Non-cancer illness code self-reported: asthma
wesleycrouse
2022-02-28
Last updated: 2022-03-03
Checks: 6 1
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 380982d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Unstaged changes:
Modified: analysis/ukb-a-232_allweights.Rmd
Modified: analysis/ukb-a-249_allweights.Rmd
Modified: analysis/ukb-a-389_allweights.Rmd
Modified: analysis/ukb-a-66_allweights.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-a-66_allweights.Rmd) and HTML (docs/ukb-a-66_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 380982d | wesleycrouse | 2022-03-01 | fixing typo in all weight reports |
| Rmd | 76fa2cd | wesleycrouse | 2022-03-01 | cleaning up all weight reports |
| html | 76fa2cd | wesleycrouse | 2022-03-01 | cleaning up all weight reports |
| html | 2509c32 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
| Rmd | 962fd16 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
trait_id <- "ukb-a-66"
trait_name <- "Non-cancer illness code self-reported: asthma"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# #print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
# twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z) > sig_thresh]
#
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes )
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)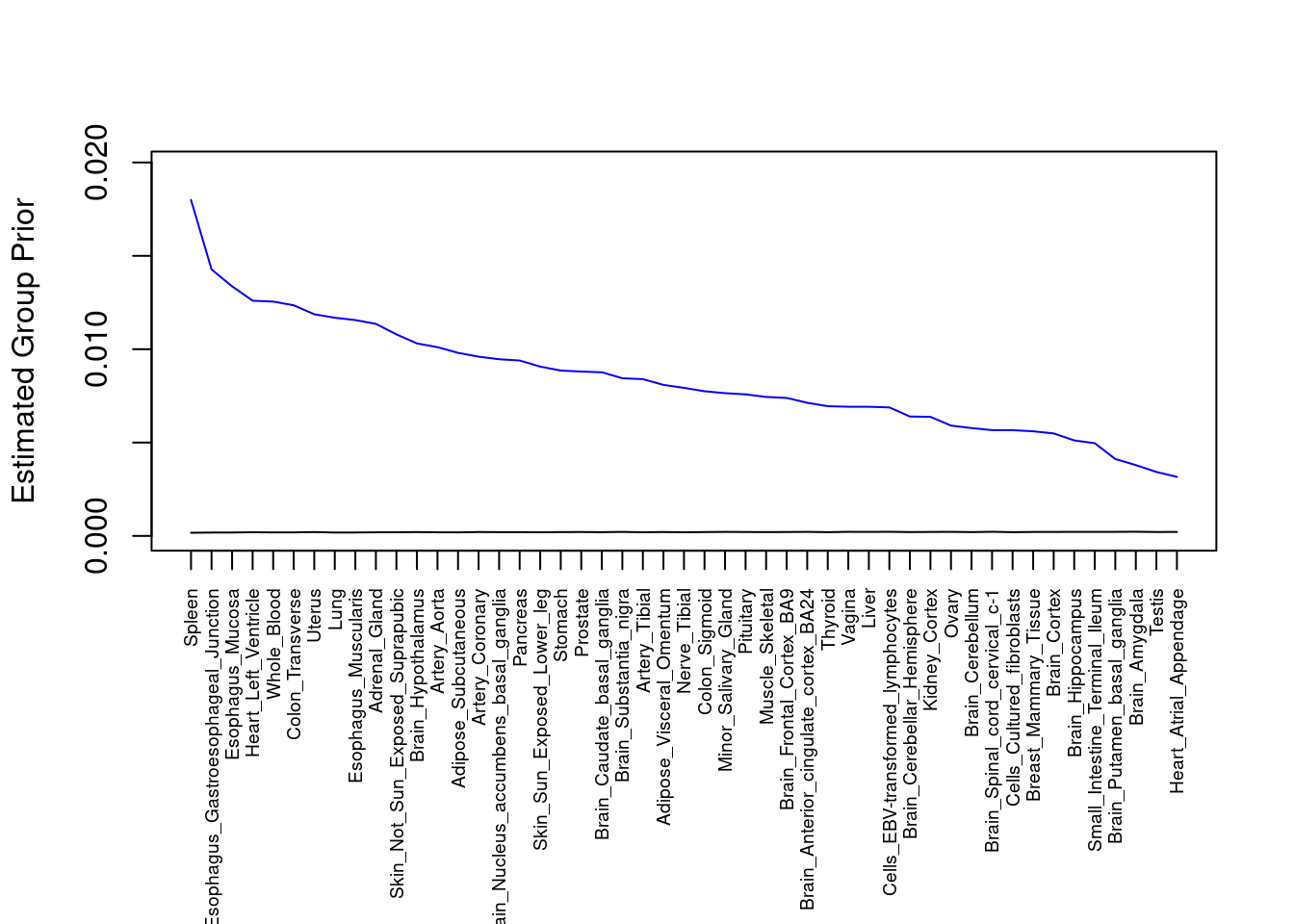
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)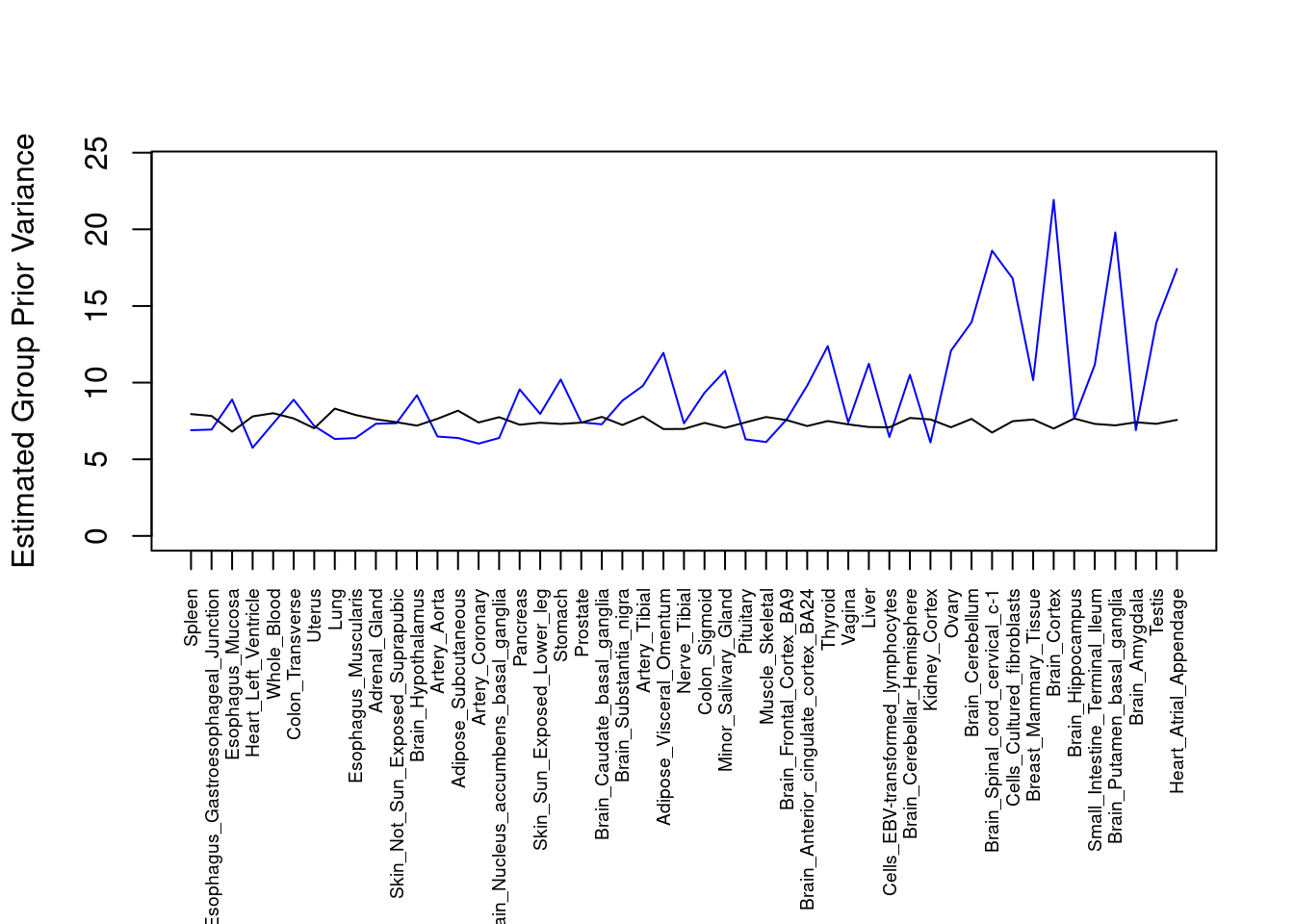
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
#plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g, output$pve_s)*1.1),
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)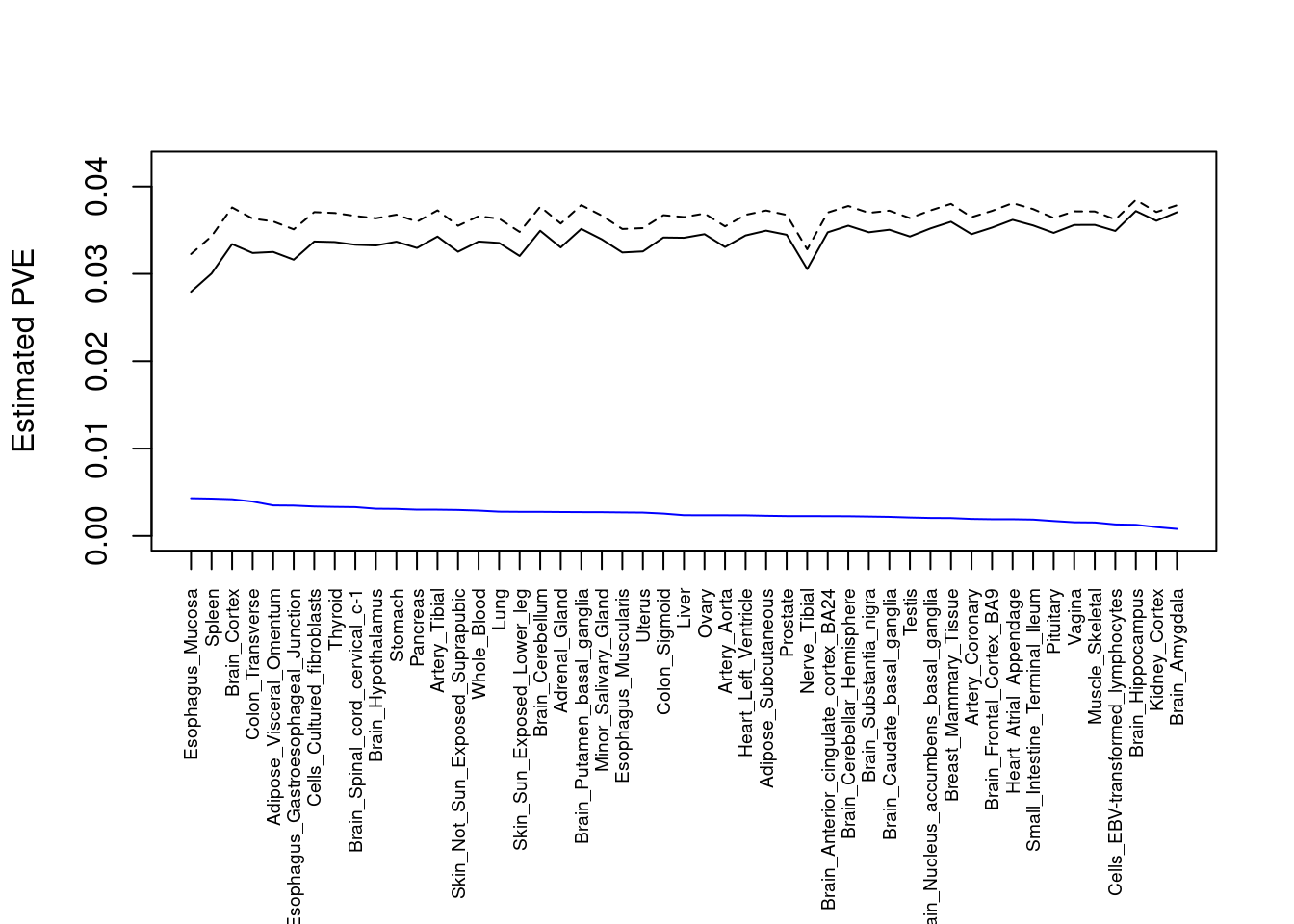
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")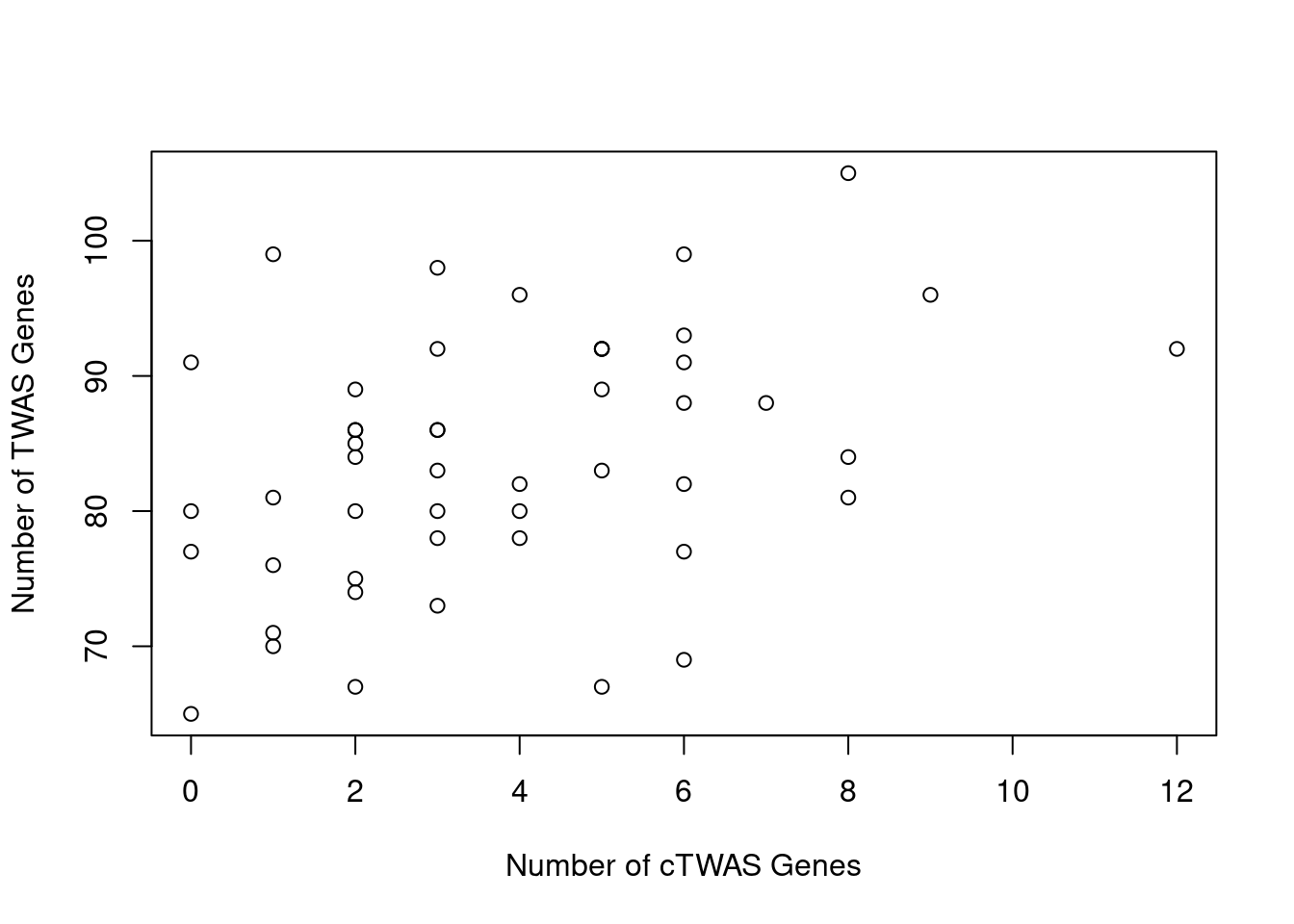
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 66#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 396Enrichment analysis for cTWAS genes
