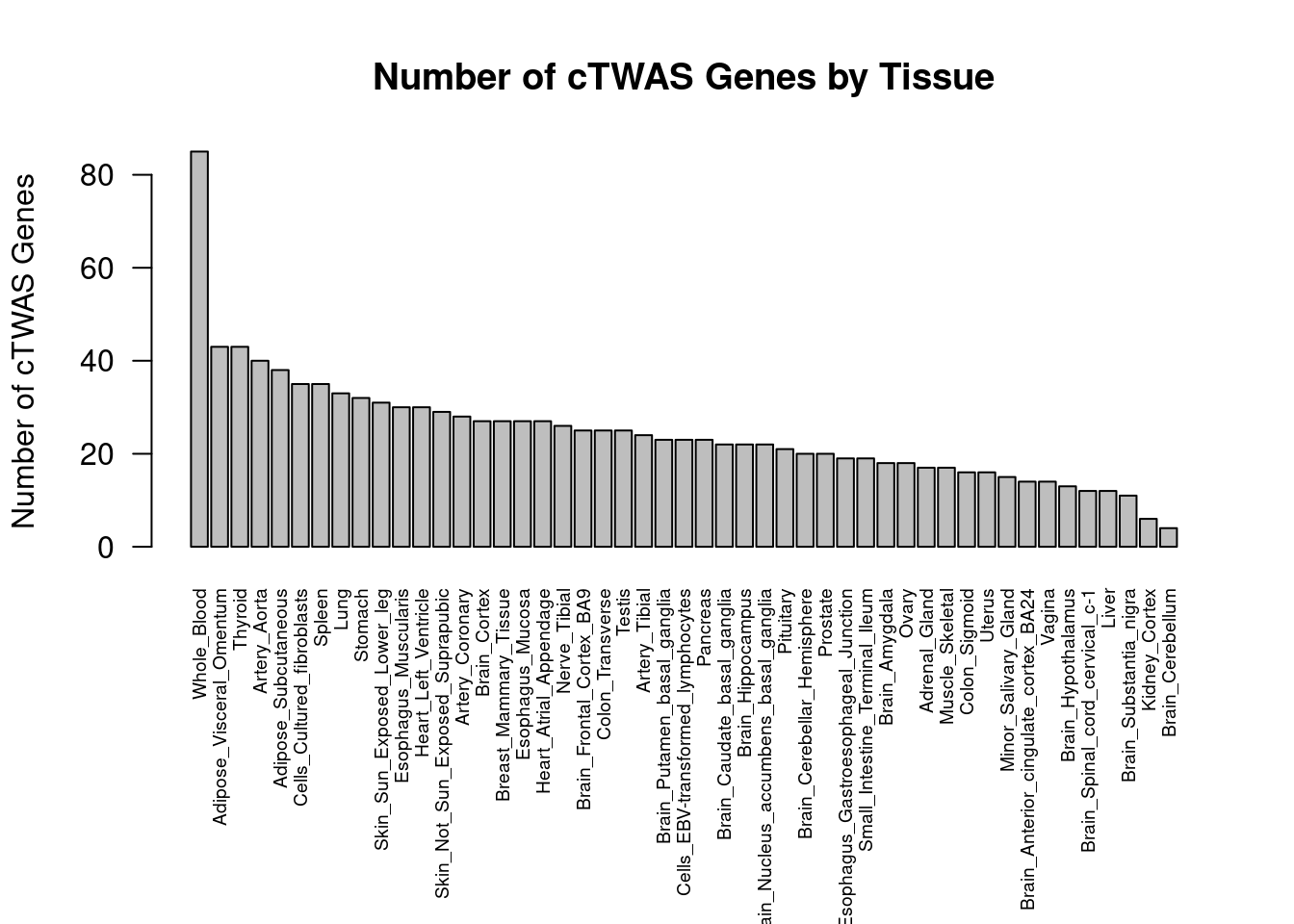
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
#barplot of number of cTWAS genes in each tissue
df_plot <- -sort(-sapply(groups[groups!="None"], function(x){length(df_group[[x]][["ctwas"]])}))
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(df_plot, las=2, ylab="Number of cTWAS Genes", main="Number of cTWAS Genes by Tissue Group")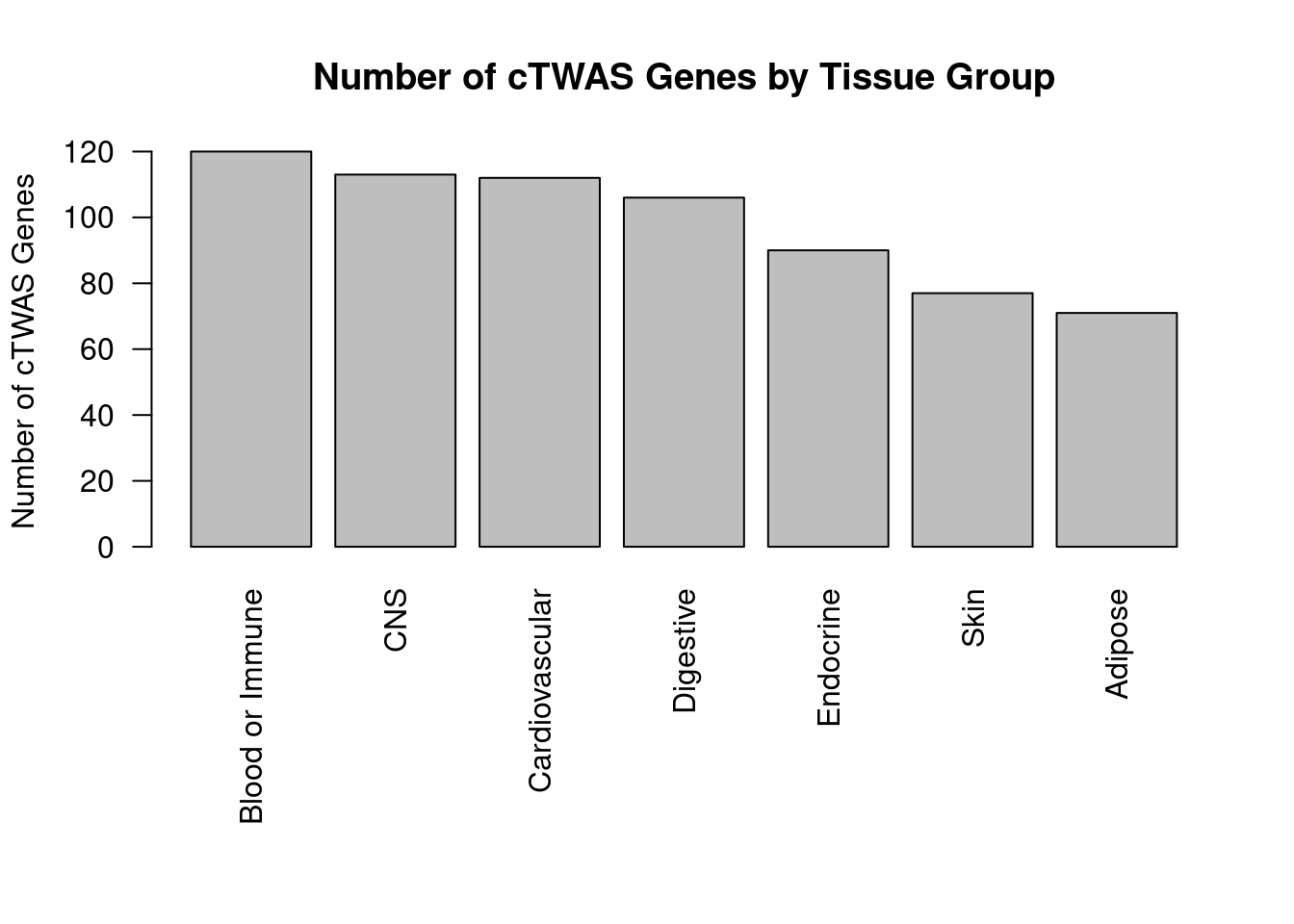
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
Enrichment analysis for cTWAS genes in each tissue group
GO
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_group, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Adipose
Number of cTWAS Genes in Tissue Group: 71
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)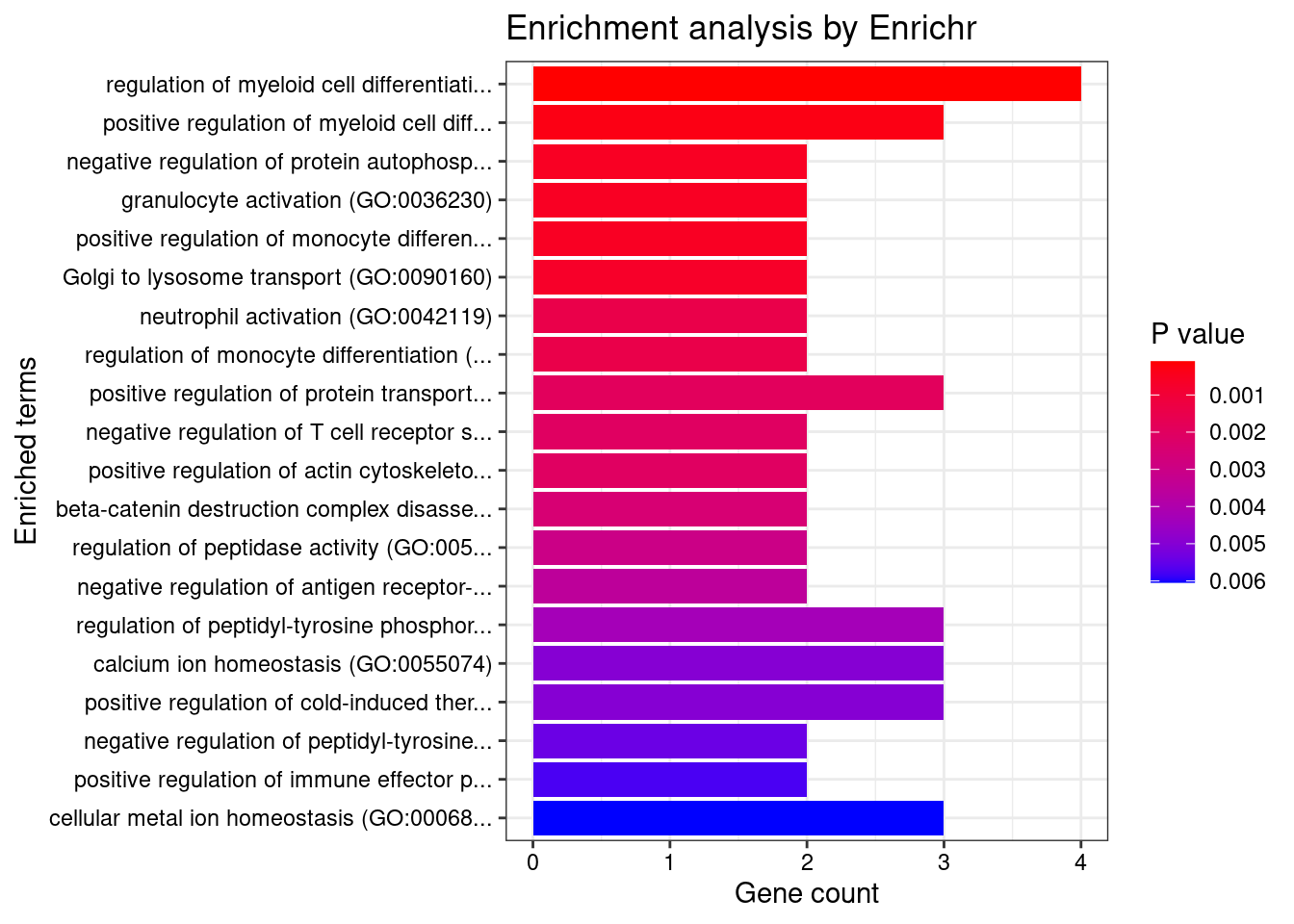
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
Endocrine
Number of cTWAS Genes in Tissue Group: 90
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)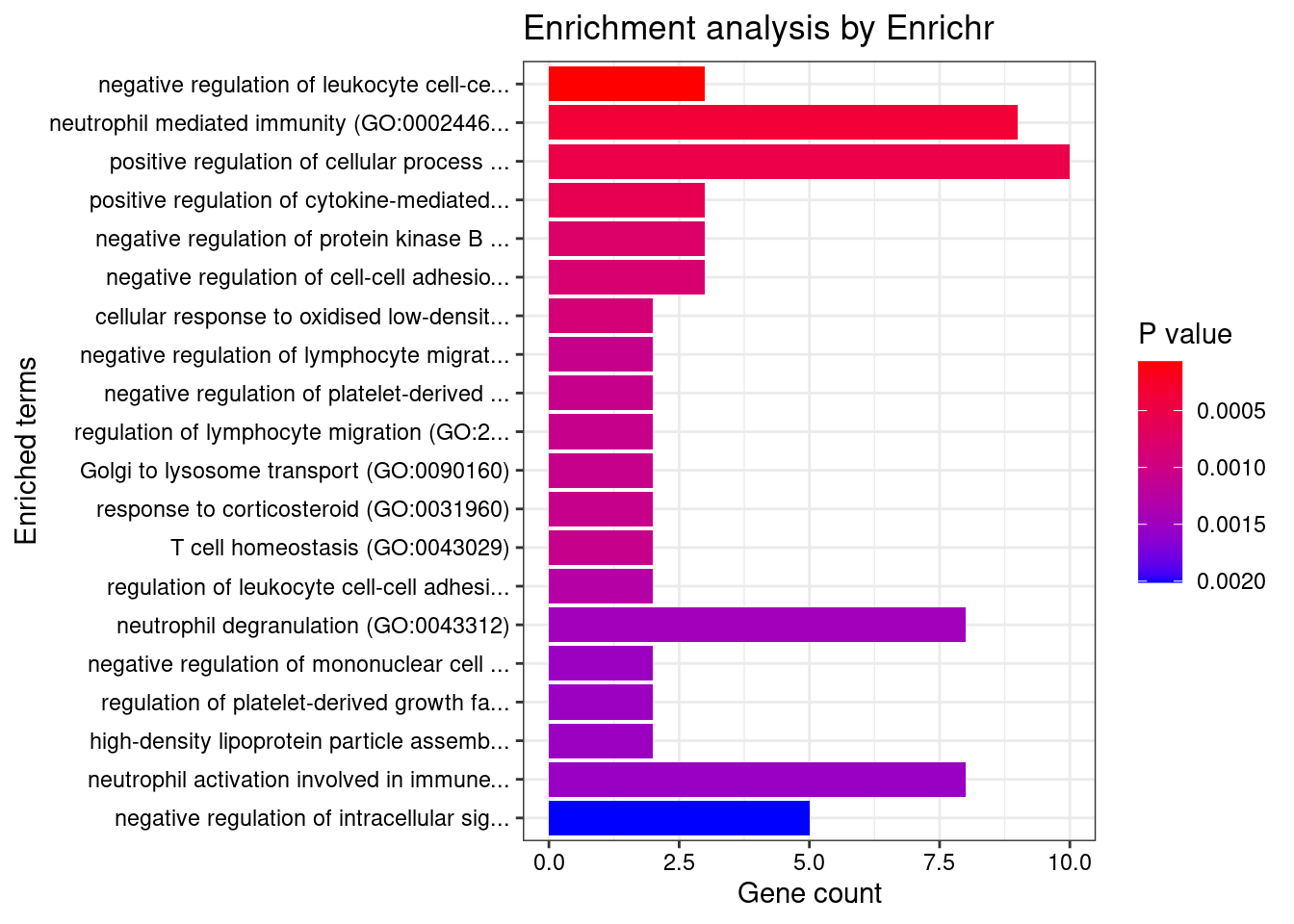
Cardiovascular
Number of cTWAS Genes in Tissue Group: 112
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)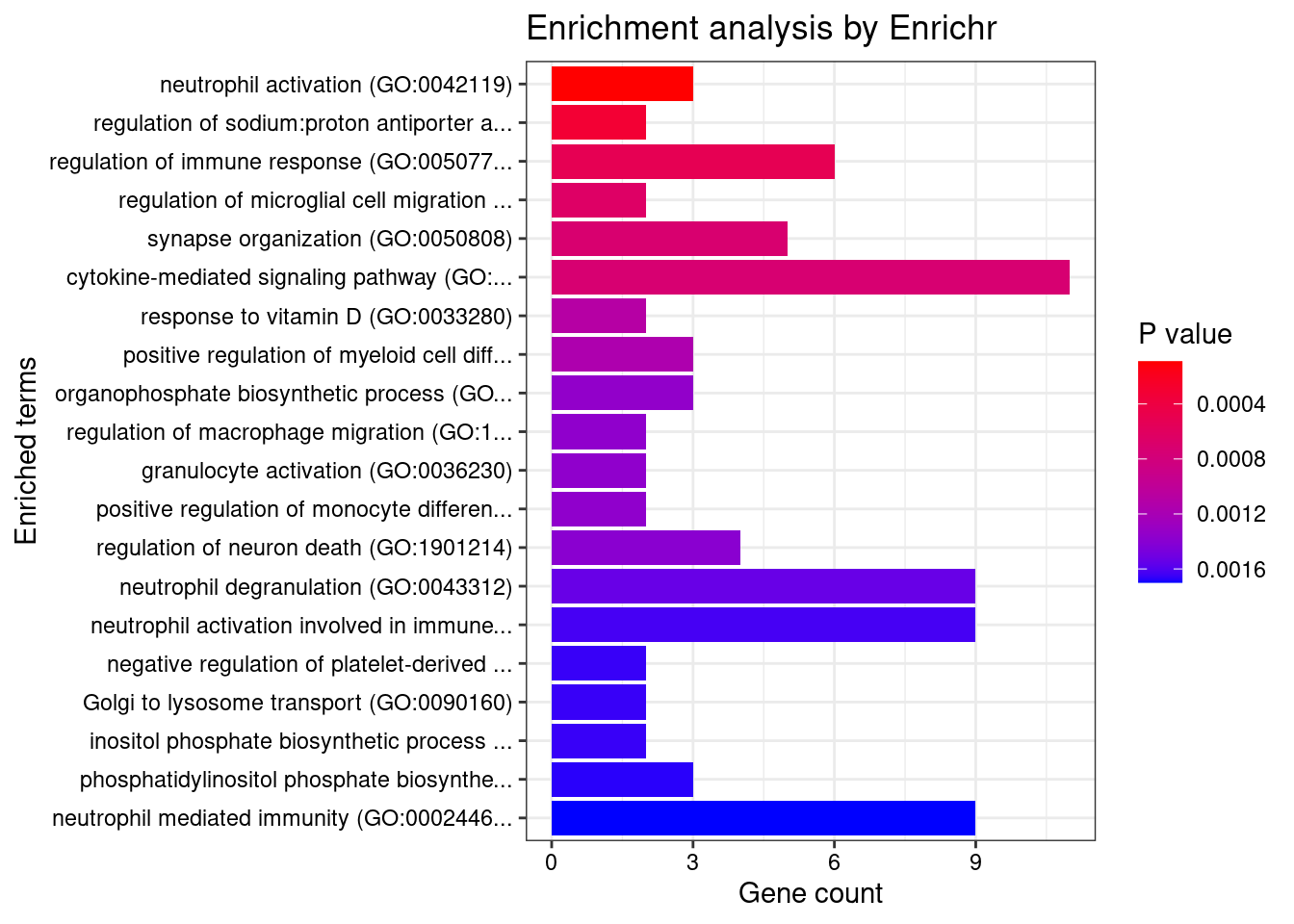
CNS
Number of cTWAS Genes in Tissue Group: 113
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of cell population proliferation (GO:0042127) 16/764 0.008206585 LYN;ACVRL1;CSF3;CADM4;CDKN2A;DOT1L;SOX11;PTPRJ;DLL1;SLC9A3R1;CYP27B1;PDF;FES;ACER3;AKT1;CD33
2 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474) 4/33 0.023286483 FCER1G;CANX;HLA-C;TAPBP
3 negative regulation of cell population proliferation (GO:0008285) 10/379 0.024288373 LYN;ACVRL1;SLC9A3R1;CYP27B1;RBP4;CDKN2A;SOX11;PTPRJ;DLL1;CD33
4 negative regulation of protein kinase B signaling (GO:0051898) 4/40 0.024288373 SLC9A3R1;PLEKHA1;AKT1;PTPRJ
5 polyol biosynthetic process (GO:0046173) 3/16 0.024288373 CYP27B1;ITPKC;ACER3
6 regulation of cell adhesion (GO:0030155) 6/133 0.024288373 LYN;ACVRL1;FES;CXCR4;PTPRJ;DLL1
7 sprouting angiogenesis (GO:0002040) 4/52 0.036341065 ACVRL1;AKT1;PIK3R3;DLL1
8 positive regulation of cellular process (GO:0048522) 12/625 0.036341065 LYN;CSF3;FCER1G;PDF;ACER3;DOT1L;AKT1;SOX11;PTK6;PTPRJ;DLL1;CXCL16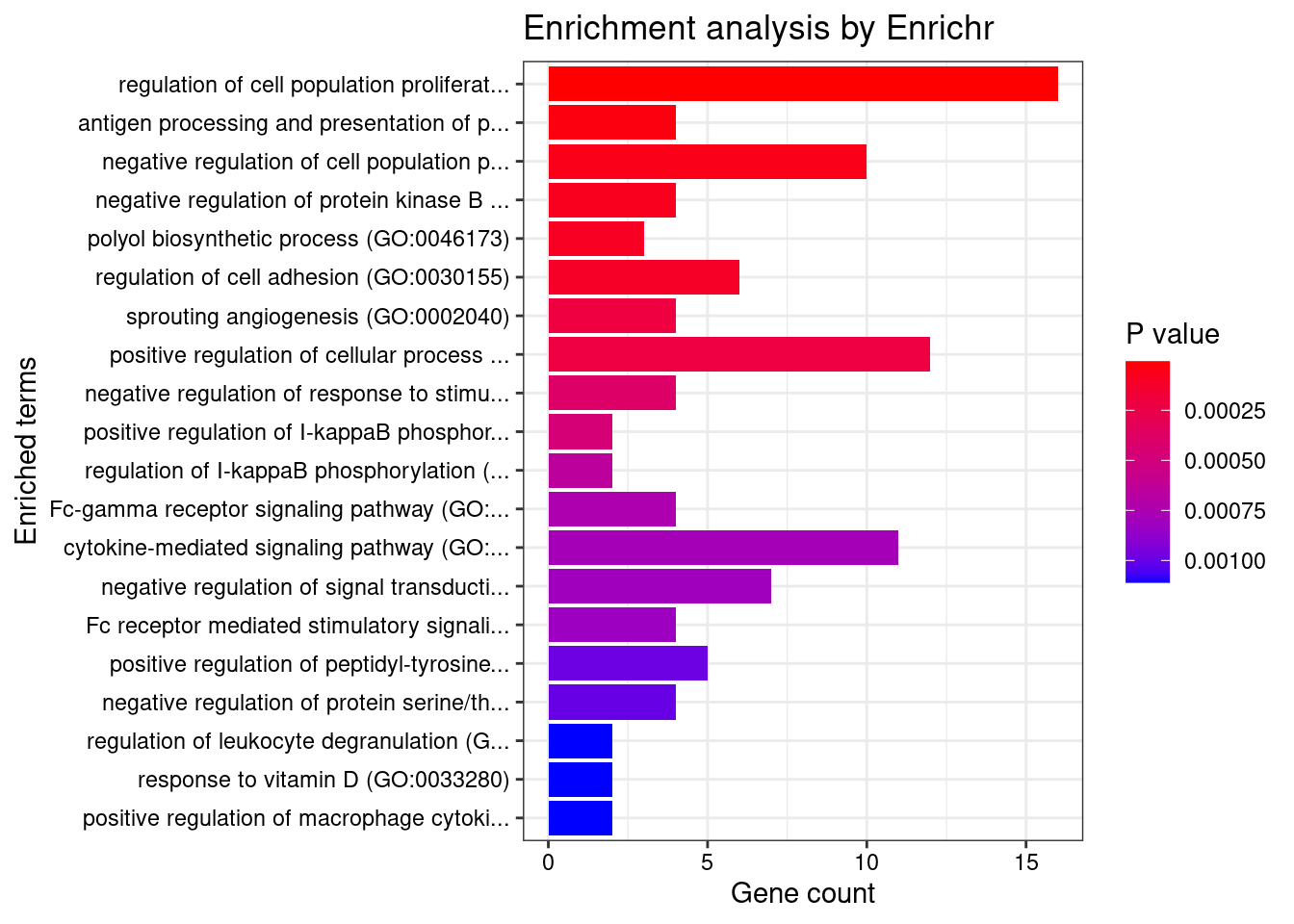
None
Number of cTWAS Genes in Tissue Group: 134
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)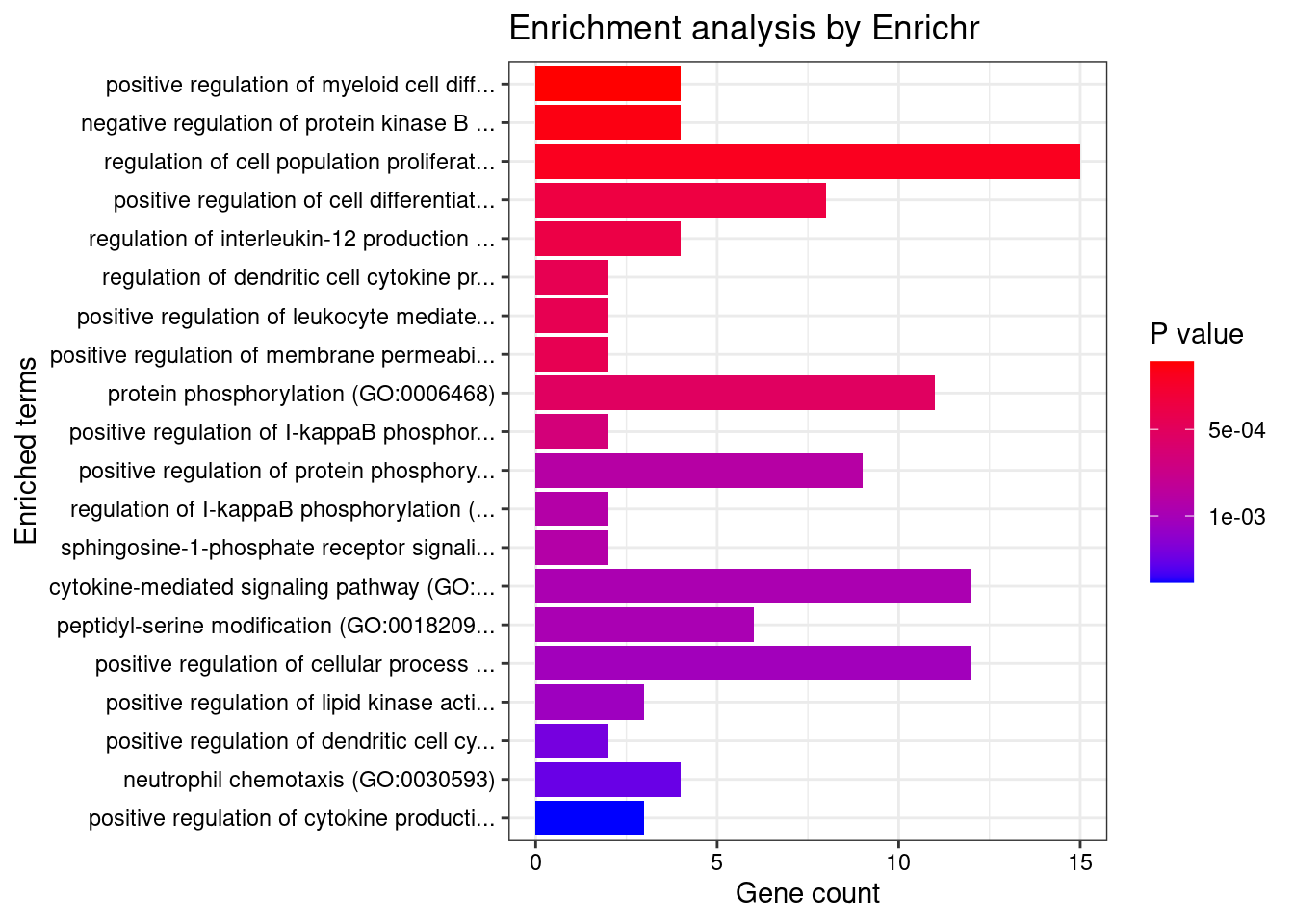
Skin
Number of cTWAS Genes in Tissue Group: 77
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of dendritic cell cytokine production (GO:0002730) 2/5 0.04748461 PLCG2;SCIMP
2 positive regulation of leukocyte mediated immunity (GO:0002705) 2/5 0.04748461 PLCG2;SCIMP
3 positive regulation of I-kappaB phosphorylation (GO:1903721) 2/6 0.04748461 PLCG2;AKT1
4 positive regulation of cytokine production involved in immune response (GO:0002720) 3/33 0.04748461 PLCG2;LAPTM5;SCIMP
5 positive regulation of interleukin-12 production (GO:0032735) 3/34 0.04748461 PLCG2;LAPTM5;SCIMP
6 regulation of I-kappaB phosphorylation (GO:1903719) 2/7 0.04748461 PLCG2;AKT1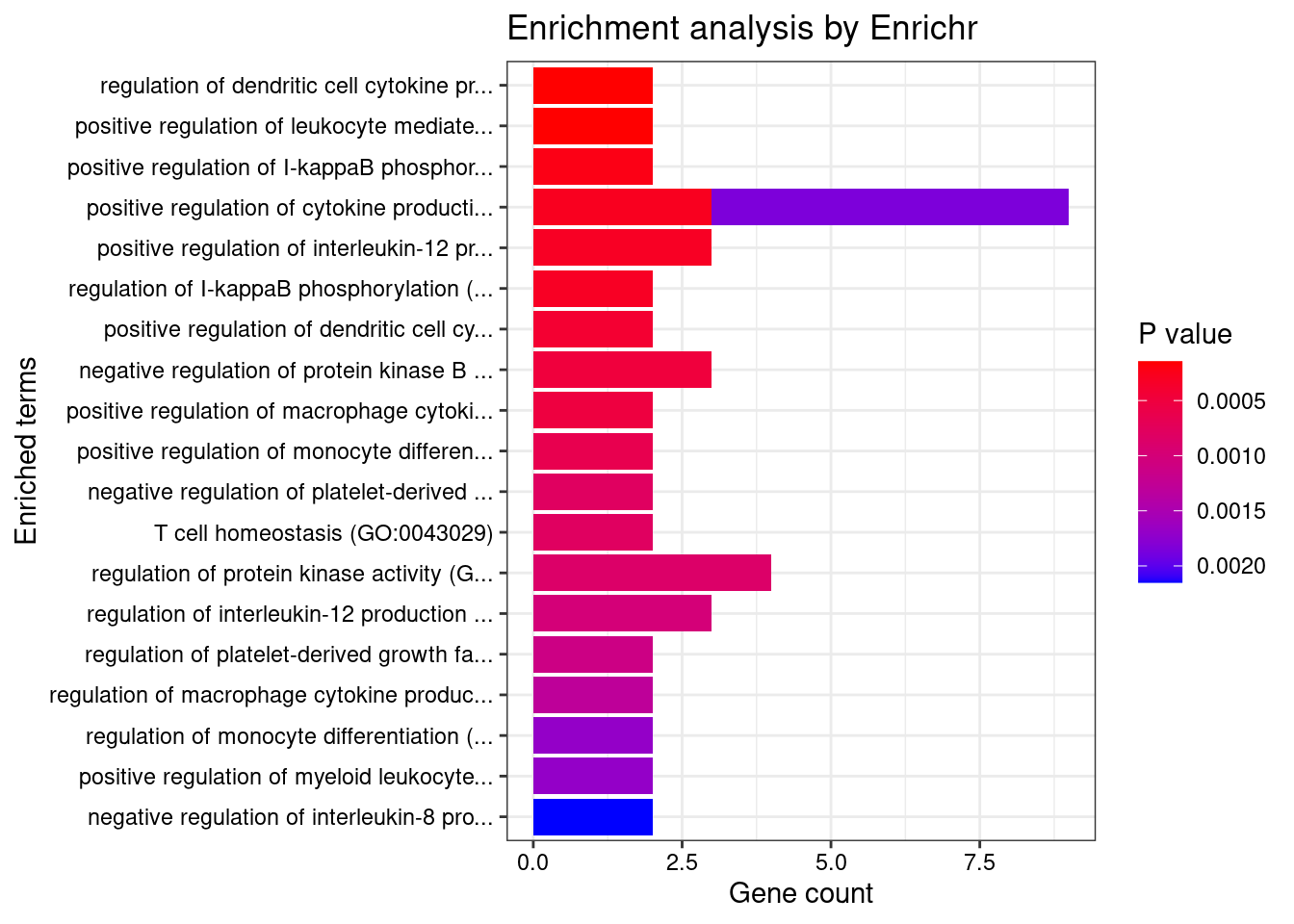
Blood or Immune
Number of cTWAS Genes in Tissue Group: 120
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)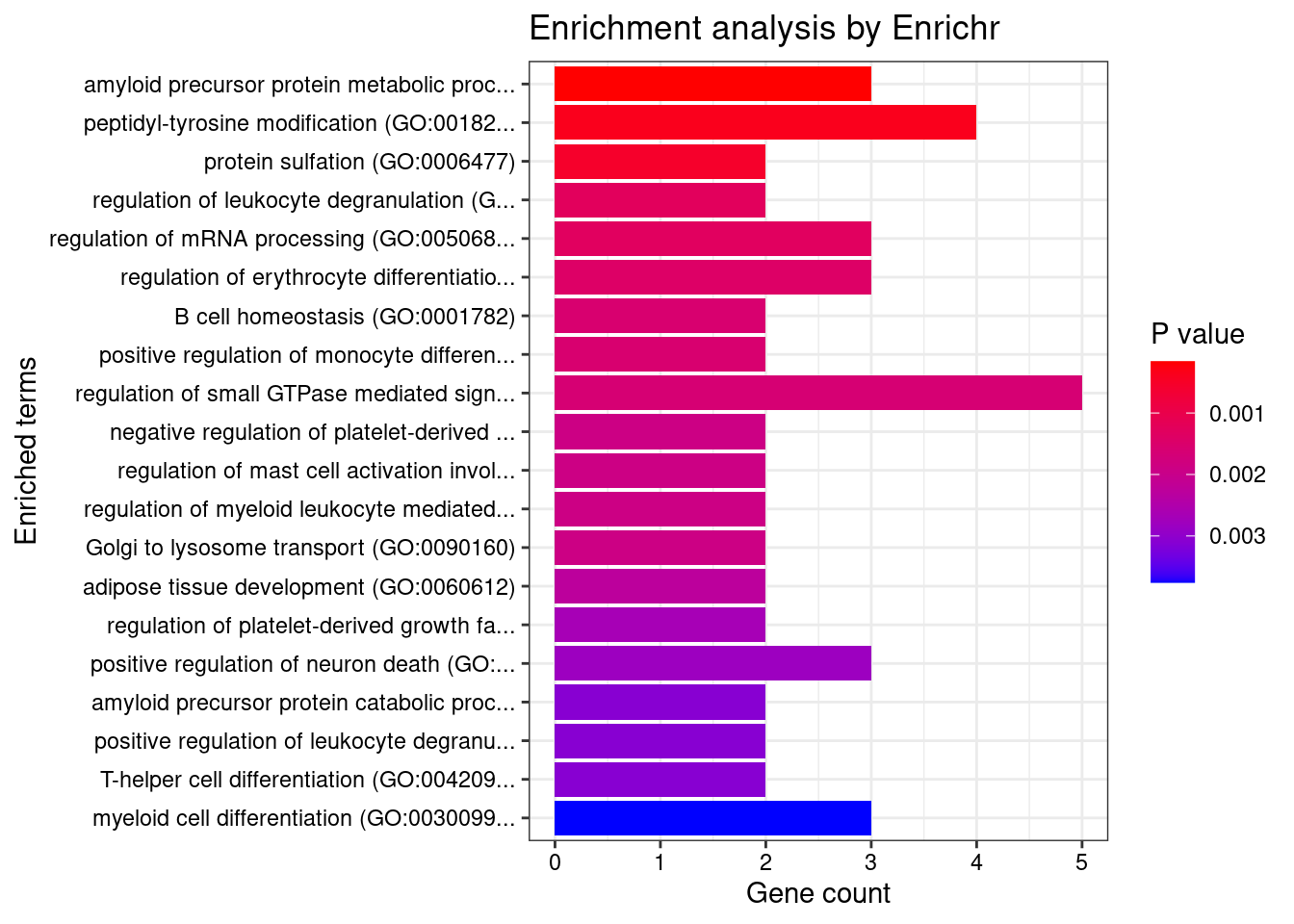
Digestive
Number of cTWAS Genes in Tissue Group: 106
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 negative regulation of protein kinase B signaling (GO:0051898) 4/40 0.03765327 SLC9A3R1;PLEKHA1;AKT1;PTPRJ
2 positive regulation of cell differentiation (GO:0045597) 8/258 0.03765327 IFITM1;REST;CSF1;FES;RPS6KA1;AKT1;SOX11;HOXA5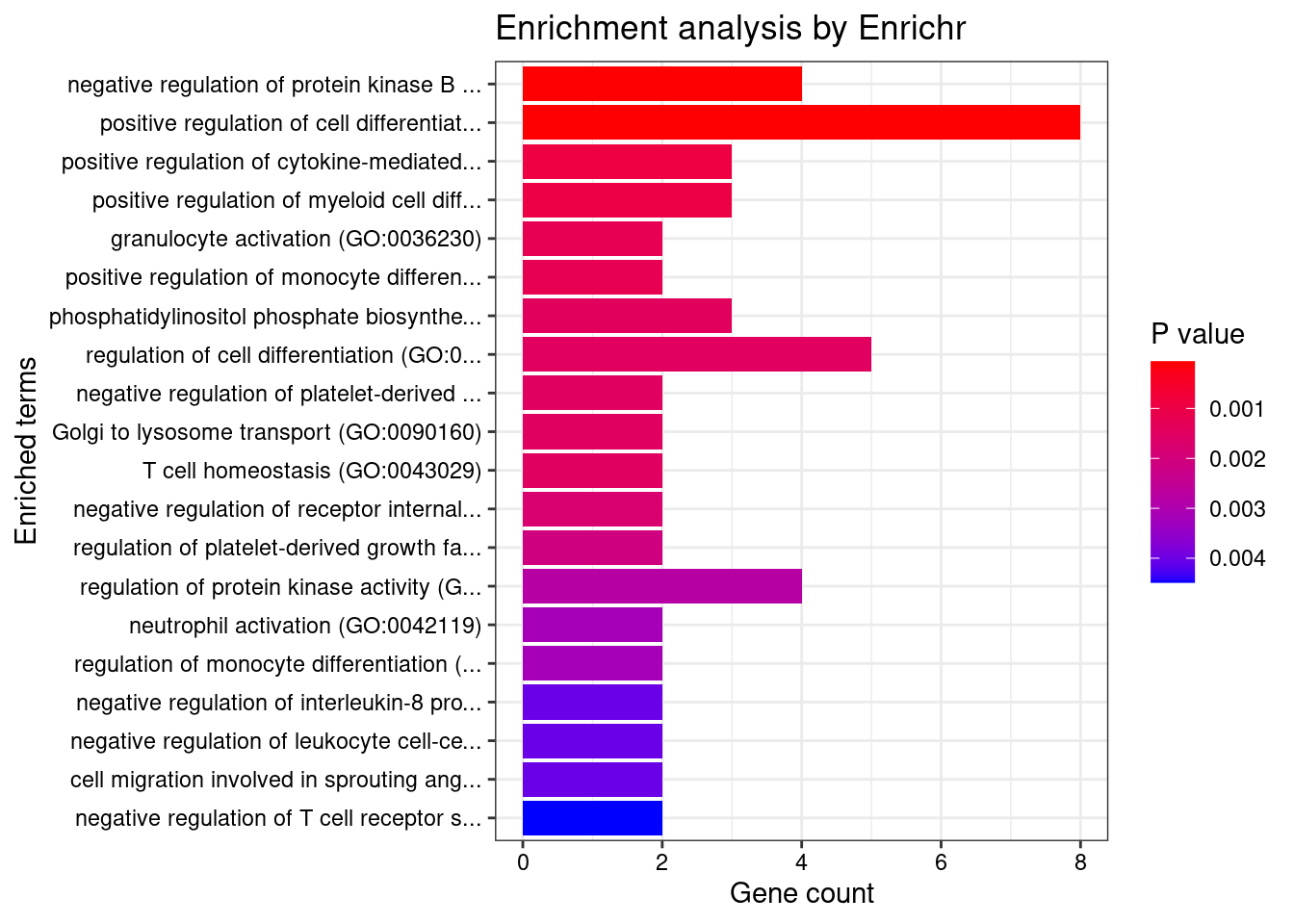
KEGG
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_group, referenceGene=background_group,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 71
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Endocrine
Number of cTWAS Genes in Tissue Group: 90
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Cardiovascular
Number of cTWAS Genes in Tissue Group: 112
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
CNS
Number of cTWAS Genes in Tissue Group: 113
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
None
Number of cTWAS Genes in Tissue Group: 134
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Skin
Number of cTWAS Genes in Tissue Group: 77
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Blood or Immune
Number of cTWAS Genes in Tissue Group: 120
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Digestive
Number of cTWAS Genes in Tissue Group: 106
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_group, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 71LINC01624 gene(s) from the input list not found in DisGeNET CURATEDGALNT1 gene(s) from the input list not found in DisGeNET CURATEDCADM4 gene(s) from the input list not found in DisGeNET CURATEDMARCH7 gene(s) from the input list not found in DisGeNET CURATEDC18orf25 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDFAM212A gene(s) from the input list not found in DisGeNET CURATEDC15orf40 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDFAM177A1 gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDFRAT1 gene(s) from the input list not found in DisGeNET CURATEDARAP2 gene(s) from the input list not found in DisGeNET CURATEDLRRC25 gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDWBP1L gene(s) from the input list not found in DisGeNET CURATEDCHP1 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDMED24 gene(s) from the input list not found in DisGeNET CURATEDRP5-894D12.5 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDRP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDSWT1 gene(s) from the input list not found in DisGeNET CURATED
Endocrine
Number of cTWAS Genes in Tissue Group: 90RP11-138A9.2 gene(s) from the input list not found in DisGeNET CURATEDRAB37 gene(s) from the input list not found in DisGeNET CURATEDAPOM gene(s) from the input list not found in DisGeNET CURATEDCTC-492K19.7 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDLRRC25 gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDSIGLEC9 gene(s) from the input list not found in DisGeNET CURATEDCD302 gene(s) from the input list not found in DisGeNET CURATEDKANK3 gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATEDRP11-50B3.2 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDCATSPER4 gene(s) from the input list not found in DisGeNET CURATEDRP11-556O15.1 gene(s) from the input list not found in DisGeNET CURATEDWIPF2 gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDPDCD7 gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDTPST1 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDGSDMA gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDLINC01623 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDDUSP14 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDTADA2A gene(s) from the input list not found in DisGeNET CURATEDPRR15L gene(s) from the input list not found in DisGeNET CURATEDMICALL2 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDRP4-796I8.1 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDRP5-894D12.5 gene(s) from the input list not found in DisGeNET CURATED
Cardiovascular
Number of cTWAS Genes in Tissue Group: 112RAB37 gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDLINC00638 gene(s) from the input list not found in DisGeNET CURATEDTMED6 gene(s) from the input list not found in DisGeNET CURATEDSLC45A4 gene(s) from the input list not found in DisGeNET CURATEDFGF11 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDKIAA1755 gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDSLFN12 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDPCDHB3 gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDJAML gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDCD302 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDKANK3 gene(s) from the input list not found in DisGeNET CURATEDIP6K1 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDRPAIN gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDPDCD7 gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDPOLR2E gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDPREX1 gene(s) from the input list not found in DisGeNET CURATEDAC005754.8 gene(s) from the input list not found in DisGeNET CURATEDTUBG2 gene(s) from the input list not found in DisGeNET CURATEDORAOV1 gene(s) from the input list not found in DisGeNET CURATEDCHP1 gene(s) from the input list not found in DisGeNET CURATEDLINC00094 gene(s) from the input list not found in DisGeNET CURATEDLINC01184 gene(s) from the input list not found in DisGeNET CURATEDNECAB2 gene(s) from the input list not found in DisGeNET CURATEDCDK11B gene(s) from the input list not found in DisGeNET CURATEDC15orf40 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDPDLIM2 gene(s) from the input list not found in DisGeNET CURATEDHOXC6 gene(s) from the input list not found in DisGeNET CURATEDMICALL2 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDRP5-894D12.5 gene(s) from the input list not found in DisGeNET CURATEDRP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDBORCS7 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATED
CNS
Number of cTWAS Genes in Tissue Group: 113ATG14 gene(s) from the input list not found in DisGeNET CURATEDCD302 gene(s) from the input list not found in DisGeNET CURATEDCYB561D1 gene(s) from the input list not found in DisGeNET CURATEDRP11-442O1.3 gene(s) from the input list not found in DisGeNET CURATEDPCDHB3 gene(s) from the input list not found in DisGeNET CURATEDDNAH10OS gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDCADM4 gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDAPOM gene(s) from the input list not found in DisGeNET CURATEDSLC5A11 gene(s) from the input list not found in DisGeNET CURATEDVKORC1L1 gene(s) from the input list not found in DisGeNET CURATEDSPPL2C gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDTTLL12 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDZNF114 gene(s) from the input list not found in DisGeNET CURATEDBTN2A2 gene(s) from the input list not found in DisGeNET CURATEDDTNB gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDAC005754.8 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDCTC-498M16.4 gene(s) from the input list not found in DisGeNET CURATEDPDF gene(s) from the input list not found in DisGeNET CURATEDFRAT1 gene(s) from the input list not found in DisGeNET CURATEDCRCP gene(s) from the input list not found in DisGeNET CURATEDRABEP1 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDPIK3R3 gene(s) from the input list not found in DisGeNET CURATEDFCRLB gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDAC144831.3 gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDCYB5D1 gene(s) from the input list not found in DisGeNET CURATEDKIAA0391 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDFGF11 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDTMED6 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDCTC-492K19.7 gene(s) from the input list not found in DisGeNET CURATEDNAA38 gene(s) from the input list not found in DisGeNET CURATEDTSPAN14 gene(s) from the input list not found in DisGeNET CURATEDZNF311 gene(s) from the input list not found in DisGeNET CURATEDMYO1G gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDUBALD2 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATED
None