#enrichment for cTWAS genes
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}[1] "GO_Biological_Process_2021"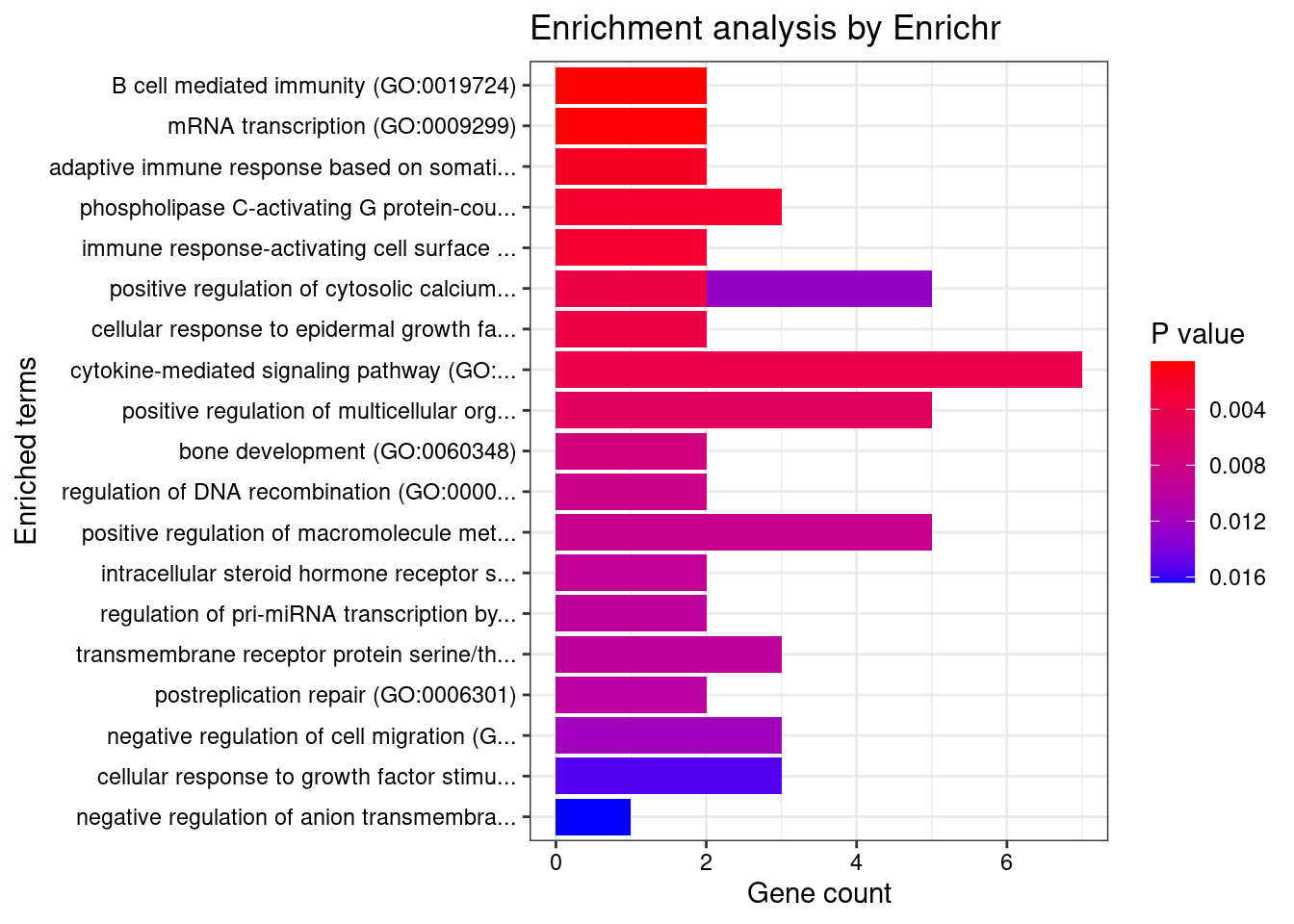
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"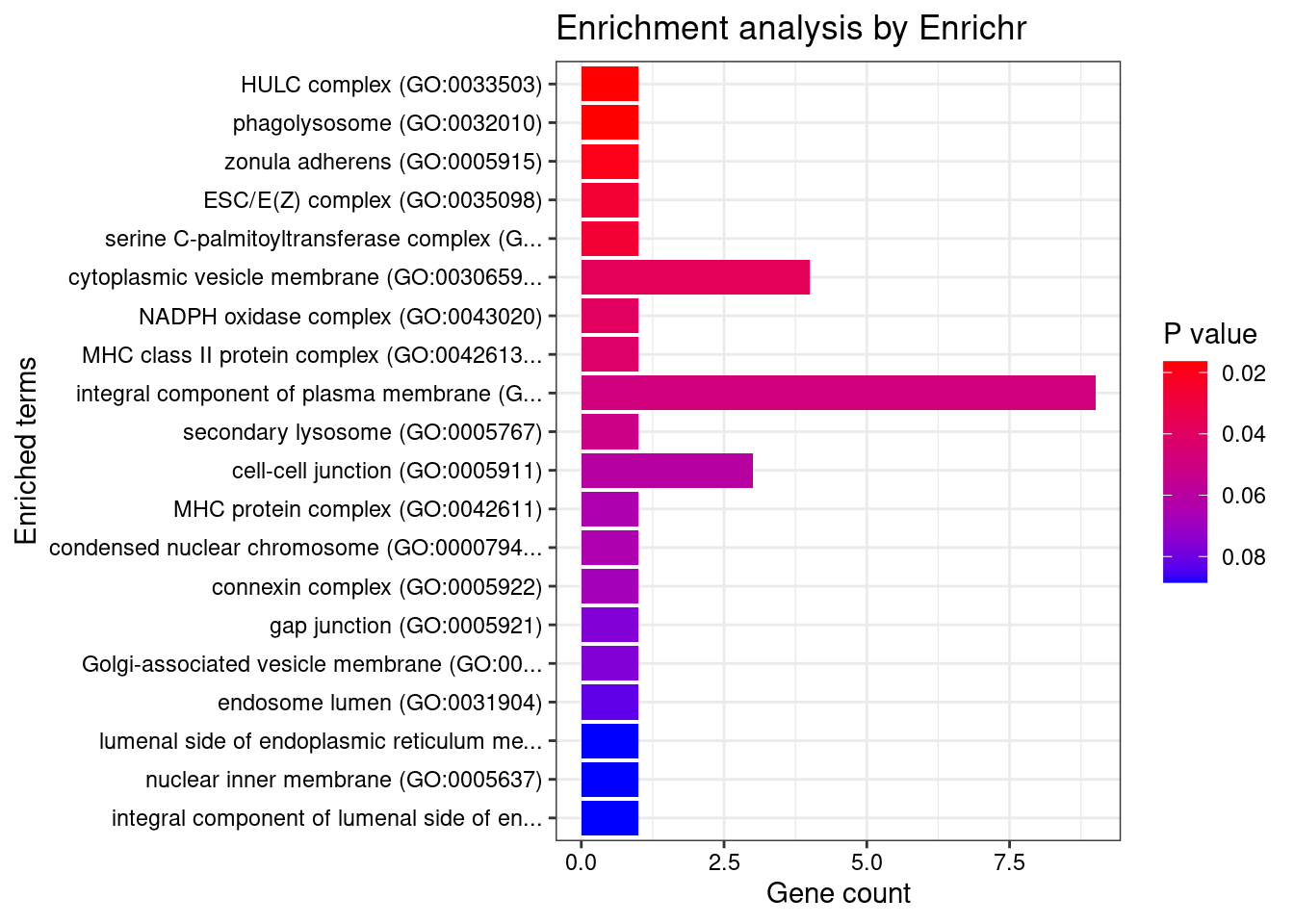
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"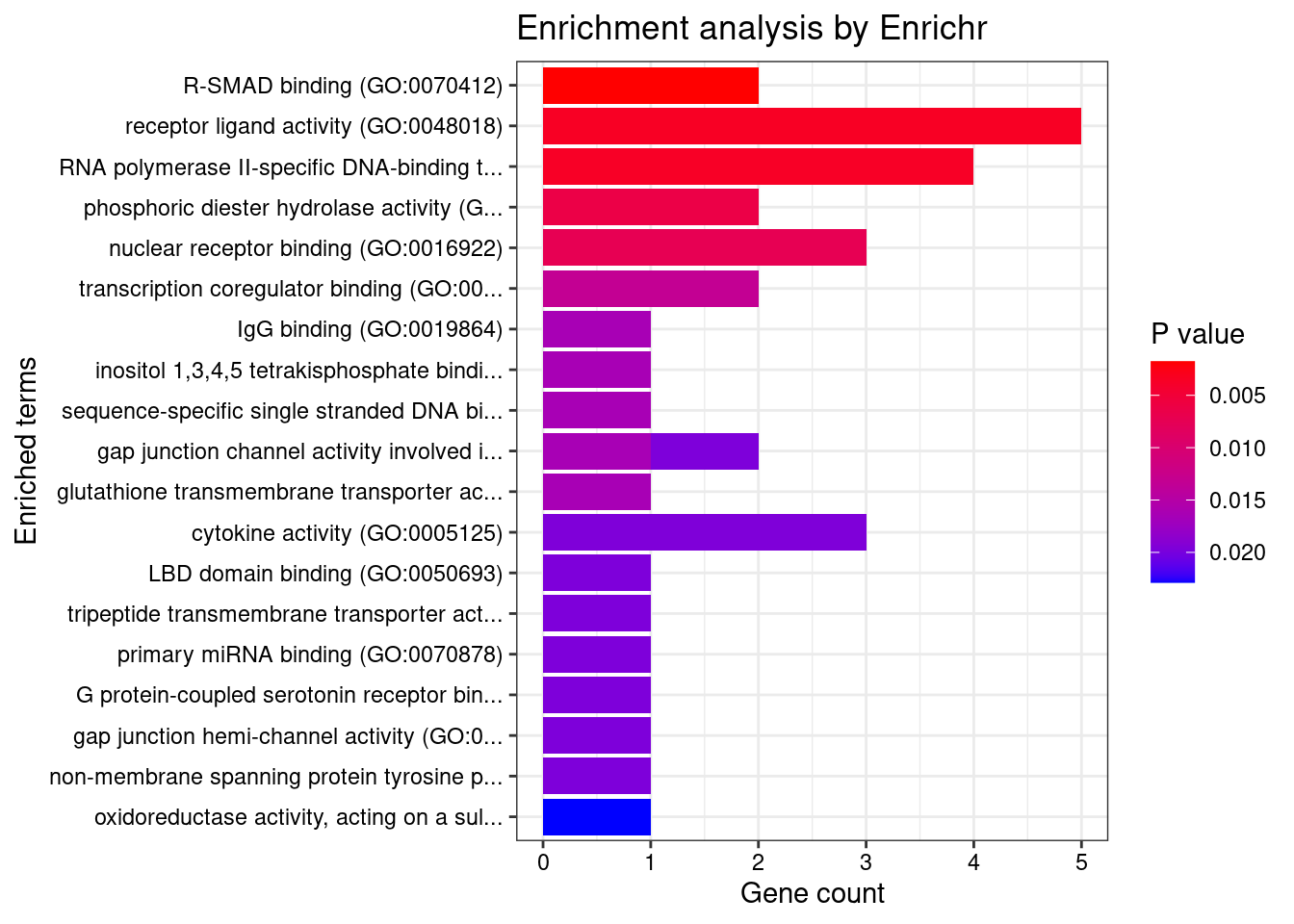
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}[1] "GO_Biological_Process_2021"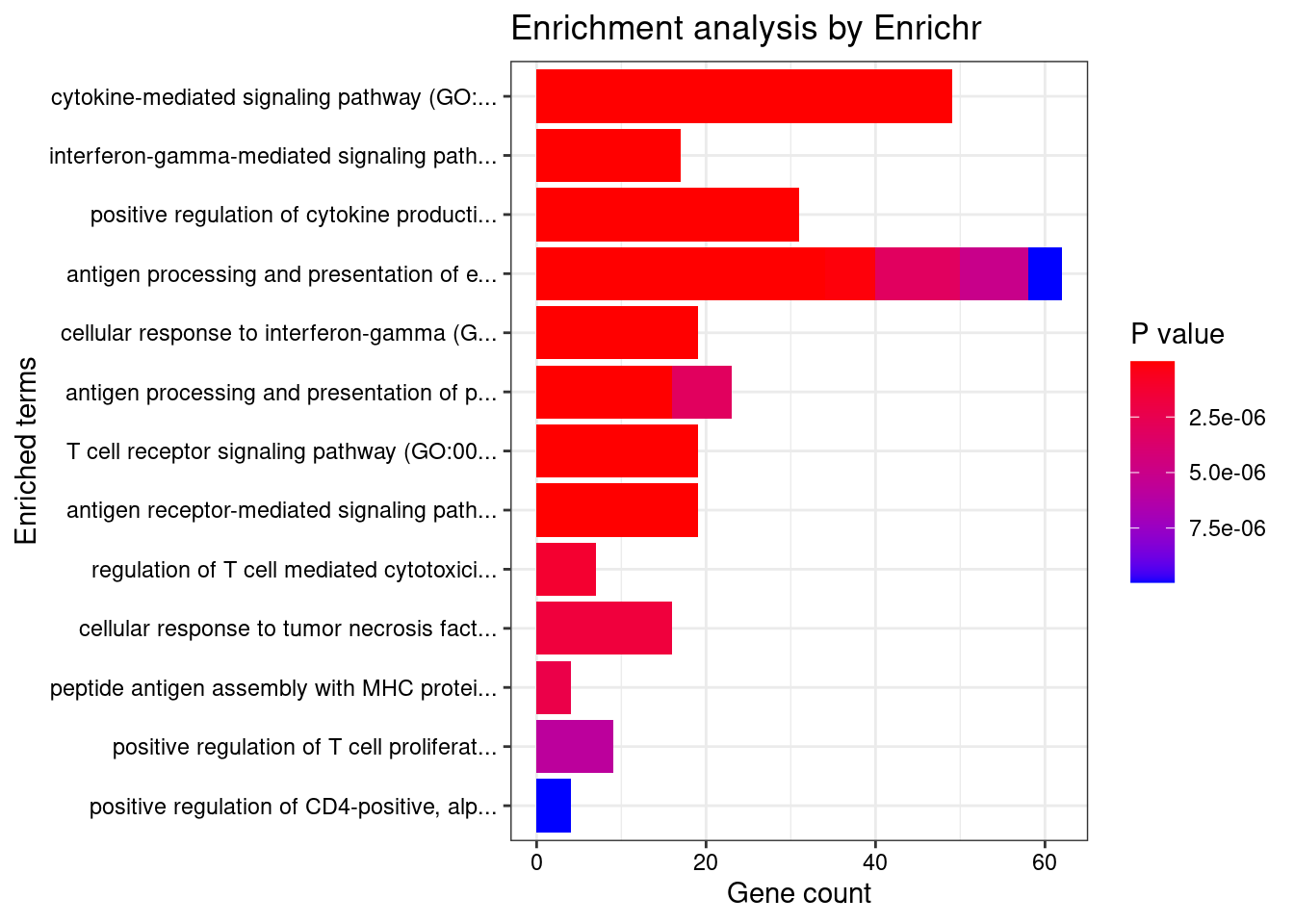
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term
1 cytokine-mediated signaling pathway (GO:0019221)
2 interferon-gamma-mediated signaling pathway (GO:0060333)
3 positive regulation of cytokine production (GO:0001819)
4 antigen processing and presentation of exogenous peptide antigen (GO:0002478)
5 cellular response to interferon-gamma (GO:0071346)
6 antigen processing and presentation of exogenous peptide antigen via MHC class II (GO:0019886)