Number of cTWAS Genes in Tissue Group: 134LRRC2 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDRP4-621B10.8 gene(s) from the input list not found in DisGeNET CURATEDHLA-DOB gene(s) from the input list not found in DisGeNET CURATEDGALNT1 gene(s) from the input list not found in DisGeNET CURATEDAPOM gene(s) from the input list not found in DisGeNET CURATEDRP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDADAM23 gene(s) from the input list not found in DisGeNET CURATEDTMCC3 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCKMT1A gene(s) from the input list not found in DisGeNET CURATEDOSCP1 gene(s) from the input list not found in DisGeNET CURATEDSUN2 gene(s) from the input list not found in DisGeNET CURATEDGPANK1 gene(s) from the input list not found in DisGeNET CURATEDIP6K1 gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDST3GAL6 gene(s) from the input list not found in DisGeNET CURATEDCDK11B gene(s) from the input list not found in DisGeNET CURATEDRP11-107M16.2 gene(s) from the input list not found in DisGeNET CURATEDTUBG2 gene(s) from the input list not found in DisGeNET CURATEDKIAA0391 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDCTC-492K19.7 gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDFAM69A gene(s) from the input list not found in DisGeNET CURATEDRP11-738E22.3 gene(s) from the input list not found in DisGeNET CURATEDMYO1G gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDNFE2L1 gene(s) from the input list not found in DisGeNET CURATEDPAPOLA gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDSWT1 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDCD302 gene(s) from the input list not found in DisGeNET CURATEDJAML gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDZNF180 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDDHRS12 gene(s) from the input list not found in DisGeNET CURATEDSCIMP gene(s) from the input list not found in DisGeNET CURATEDCNOT11 gene(s) from the input list not found in DisGeNET CURATEDMICALL2 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDC5orf64-AS1 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDPIK3R3 gene(s) from the input list not found in DisGeNET CURATEDRP11-219G17.9 gene(s) from the input list not found in DisGeNET CURATEDCYB5D1 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDMED24 gene(s) from the input list not found in DisGeNET CURATEDTMED6 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDADGRF5 gene(s) from the input list not found in DisGeNET CURATED
Skin
Number of cTWAS Genes in Tissue Group: 77CD302 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDGUCY1B3 gene(s) from the input list not found in DisGeNET CURATEDRPN1 gene(s) from the input list not found in DisGeNET CURATEDEMILIN3 gene(s) from the input list not found in DisGeNET CURATEDNUDC gene(s) from the input list not found in DisGeNET CURATEDC6orf47 gene(s) from the input list not found in DisGeNET CURATEDCDK18 gene(s) from the input list not found in DisGeNET CURATEDAPOM gene(s) from the input list not found in DisGeNET CURATEDC18orf25 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDSENP3 gene(s) from the input list not found in DisGeNET CURATEDRNF2 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDSCIMP gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDRP11-672A2.4 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDRP11-387H17.6 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDMEX3A gene(s) from the input list not found in DisGeNET CURATEDCRCP gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDTUBG2 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDFGF11 gene(s) from the input list not found in DisGeNET CURATEDPRR15L gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDRP11-20J15.3 gene(s) from the input list not found in DisGeNET CURATEDRAP1GAP2 gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDTADA2A gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
159 Stomatitis 0.03041097 2/42 4/9703
480 Oral Mucositis 0.03041097 2/42 4/9703
Blood or Immune
Number of cTWAS Genes in Tissue Group: 120OSTF1 gene(s) from the input list not found in DisGeNET CURATEDGPR4 gene(s) from the input list not found in DisGeNET CURATEDTTC39C gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDBAZ2B gene(s) from the input list not found in DisGeNET CURATEDCTC-492K19.7 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDSUMO3 gene(s) from the input list not found in DisGeNET CURATEDRP11-45M22.5 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDCITED4 gene(s) from the input list not found in DisGeNET CURATEDZNF114 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDUBE2F gene(s) from the input list not found in DisGeNET CURATEDTRMT12 gene(s) from the input list not found in DisGeNET CURATEDCA11 gene(s) from the input list not found in DisGeNET CURATEDMED11 gene(s) from the input list not found in DisGeNET CURATEDGLG1 gene(s) from the input list not found in DisGeNET CURATEDLRRC25 gene(s) from the input list not found in DisGeNET CURATEDRNF181 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDCDC42BPA gene(s) from the input list not found in DisGeNET CURATEDARAP2 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDRABEP1 gene(s) from the input list not found in DisGeNET CURATEDMTMR12 gene(s) from the input list not found in DisGeNET CURATEDUQCRC1 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDTPST1 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDKIAA0391 gene(s) from the input list not found in DisGeNET CURATEDTBC1D14 gene(s) from the input list not found in DisGeNET CURATEDTADA1 gene(s) from the input list not found in DisGeNET CURATEDDDX60L gene(s) from the input list not found in DisGeNET CURATEDCCDC9 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDUBOX5 gene(s) from the input list not found in DisGeNET CURATEDZFPM1 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDPDLIM2 gene(s) from the input list not found in DisGeNET CURATEDZFP36L1 gene(s) from the input list not found in DisGeNET CURATEDMICALL2 gene(s) from the input list not found in DisGeNET CURATEDTPST2 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDHDHD5 gene(s) from the input list not found in DisGeNET CURATED
Digestive
Number of cTWAS Genes in Tissue Group: 106CRCP gene(s) from the input list not found in DisGeNET CURATEDC19orf35 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDRP11-45M22.5 gene(s) from the input list not found in DisGeNET CURATEDPCDHB2 gene(s) from the input list not found in DisGeNET CURATEDCD101 gene(s) from the input list not found in DisGeNET CURATEDUBASH3B gene(s) from the input list not found in DisGeNET CURATEDLAMP1 gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDTRMT12 gene(s) from the input list not found in DisGeNET CURATEDJAML gene(s) from the input list not found in DisGeNET CURATEDEHD4 gene(s) from the input list not found in DisGeNET CURATEDFRAT2 gene(s) from the input list not found in DisGeNET CURATEDTMCC3 gene(s) from the input list not found in DisGeNET CURATEDCSNK1G1 gene(s) from the input list not found in DisGeNET CURATEDAP000350.5 gene(s) from the input list not found in DisGeNET CURATEDTMEM99 gene(s) from the input list not found in DisGeNET CURATEDFAM102A gene(s) from the input list not found in DisGeNET CURATEDCDC42BPA gene(s) from the input list not found in DisGeNET CURATEDRPUSD4 gene(s) from the input list not found in DisGeNET CURATEDSCG3 gene(s) from the input list not found in DisGeNET CURATEDATG14 gene(s) from the input list not found in DisGeNET CURATEDVWA7 gene(s) from the input list not found in DisGeNET CURATEDZBTB22 gene(s) from the input list not found in DisGeNET CURATEDPKN3 gene(s) from the input list not found in DisGeNET CURATEDBPI gene(s) from the input list not found in DisGeNET CURATEDLSM4 gene(s) from the input list not found in DisGeNET CURATEDAP1M2 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDRP11-138A9.1 gene(s) from the input list not found in DisGeNET CURATEDATP13A1 gene(s) from the input list not found in DisGeNET CURATEDFES gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDPIK3R3 gene(s) from the input list not found in DisGeNET CURATEDTADA1 gene(s) from the input list not found in DisGeNET CURATEDNAA38 gene(s) from the input list not found in DisGeNET CURATEDCPSF4 gene(s) from the input list not found in DisGeNET CURATEDCDC37 gene(s) from the input list not found in DisGeNET CURATEDTUBG2 gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDSLC2A6 gene(s) from the input list not found in DisGeNET CURATEDSLFN13 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDLINC01184 gene(s) from the input list not found in DisGeNET CURATEDTMEM150A gene(s) from the input list not found in DisGeNET CURATEDGGT5 gene(s) from the input list not found in DisGeNET CURATEDNECAB2 gene(s) from the input list not found in DisGeNET CURATEDDUSP14 gene(s) from the input list not found in DisGeNET CURATEDC15orf40 gene(s) from the input list not found in DisGeNET CURATEDBORCS7 gene(s) from the input list not found in DisGeNET CURATEDHOXC6 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDRAI14 gene(s) from the input list not found in DisGeNET CURATEDTHOC7 gene(s) from the input list not found in DisGeNET CURATEDMARCH7 gene(s) from the input list not found in DisGeNET CURATEDGene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_group=ctwas_genes_group)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_group]})
#hypergeometric test
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_group)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
#multiple testing correction
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 71
gene_set nset ngenes percent padj
1 gwascatalog 4753 42 0.008836524 6.741585e-06
2 mgi_essential 1784 16 0.008968610 1.930904e-02
3 fda_approved_drug_targets 267 4 0.014981273 6.168503e-02
4 clinvar_path_likelypath 2206 12 0.005439710 4.257896e-01
5 core_essentials_hart 215 0 0.000000000 1.000000e+00
Endocrine
Number of cTWAS Genes in Tissue Group: 90
gene_set nset ngenes percent padj
1 gwascatalog 5520 52 0.009420290 2.847080e-08
2 mgi_essential 2078 12 0.005774783 4.249481e-01
3 fda_approved_drug_targets 307 3 0.009771987 4.249481e-01
4 clinvar_path_likelypath 2540 12 0.004724409 6.516744e-01
5 core_essentials_hart 240 1 0.004166667 6.750012e-01
Cardiovascular
Number of cTWAS Genes in Tissue Group: 112
gene_set nset ngenes percent padj
1 gwascatalog 5286 61 0.011539917 2.279009e-06
2 fda_approved_drug_targets 288 6 0.020833333 3.260250e-02
3 mgi_essential 2013 19 0.009438649 1.288971e-01
4 core_essentials_hart 245 2 0.008163265 6.138377e-01
5 clinvar_path_likelypath 2449 13 0.005308289 8.547418e-01
CNS
Number of cTWAS Genes in Tissue Group: 113
gene_set nset ngenes percent padj
1 gwascatalog 5500 63 0.011454545 8.325974e-08
2 fda_approved_drug_targets 321 5 0.015576324 1.265855e-01
3 mgi_essential 2120 18 0.008490566 1.791109e-01
4 core_essentials_hart 246 2 0.008130081 4.552458e-01
5 clinvar_path_likelypath 2567 17 0.006622517 4.552458e-01
None
Number of cTWAS Genes in Tissue Group: 134
gene_set nset ngenes percent padj
1 gwascatalog 5703 77 0.013501666 2.358511e-10
2 mgi_essential 2172 29 0.013351750 1.372137e-03
3 fda_approved_drug_targets 323 8 0.024767802 3.615244e-03
4 core_essentials_hart 257 3 0.011673152 3.438471e-01
5 clinvar_path_likelypath 2641 17 0.006436956 7.046695e-01
Skin
Number of cTWAS Genes in Tissue Group: 77
gene_set nset ngenes percent padj
1 gwascatalog 5216 41 0.007860429 0.0002625334
2 mgi_essential 1980 16 0.008080808 0.0443589772
3 fda_approved_drug_targets 281 3 0.010676157 0.2352818603
4 clinvar_path_likelypath 2399 14 0.005835765 0.2663500041
5 core_essentials_hart 240 1 0.004166667 0.6741040032
Blood or Immune
Number of cTWAS Genes in Tissue Group: 120
gene_set nset ngenes percent padj
1 gwascatalog 4912 65 0.013232899 1.279969e-06
2 mgi_essential 1858 21 0.011302476 1.180551e-01
3 fda_approved_drug_targets 262 4 0.015267176 2.425268e-01
4 core_essentials_hart 225 1 0.004444444 8.279801e-01
5 clinvar_path_likelypath 2260 16 0.007079646 8.279801e-01
Digestive
Number of cTWAS Genes in Tissue Group: 106
gene_set nset ngenes percent padj
1 gwascatalog 5483 63 0.011490060 6.072635e-09
2 mgi_essential 2089 20 0.009573959 5.558586e-02
3 fda_approved_drug_targets 311 5 0.016077170 6.510390e-02