7 antigen processing and presentation of peptide antigen via MHC class II (GO:0002495)
8 T cell receptor signaling pathway (GO:0050852)
9 antigen receptor-mediated signaling pathway (GO:0050851)
10 antigen processing and presentation of endogenous peptide antigen (GO:0002483)
11 regulation of T cell mediated cytotoxicity (GO:0001914)
12 cellular response to tumor necrosis factor (GO:0071356)
13 peptide antigen assembly with MHC protein complex (GO:0002501)
14 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474)
15 antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590)
16 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway (GO:0002484)
17 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent (GO:0002486)
18 positive regulation of T cell proliferation (GO:0042102)
19 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent (GO:0002480)
20 positive regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043372)
21 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (GO:0002479)
22 positive regulation of interferon-gamma production (GO:0032729)
23 positive regulation of T cell activation (GO:0050870)
24 regulation of T cell proliferation (GO:0042129)
25 positive regulation of immune effector process (GO:0002699)
26 cellular response to cytokine stimulus (GO:0071345)
27 positive regulation of interleukin-13 production (GO:0032736)
28 regulation of cytokine production (GO:0001817)
29 positive regulation of T cell cytokine production (GO:0002726)
30 regulation of macromolecule metabolic process (GO:0060255)
31 regulation of interferon-gamma production (GO:0032649)
32 regulation of immune response (GO:0050776)
33 regulation of inflammatory response (GO:0050727)
34 regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043370)
35 macrophage activation (GO:0042116)
36 positive regulation of T cell mediated immunity (GO:0002711)
37 positive regulation of CD4-positive, alpha-beta T cell activation (GO:2000516)