4 core_essentials_hart 250 2 0.008000000 5.533058e-01
5 clinvar_path_likelypath 2537 12 0.004729996 8.480962e-01Analysis of TWAS False Positives by Region
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_tag, function(x){unlist(region_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from region total to get combined pip for other genes in region
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip < 0.5)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip < 0.5)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
| Version | Author | Date |
|---|---|---|
| 95e0f8e | wesleycrouse | 2022-04-07 |
Analysis of TWAS False Positives by Credible Set
cTWAS is using susie settings that mask credible sets consisting of variables with minimum pairwise correlations below a specified threshold. The default threshold is 0.5. I think this is intended to mask credible sets with “diffuse” support. As a consequence, many of the genes considered here (TWAS false positives; significant z score but low PIP) are not assigned to a credible set (have cs_index=0). For this reason, the first figure is not really appropriate for answering the question “are TWAS false positives due to SNPs or genes”.
The second figure includes only TWAS genes that are assigned to a reported causal set (i.e. they are in a “pure” causal set with high pairwise correlations). I think that this figure is closer to the intended analysis. However, it may be biased in some way because we have excluded many TWAS false positive genes that are in “impure” credible sets.
Some alternatives to these figures include the region-based analysis in the previous section; or re-analysis with lower/no minimum pairwise correlation threshold (“min_abs_corr” option in susie_get_cs) for reporting credible sets.
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
| Version | Author | Date |
|---|---|---|
| 95e0f8e | wesleycrouse | 2022-04-07 |
####################
#using only genes assigned to a credible set
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
#exclude genes that are not assigned to a credible set, cs_index==0
gene_pips <- gene_pips[as.numeric(sapply(gene_pips$region_cs_tag, function(x){rev(unlist(strsplit(x, "_")))[1]}))!=0,]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
cTWAS genes without genome-wide significant SNP nearby
novel_genes <- data.frame(genename=as.character(), weight=as.character(), susie_pip=as.numeric(), snp_maxz=as.numeric())
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$ctwas,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, sapply(gene_pips$region_tag, function(x){region_pips[x,"snp_maxz"]}))
names(gene_pips)[ncol(gene_pips)] <- "snp_maxz"
if (nrow(gene_pips)>0){
gene_pips$weight <- names(df)[i]
gene_pips <- gene_pips[gene_pips$snp_maxz < qnorm(1-(5E-8/2), lower=T),c("genename", "weight", "susie_pip", "snp_maxz")]
novel_genes <- rbind(novel_genes, gene_pips)
}
}
novel_genes_summary <- data.frame(genename=unique(novel_genes$genename))
novel_genes_summary$nweights <- sapply(novel_genes_summary$genename, function(x){length(novel_genes$weight[novel_genes$genename==x])})
novel_genes_summary$weights <- sapply(novel_genes_summary$genename, function(x){paste(novel_genes$weight[novel_genes$genename==x],collapse=", ")})
novel_genes_summary <- novel_genes_summary[order(-novel_genes_summary$nweights),]
novel_genes_summary[,c("genename","nweights")] genename nweights
21 RP11-394I13.2 18
38 KIF18B 12
22 AKAP10 11
44 THOC7 11
19 PPP5C 8
20 UBOX5 8
30 EEF1A2 8
34 RAI14 8
43 SOX11 8
5 ACHE 7
10 RAB21 7
9 RP11-672A2.4 6
58 FASTKD5 6
2 SPATS2L 5
3 ATXN7 5
15 ITPKC 5
17 CKMT1A 5
45 C19orf35 5
46 ST3GAL4 5
47 PLCG2 5
48 CTC-492K19.7 5
52 KIAA0040 5
70 TAL1 5
75 ADAM33 5
24 FGD5 4
31 NR5A2 4
42 PTK6 4
51 MAP2K5 4
64 NNT 4
85 MICALL2 4
6 KMT2E 3
8 PLEKHA1 3
11 C15orf40 3
23 KANK3 3
26 TLE4 3
37 SLC4A1 3
40 UBE2F 3
1 SWT1 2
4 LCORL 2
16 KCNQ3 2
18 GALNT1 2
27 HOXC6 2
28 NECAB2 2
32 RP11-373D23.3 2
33 HS6ST1 2
35 LINC01184 2
39 CDK11B 2
55 F5 2
60 BMP1 2
62 PAPOLA 2
67 OSGIN1 2
72 CDC14A 2
76 HNRNPAB 2
81 ITGA2 2
83 PDCD7 2
90 TMCC3 2
92 BCL2L2 2
103 TRMT12 2
7 CAV1 1
12 MYH10 1
13 RP11-322E11.5 1
14 CD320 1
25 CSGALNACT1 1
29 KIAA1755 1
36 SLFN12 1
41 TIRAP 1
49 SLC5A11 1
50 DOT1L 1
53 CXCR4 1
54 ACER3 1
56 TSPAN14 1
57 CYP27B1 1
59 CTC-498M16.4 1
61 TMPO 1
63 GUCY1B3 1
65 MYLK4 1
66 PNLIPRP1 1
68 EMILIN3 1
69 MLLT1 1
71 KRT10 1
73 SCG3 1
74 SLFN13 1
77 TACR2 1
78 TMEM99 1
79 RPUSD4 1
80 BPI 1
82 EYA4 1
84 SMG1 1
86 HOMEZ 1
87 VPS26A 1
88 ST3GAL6 1
89 ADAM10 1
91 ADAM23 1
93 EML4 1
94 C5orf64-AS1 1
95 BCKDHA 1
96 JAM3 1
97 COQ8B 1
98 RP11-107M16.2 1
99 SUN2 1
100 RNF2 1
101 CEP85L 1
102 SHANK3 1
104 MAP3K5 1
105 PDLIM2 1
106 ITGB2 1
107 SUMO3 1
108 TPST2 1
109 ADTRP 1
110 FZD6 1
111 RP4-796I8.1 1
112 RP11-50B3.2 1
113 PYGL 1
114 CD70 1
115 TGM2 1
116 RORC 1
117 SELL 1
118 TRAF3IP3 1
119 PAQR9 1
120 DDX60L 1
121 NPR3 1
122 MTMR12 1
123 ADAM19 1
124 OSTF1 1
125 GLG1 1
126 RALBP1 1
127 GPR4 1
128 SLC25A1 1Tissue-specificity for cTWAS genes
gene_pips_by_weight <- data.frame(genename=as.character(ctwas_genes))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips
gene_pips <- gene_pips[match(ctwas_genes, gene_pips$genename),,drop=F]
#gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- -1
gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- 0 #missing values coded as PIP=0
gene_pips_by_weight <- cbind(gene_pips_by_weight, gene_pips$susie_pip)
names(gene_pips_by_weight)[ncol(gene_pips_by_weight)] <- names(df)[i]
}
gene_pips_by_weight <- as.matrix(gene_pips_by_weight[,-1])
rownames(gene_pips_by_weight) <- ctwas_genes
#number of tissues with PIP>0.5 for cTWAS genes
ctwas_frequency <- rowSums(gene_pips_by_weight>0.5)
hist(ctwas_frequency, col="grey", breaks=0:max(ctwas_frequency), xlim=c(0,ncol(gene_pips_by_weight)),
xlab="Number of Tissues with PIP>0.5",
ylab="Number of cTWAS Genes",
main="Tissue Specificity for cTWAS Genes")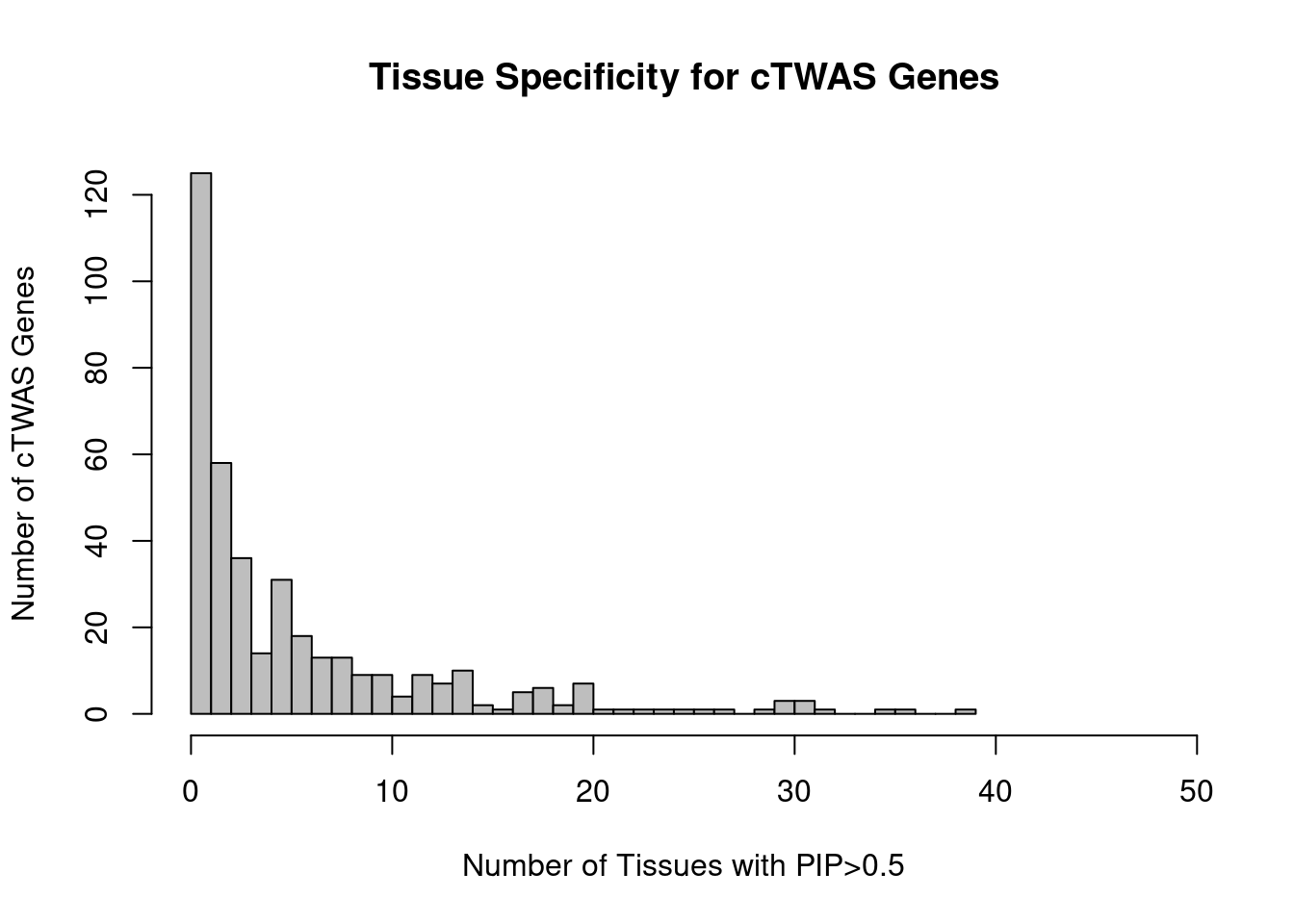
#heatmap of gene PIPs
cluster_ctwas_genes <- hclust(dist(gene_pips_by_weight))
cluster_ctwas_weights <- hclust(dist(t(gene_pips_by_weight)))
plot(cluster_ctwas_weights, cex=0.6)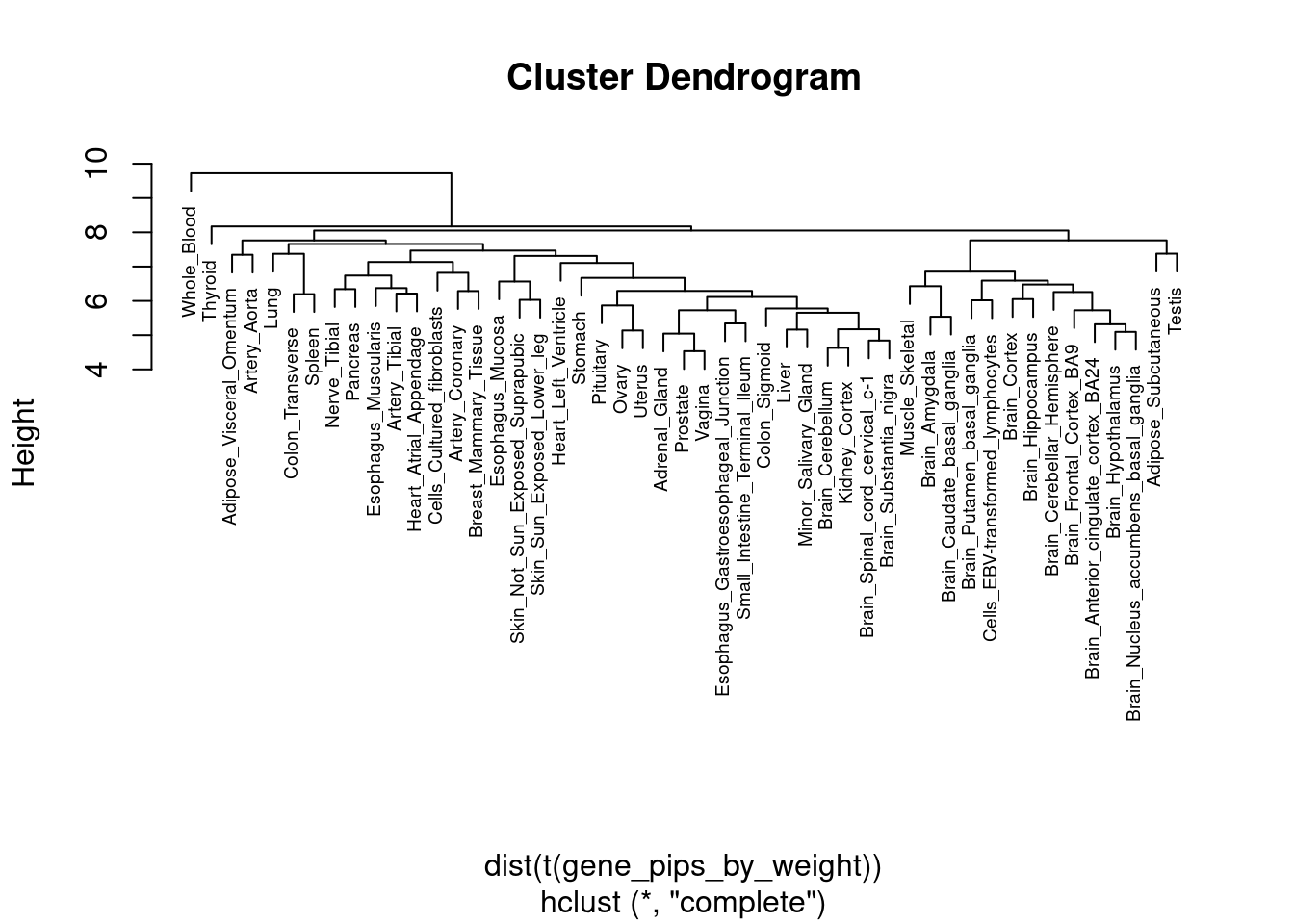
plot(cluster_ctwas_genes, cex=0.6, labels=F)
par(mar=c(14.1, 4.1, 4.1, 2.1))
image(t(gene_pips_by_weight[rev(cluster_ctwas_genes$order),rev(cluster_ctwas_weights$order)]),
axes=F)
mtext(text=colnames(gene_pips_by_weight)[cluster_ctwas_weights$order], side=1, line=0.3, at=seq(0,1,1/(ncol(gene_pips_by_weight)-1)), las=2, cex=0.8)
mtext(text=rownames(gene_pips_by_weight)[cluster_ctwas_genes$order], side=2, line=0.3, at=seq(0,1,1/(nrow(gene_pips_by_weight)-1)), las=1, cex=0.4)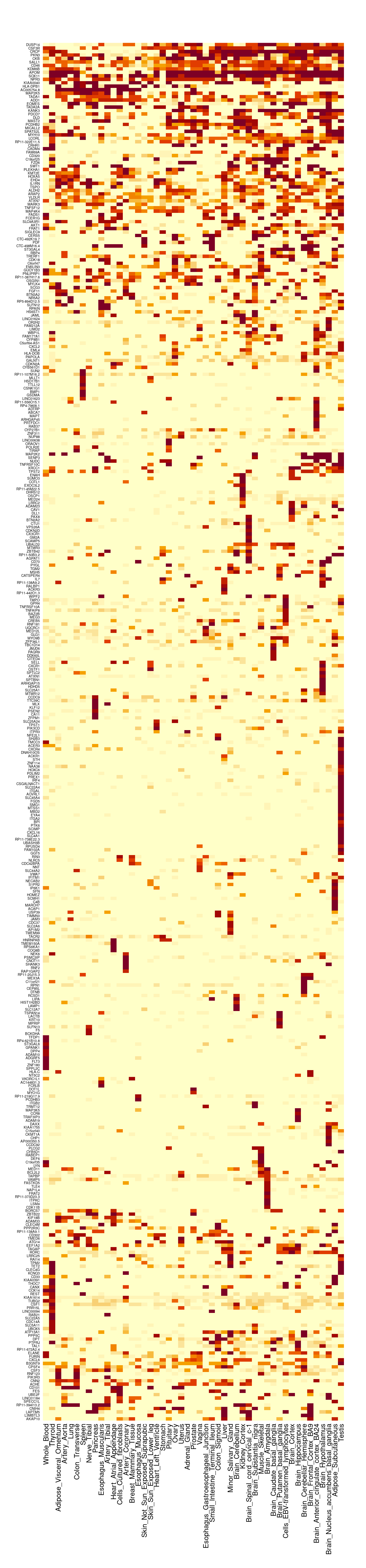
cTWAS genes with highest proportion of total PIP on a single tissue
#genes with highest porportion of PIP on a single tissue
gene_pips_proportion <- gene_pips_by_weight/rowSums(gene_pips_by_weight)
proportion_table <- data.frame(genename=as.character(rownames(gene_pips_proportion)))
proportion_table$max_pip_prop <- apply(gene_pips_proportion,1,max)
proportion_table$max_weight <- colnames(gene_pips_proportion)[apply(gene_pips_proportion,1,which.max)]
proportion_table[order(-proportion_table$max_pip_prop),] genename max_pip_prop max_weight
48 LINC01624 1.00000000 Adipose_Visceral_Omentum
148 DLL1 1.00000000 Brain_Caudate_basal_ganglia
332 RP11-556O15.1 1.00000000 Testis
348 RP11-50B3.2 1.00000000 Thyroid
286 C5orf64-AS1 1.00000000 Nerve_Tibial
337 GSDMA 0.99999957 Testis
344 AGPAT1 0.99999106 Thyroid
159 HLA-C 0.99989447 Brain_Cerebellar_Hemisphere
45 OR2H2 0.99983693 Adipose_Visceral_Omentum
77 WIPF2 0.99942162 Adrenal_Gland
334 LINC01623 0.99856015 Testis
381 ZFP36L1 0.99360519 Whole_Blood
165 SPPL2C 0.98808999 Brain_Cerebellar_Hemisphere
271 TFDP1 0.97585854 Lung
378 CREB5 0.97303264 Whole_Blood
361 RNF181 0.97098027 Whole_Blood
367 UQCRC1 0.96935260 Whole_Blood
209 C6orf47 0.94053335 Cells_Cultured_fibroblasts
318 SENP3 0.93377044 Skin_Sun_Exposed_Lower_leg
304 RP11-107M16.2 0.92869514 Prostate
238 SLC2A6 0.90646505 Esophagus_Mucosa
329 VWA7 0.89876141 Stomach
188 ZNF311 0.89673179 Brain_Hippocampus
31 CRHR1 0.89434303 Adipose_Subcutaneous
390 MYO9B 0.87900466 Whole_Blood
254 MBD2 0.87742831 Heart_Atrial_Appendage
310 RP11-20J15.3 0.86952385 Skin_Not_Sun_Exposed_Suprapubic
330 SLC44A2 0.85735072 Stomach
210 TRERF1 0.83067466 Cells_Cultured_fibroblasts
368 PAQR9 0.81666543 Whole_Blood
86 IRF4 0.80137055 Artery_Aorta
341 RP4-796I8.1 0.79531785 Testis
389 TTC39C 0.76861256 Whole_Blood
319 MAP2K2 0.73455759 Skin_Sun_Exposed_Lower_leg
323 ENAH 0.72784540 Spleen
285 CXCL2 0.72452648 Nerve_Tibial
265 RP4-621B10.8 0.72335360 Lung
364 CXCR1 0.72021581 Whole_Blood
100 PREX1 0.71083645 Artery_Aorta
387 JMJD6 0.71014206 Whole_Blood
351 CD70 0.69930435 Thyroid
369 MED12L 0.69903283 Whole_Blood
371 TBC1D14 0.69673218 Whole_Blood
126 LINC00638 0.69448123 Artery_Tibial
305 SUN2 0.66402174 Prostate
338 MAPT 0.66379255 Testis
111 RPAIN 0.63484241 Artery_Coronary
357 SELL 0.63095008 Whole_Blood
241 CDC37 0.62524509 Esophagus_Mucosa
242 AP1M2 0.62260471 Esophagus_Mucosa
355 CITED4 0.61812932 Whole_Blood
263 CDKN2D 0.61005864 Heart_Left_Ventricle
219 MLLT1 0.60918648 Cells_EBV-transformed_lymphocytes
278 OSCP1 0.60900921 Muscle_Skeletal
345 MSH5 0.60767732 Thyroid
333 ADTRP 0.59462471 Testis
339 ABCA7 0.59248432 Testis
204 CDK18 0.59139694 Cells_Cultured_fibroblasts
384 GLG1 0.57916991 Whole_Blood
145 PAX8 0.57841895 Brain_Caudate_basal_ganglia
362 ARHGAP15 0.57299462 Whole_Blood
372 DDX60L 0.56499367 Whole_Blood
386 MLX 0.55776044 Whole_Blood
282 DHRS12 0.54725251 Muscle_Skeletal
42 FAM212A 0.54648465 Adipose_Visceral_Omentum
268 GPANK1 0.54596039 Lung
32 RP11-322E11.5 0.54539006 Adipose_Subcutaneous
170 ACKR1 0.54461980 Brain_Cortex
198 CDKN2A 0.53713831 Brain_Spinal_cord_cervical_c-1
162 NT5C2 0.53308727 Brain_Cerebellar_Hemisphere
222 MPRIP 0.53062948 Colon_Sigmoid
306 MEX3A 0.52668948 Skin_Not_Sun_Exposed_Suprapubic
340 ARHGAP45 0.51353141 Testis
363 BAZ2B 0.50835971 Whole_Blood
147 BTN2A2 0.50369528 Brain_Caudate_basal_ganglia
174 CXCR4 0.50226329 Brain_Cortex
189 CYP27B1 0.50122916 Brain_Hippocampus
181 STH 0.49834780 Brain_Putamen_basal_ganglia
284 EML4 0.48910892 Nerve_Tibial
37 CADM4 0.48558039 Adipose_Subcutaneous
113 SLFN12 0.47694438 Artery_Coronary
380 KLF12 0.47656255 Whole_Blood
287 HLA-DOB 0.45808982 Nerve_Tibial
336 PRTFDC1 0.45062034 Testis
214 RP11-387H17.6 0.44941613 Cells_Cultured_fibroblasts
205 GUCY1B3 0.43740751 Cells_Cultured_fibroblasts
166 AC144831.3 0.43644725 Brain_Cerebellar_Hemisphere
62 MED24 0.43213471 Muscle_Skeletal
96 ITGAL 0.42781046 Artery_Aorta
382 SPTLC2 0.41931565 Whole_Blood
251 MTSS1 0.41058006 Heart_Atrial_Appendage
124 ORAOV1 0.41049201 Artery_Tibial
377 ATXN1 0.40687675 Whole_Blood
230 EXOC3L2 0.40381867 Spleen
312 RAP1GAP2 0.40012373 Skin_Not_Sun_Exposed_Suprapubic
216 EMILIN3 0.39561583 Cells_Cultured_fibroblasts
130 POLR2E 0.39459362 Artery_Tibial
89 CSGALNACT1 0.39141419 Artery_Aorta
315 NUDC 0.38862090 Skin_Sun_Exposed_Lower_leg
346 RP11-138A9.2 0.37515508 Thyroid
227 CSNK1G1 0.37385737 Colon_Transverse
311 C11orf21 0.37212088 Skin_Not_Sun_Exposed_Suprapubic
267 ST3GAL6 0.37113352 Lung
325 COTL1 0.36444515 Spleen
250 EYA4 0.36182250 Heart_Atrial_Appendage
98 RAB37 0.36162289 Testis
104 HS6ST1 0.36125023 Whole_Blood
243 FAM102A 0.35747026 Esophagus_Muscularis
240 TMEM99 0.35006247 Esophagus_Mucosa
374 MTMR12 0.34850402 Whole_Blood
218 HSD17B1 0.34731150 Cells_EBV-transformed_lymphocytes
266 DPP4 0.33863731 Lung
269 ADGRF5 0.33851066 Lung
184 HLA-DPB1 0.33817787 Brain_Putamen_basal_ganglia
342 CATSPER4 0.33762203 Thyroid
395 HDHD5 0.33499499 Whole_Blood
18 KMT2E 0.33170465 Adipose_Subcutaneous
276 SH2B3 0.33074016 Minor_Salivary_Gland
158 FCRLB 0.33072375 Brain_Cerebellar_Hemisphere
193 BMP1 0.33030520 Colon_Transverse
257 GM2A 0.32866686 Heart_Left_Ventricle