38 tumor necrosis factor-mediated signaling pathway (GO:0033209)
39 positive regulation of lymphocyte proliferation (GO:0050671)
40 positive regulation of DNA-binding transcription factor activity (GO:0051091)
41 positive regulation of interleukin-6 production (GO:0032755)
42 positive regulation of T cell mediated cytotoxicity (GO:0001916)
43 regulation of chemokine production (GO:0032642)
44 regulation of extracellular matrix organization (GO:1903053)
45 positive regulation of leukocyte mediated cytotoxicity (GO:0001912)
46 positive regulation of T cell differentiation (GO:0045582)
47 regulation of interleukin-13 production (GO:0032656)
48 negative regulation of natural killer cell mediated cytotoxicity (GO:0045953)
49 positive regulation of NF-kappaB transcription factor activity (GO:0051092)
50 regulation of interleukin-6 production (GO:0032675)
51 regulation of interleukin-10 production (GO:0032653)
52 antigen processing and presentation of endogenous peptide antigen via MHC class I (GO:0019885)
53 regulation of apoptotic cell clearance (GO:2000425)
54 regulation of CD4-positive, alpha-beta T cell activation (GO:2000514)
55 positive regulation of apoptotic cell clearance (GO:2000427)
56 cellular response to interleukin-7 (GO:0098761)
57 regulation of neuroinflammatory response (GO:0150077)
58 interleukin-7-mediated signaling pathway (GO:0038111)
59 positive regulation of alpha-beta T cell activation (GO:0046635)
60 positive regulation of memory T cell differentiation (GO:0043382)
61 regulation of immune effector process (GO:0002697)
62 positive regulation of chemokine production (GO:0032722)
63 positive regulation of phagocytosis (GO:0050766)
64 inflammatory response (GO:0006954)
65 regulation of memory T cell differentiation (GO:0043380)
66 microglial cell activation (GO:0001774)
67 regulation of viral entry into host cell (GO:0046596)
68 regulation of interleukin-4 production (GO:0032673)