394 CCDC9 0.32613019 Whole_Blood
228 RP11-45M22.5 0.32598478 Spleen
375 TNFAIP8 0.32566360 Whole_Blood
202 PAPOLA 0.32551682 Breast_Mammary_Tissue
8 IL1RN 0.32533267 Brain_Hippocampus
270 FLT3 0.32496720 Lung
360 SPTBN1 0.32459285 Whole_Blood
261 SCAMP5 0.32137054 Heart_Left_Ventricle
383 MEG3 0.32034494 Whole_Blood
301 TPST1 0.31434840 Whole_Blood
52 WBP1L 0.30820669 Adipose_Visceral_Omentum
343 CYP8B1 0.30625388 Thyroid
379 OSTF1 0.30530564 Whole_Blood
109 JAML 0.30505156 Artery_Coronary
211 PNLIPRP1 0.30309850 Cells_Cultured_fibroblasts
161 VKORC1L1 0.30193898 Brain_Cerebellar_Hemisphere
12 ARAP2 0.29936571 Whole_Blood
160 MYO1G 0.29536506 Brain_Cerebellar_Hemisphere
298 COQ8B 0.29490524 Pancreas
328 TPST2 0.29470155 Spleen
385 ZFPM1 0.29443753 Whole_Blood
94 IFITM1 0.29347535 Stomach
353 TGM2 0.29148062 Thyroid
295 NEK6 0.28832140 Whole_Blood
308 RPN1 0.28814766 Skin_Not_Sun_Exposed_Suprapubic
239 TACR2 0.28660948 Esophagus_Mucosa
245 RPUSD4 0.28442608 Esophagus_Muscularis
256 CX3CR1 0.28430166 Heart_Left_Ventricle
186 RP11-442O1.3 0.28400012 Brain_Frontal_Cortex_BA9
347 IL7 0.28307841 Thyroid
212 OSGIN1 0.27568416 Cells_Cultured_fibroblasts
303 MTMR3 0.27564164 Pituitary
359 PSEN2 0.27512210 Whole_Blood
179 LACTB 0.27436645 Brain_Cortex
49 RP5-894D12.5 0.27434643 Adipose_Visceral_Omentum
307 RNF2 0.27296075 Skin_Not_Sun_Exposed_Suprapubic
349 PYGL 0.27148954 Thyroid
331 SLC25A24 0.27035748 Whole_Blood
26 ALDH2 0.27010682 Whole_Blood
128 NUP88 0.26795417 Artery_Tibial
41 MARCH7 0.26151294 Esophagus_Mucosa
235 RPS6KA1 0.26129944 Lung
185 TSPAN14 0.26022923 Brain_Frontal_Cortex_BA9
291 TNFRSF10C 0.25923007 Skin_Sun_Exposed_Lower_leg
143 UBALD2 0.25878902 Brain_Anterior_cingulate_cortex_BA24
157 CTU1 0.25695581 Brain_Caudate_basal_ganglia
393 CA11 0.25413775 Whole_Blood
177 ACER3 0.25229059 Brain_Cortex
30 MYH10 0.25113711 Adipose_Subcutaneous
255 ACKR3 0.25006916 Heart_Left_Ventricle
226 RIN3 0.24951359 Colon_Transverse
168 ACVRL1 0.24691477 Brain_Cerebellum
280 LRRC2 0.24658024 Muscle_Skeletal
82 IP6K1 0.24646762 Artery_Aorta
396 SLC25A1 0.24595477 Whole_Blood
297 PSMC3IP 0.24555876 Thyroid
197 CYB561D1 0.24131957 Brain_Spinal_cord_cervical_c-1
90 PDLIM2 0.24012006 Artery_Aorta
125 TIRAP 0.23968785 Artery_Tibial
289 FAM69A 0.23927135 Ovary
302 ZBTB42 0.23671061 Pituitary
272 ADAM10 0.23646566 Lung
91 SLC45A4 0.23636328 Artery_Aorta
201 TTLL12 0.23613202 Brain_Substantia_nigra
64 LIMD2 0.23023256 Adipose_Visceral_Omentum
299 PIK3CD 0.22659108 Pituitary
95 HOXC6 0.22521601 Artery_Aorta
102 NR5A2 0.22519401 Artery_Coronary
65 GALNT1 0.22375901 Breast_Mammary_Tissue
178 DNAH10OS 0.22173000 Brain_Cortex
366 CCR8 0.22172256 Whole_Blood
56 FAM177A1 0.22126403 Adipose_Visceral_Omentum
354 ITPR3 0.21967730 Vagina
252 PDCD7 0.21668280 Heart_Atrial_Appendage
224 CSF3R 0.21421047 Stomach
67 S1PR2 0.21263494 Adipose_Visceral_Omentum
275 TMCC3 0.21220321 Small_Intestine_Terminal_Ileum
46 C4B 0.21170829 Brain_Spinal_cord_cervical_c-1
200 TNFSF12 0.20746094 Heart_Left_Ventricle
11 LCORL 0.20652087 Adipose_Subcutaneous
112 FGF11 0.20415792 Artery_Coronary
277 NFE2L1 0.20085983 Minor_Salivary_Gland
223 KRT10 0.20037221 Colon_Sigmoid
27 EHD4 0.19977436 Brain_Frontal_Cortex_BA9
137 TNFRSF10A 0.19964964 Brain_Amygdala
237 HNRNPAB 0.19673729 Esophagus_Mucosa
350 SALL1 0.19572639 Thyroid
259 HOMEZ 0.19457005 Heart_Left_Ventricle
84 SLC22A4 0.19432874 Artery_Aorta
199 TMPO 0.19285512 Brain_Substantia_nigra
253 SMG1 0.19135681 Heart_Atrial_Appendage
236 TMEM150A 0.19109125 Esophagus_Mucosa
58 CKMT1A 0.18949930 Adipose_Visceral_Omentum
327 SUMO3 0.18666878 Spleen
260 MARK3 0.18664863 Heart_Left_Ventricle
183 F5 0.18370320 Brain_Frontal_Cortex_BA9
314 SHANK3 0.17956256 Skin_Not_Sun_Exposed_Suprapubic
290 RP11-738E22.3 0.17859461 Ovary
19 CAV1 0.17839243 Adipose_Subcutaneous
388 RALBP1 0.17695255 Whole_Blood
81 FGD5 0.17626739 Artery_Aorta
155 CTC-492K19.7 0.17549832 Thyroid
279 ADAM23 0.17480422 Muscle_Skeletal
97 NECAB2 0.17238676 Artery_Aorta
196 RABEP1 0.17061513 Brain_Putamen_basal_ganglia
309 CEP85L 0.17038832 Skin_Not_Sun_Exposed_Suprapubic
320 XRCC1 0.16748403 Skin_Sun_Exposed_Lower_leg
114 SLC4A1 0.16655351 Heart_Atrial_Appendage
246 NLRC5 0.16513432 Whole_Blood
273 RP11-219G17.9 0.16381536 Lung
192 CTC-498M16.4 0.16211204 Brain_Nucleus_accumbens_basal_ganglia
358 TRAF3IP3 0.15970605 Whole_Blood
294 CNOT11 0.15665518 Pancreas
208 BTN3A2 0.15444270 Cells_Cultured_fibroblasts
376 ADAM19 0.15168727 Whole_Blood
153 PLCG2 0.15070986 Skin_Not_Sun_Exposed_Suprapubic
167 DOT1L 0.14975782 Brain_Cerebellar_Hemisphere
249 ITGA2 0.14920574 Heart_Atrial_Appendage
54 CCDC92 0.14755446 Skin_Not_Sun_Exposed_Suprapubic
206 NNT 0.14527044 Cells_Cultured_fibroblasts
391 TIMM50 0.14439665 Whole_Blood
75 AKT1 0.14434741 Cells_Cultured_fibroblasts
107 AC005754.8 0.14430991 Brain_Frontal_Cortex_BA9
313 SCIMP 0.14310181 Uterus
195 USP39 0.14302023 Skin_Sun_Exposed_Lower_leg
316 SFN 0.14293405 Skin_Sun_Exposed_Lower_leg
40 CD46 0.14272662 Adipose_Visceral_Omentum
233 SLFN13 0.14255038 Esophagus_Gastroesophageal_Junction
136 APOM 0.14214213 Brain_Amygdala
57 CHP1 0.14109949 Adipose_Visceral_Omentum
22 PLEKHA1 0.14013860 Stomach
392 GPR4 0.13977148 Whole_Blood
231 CDC42BPA 0.13959999 Esophagus_Muscularis
44 HIST1H2BD 0.13925426 Whole_Blood
248 GGT5 0.13843450 Esophagus_Muscularis
10 ATXN7 0.13767843 Adipose_Subcutaneous
61 KDM6B 0.13707911 Adipose_Visceral_Omentum
28 C15orf40 0.13688003 Adipose_Subcutaneous
135 PCDHB3 0.13676817 Brain_Caudate_basal_ganglia
194 PDF 0.13664663 Brain_Nucleus_accumbens_basal_ganglia
264 VPS26A 0.13565232 Liver
203 CLEC4M 0.13510641 Thyroid
151 ST3GAL4 0.13484000 Prostate
274 ZNF180 0.13332581 Lung
87 DLD 0.13325621 Nerve_Tibial
321 TRMT12 0.13150871 Small_Intestine_Terminal_Ileum
296 JAM3 0.13026507 Pancreas
79 SCMH1 0.13023939 Artery_Aorta
180 NAA38 0.12879717 Stomach
154 CXCL16 0.12864899 Brain_Caudate_basal_ganglia
35 CD320 0.12808650 Adipose_Subcutaneous
20 VLDLR 0.12773763 Whole_Blood
232 SCG3 0.12731172 Esophagus_Gastroesophageal_Junction
258 MICALL2 0.12688510 Spleen
217 ACAP1 0.12683512 Whole_Blood
16 HOXA5 0.12466202 Esophagus_Muscularis
191 DEF6 0.12255166 Brain_Putamen_basal_ganglia
25 RAB21 0.12231616 Testis
163 CERS5 0.12210828 Brain_Cerebellar_Hemisphere
133 SOX11 0.12199790 Small_Intestine_Terminal_Ileum
247 BPI 0.12109659 Esophagus_Muscularis
121 DAXX 0.12019556 Lung
182 ZNF114 0.11952620 Brain_Cortex
213 TADA2A 0.11944377 Cells_Cultured_fibroblasts
234 ADAM33 0.11790401 Uterus
150 FADS1 0.11551065 Stomach
122 ZBTB22 0.11515579 Artery_Tibial
352 SIGLEC9 0.11515125 Thyroid
93 LIPA 0.11449964 Whole_Blood
283 BCL2L2 0.11381604 Muscle_Skeletal
123 LINC00094 0.11356889 Artery_Tibial
288 TSPO 0.11353714 Nerve_Tibial
108 BORCS7 0.11261517 Heart_Atrial_Appendage
55 LAMP1 0.11248137 Whole_Blood
244 UBASH3B 0.11236080 Esophagus_Muscularis
131 PTK6 0.11146639 Thyroid
72 VAMP5 0.10820754 Cells_Cultured_fibroblasts
33 C18orf25 0.10709181 Adipose_Subcutaneous
71 AP000350.5 0.10523489 Skin_Not_Sun_Exposed_Suprapubic
2 MAST2 0.10512746 Heart_Atrial_Appendage
171 RCSD1 0.10165325 Pancreas
116 LSM4 0.10158347 Small_Intestine_Terminal_Ileum
335 FZD6 0.10154442 Testis
207 MYLK4 0.10051209 Cells_Cultured_fibroblasts
36 ITPKC 0.09988538 Small_Intestine_Terminal_Ileum
13 SLC12A7 0.09955605 Adipose_Visceral_Omentum
6 SWT1 0.09955583 Adipose_Subcutaneous
141 CKB 0.09945771 Brain_Cortex
99 KIAA1755 0.09921705 Artery_Aorta
15 TAPBP 0.09884438 Brain_Frontal_Cortex_BA9
229 DUSP14 0.09802794 Esophagus_Mucosa
292 BCKDHA 0.09795135 Ovary
326 ITGB2 0.09699306 Spleen
14 SLC22A5 0.09652877 Lung
175 CRCP 0.09639742 Brain_Hippocampus
324 MAP3K5 0.09603741 Spleen
144 C19orf35 0.09428049 Brain_Nucleus_accumbens_basal_ganglia
74 FRAT2 0.09269468 Spleen
140 LYN 0.09083061 Spleen
47 TAGAP 0.09072957 Skin_Sun_Exposed_Lower_leg
149 RBP4 0.08978528 Brain_Hippocampus
146 CD302 0.08743323 Brain_Putamen_basal_ganglia
187 CYB5D1 0.08637018 Brain_Frontal_Cortex_BA9
3 CSF1 0.08556112 Cells_Cultured_fibroblasts
215 PRR15L 0.08502358 Thyroid
129 TUBG2 0.08429790 Cells_Cultured_fibroblasts
152 ATG14 0.08423849 Brain_Hippocampus
101 EEF1A2 0.08400048 Artery_Coronary
365 EOMES 0.08399100 Whole_Blood
370 ADD1 0.08357596 Whole_Blood
120 PCDHB2 0.08350672 Pituitary
115 KIF18B 0.08254946 Whole_Blood
142 SLC9A3R1 0.08168501 Cells_Cultured_fibroblasts
21 FRAT1 0.08140854 Adipose_Visceral_Omentum
9 SPATS2L 0.08075678 Spleen
60 MED11 0.08075508 Nerve_Tibial
293 MAP4K4 0.08015466 Pancreas
173 DTNB 0.07867398 Brain_Cortex
78 KANK3 0.07834727 Heart_Left_Ventricle
176 PKN3 0.07808787 Stomach
169 MAP2K5 0.07808517 Brain_Putamen_basal_ganglia
110 PPP2R3C 0.07797272 Cells_Cultured_fibroblasts
322 TADA1 0.07728038 Stomach
5 KIAA1614 0.07648871 Breast_Mammary_Tissue
68 ATP13A1 0.07577582 Whole_Blood
80 FCER1G 0.07558057 Nerve_Tibial
88 RP11-138A9.1 0.07397046 Esophagus_Muscularis
92 TLE4 0.07171713 Breast_Mammary_Tissue
134 RNF123 0.06993441 Brain_Amygdala
373 NPR3 0.06952557 Whole_Blood
70 PPP5C 0.06859112 Esophagus_Mucosa
103 RP11-373D23.3 0.06814858 Artery_Coronary
132 PIK3R3 0.06640586 Brain_Putamen_basal_ganglia
317 NAP1L4 0.06523116 Skin_Sun_Exposed_Lower_leg
85 CANX 0.06451963 Artery_Tibial
127 TMED6 0.06423617 Brain_Hippocampus
83 REST 0.06409781 Artery_Aorta
119 TET2 0.06286641 Whole_Blood
190 FASTKD5 0.06256677 Pituitary
59 FURIN 0.06157816 Adipose_Visceral_Omentum
43 CXCL6 0.06115421 Brain_Cortex
105 RAI14 0.06049921 Brain_Amygdala
172 KIAA0040 0.06016279 Lung
221 TAL1 0.05885223 Colon_Sigmoid
156 CD33 0.05737688 Skin_Not_Sun_Exposed_Suprapubic
139 THOC7 0.05715536 Liver
24 RP11-672A2.4 0.05653036 Artery_Aorta
356 RORC 0.05615313 Whole_Blood
53 COX14 0.05598498 Adipose_Visceral_Omentum
225 CDC14A 0.05594325 Whole_Blood
39 DPT 0.05569325 Skin_Sun_Exposed_Lower_leg
138 KIAA0391 0.05484068 Minor_Salivary_Gland
66 ELANE 0.05460543 Adipose_Visceral_Omentum
34 CNN2 0.05338176 Brain_Frontal_Cortex_BA9
117 CDK11B 0.05331475 Pancreas
23 PTPRJ 0.05326307 Brain_Cortex
220 CLEC4G 0.05205727 Cells_EBV-transformed_lymphocytes
51 KCNQ3 0.05129536 Skin_Not_Sun_Exposed_Suprapubic
281 TPM2 0.05111167 Skin_Not_Sun_Exposed_Suprapubic
69 LRRC25 0.04892774 Thyroid
164 SLC5A11 0.04681905 Brain_Cerebellar_Hemisphere
63 CSF3 0.04671120 Brain_Cerebellar_Hemisphere
38 UBOX5 0.04655677 Adipose_Subcutaneous
76 AKAP10 0.04448909 Nerve_Tibial
118 UBE2F 0.04255995 Whole_Blood
397 SPECC1L 0.04179855 Whole_Blood
50 CPSF4 0.03991015 Whole_Blood
300 L3MBTL3 0.03836540 Pituitary
106 LINC01184 0.03753389 Stomach
262 B3GNT9 0.03633381 Heart_Left_Ventricle
1 LAPTM5 0.03536533 Artery_Tibial
73 RP11-394I13.2 0.03493873 Skin_Sun_Exposed_Lower_leg
7 CNIH4 0.03283872 Cells_Cultured_fibroblasts
17 ACHE 0.03138949 Whole_Blood
29 FES 0.02775300 Cells_Cultured_fibroblasts
4 CD101 0.02650496 Esophagus_Mucosa
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.5 disgenet2r_0.99.2 WebGestaltR_0.4.4 enrichR_3.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.8 reshape2_1.4.3 purrr_0.3.4 lattice_0.20-38 colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 utf8_1.2.1 rlang_0.4.11 later_0.8.0 pillar_1.6.1 withr_2.4.1 glue_1.4.2 DBI_1.1.1 gdtools_0.1.9 rngtools_1.5 doRNG_1.8.2 plyr_1.8.4 foreach_1.5.1 lifecycle_1.0.0 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0 workflowr_1.6.2 codetools_0.2-16 evaluate_0.14 labeling_0.3 knitr_1.23 doParallel_1.0.16 httpuv_1.5.1 curl_3.3 parallel_3.6.1 fansi_0.5.0 Rcpp_1.0.6 readr_1.4.0 promises_1.0.1 scales_1.1.0 jsonlite_1.6 apcluster_1.4.8 farver_2.1.0 fs_1.3.1 hms_1.1.0 rjson_0.2.20 digest_0.6.20 stringi_1.4.3 dplyr_1.0.7 grid_3.6.1 rprojroot_2.0.2 tools_3.6.1 magrittr_2.0.1 tibble_3.1.2 crayon_1.4.1
[56] whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2 Matrix_1.2-18 svglite_1.2.2 rmarkdown_1.13 httr_1.4.1 iterators_1.0.13 R6_2.5.0 igraph_1.2.4.1 git2r_0.26.1 compiler_3.6.1