69 regulation of interleukin-8 production (GO:0032677)
70 immune response-activating cell surface receptor signaling pathway (GO:0002429)
71 T cell differentiation (GO:0030217)
72 regulation of myoblast fusion (GO:1901739)
73 negative regulation of gene silencing by miRNA (GO:0060965)
74 natural killer cell activation (GO:0030101)
75 regulation of regulatory T cell differentiation (GO:0045589)
Overlap Adjusted.P.value
1 49/621 2.918874e-13
2 17/68 1.530110e-11
3 31/335 8.214262e-10
4 18/103 9.581442e-10
5 19/121 1.343191e-09
6 16/98 3.237513e-08
7 16/100 3.800914e-08
8 19/158 9.845743e-08
9 19/185 1.276369e-06
10 6/14 3.320737e-05
11 7/29 2.402528e-04
12 16/194 3.092217e-04
13 4/6 3.690069e-04
14 7/33 4.560041e-04
15 10/78 4.560041e-04
16 4/7 6.481172e-04
17 4/7 6.481172e-04
18 9/66 7.066178e-04
19 4/8 1.084560e-03
20 4/8 1.084560e-03
21 9/73 1.406255e-03
22 8/57 1.543449e-03
23 9/75 1.604264e-03
24 9/76 1.713860e-03
25 6/32 2.964061e-03
26 24/482 3.098674e-03
27 4/11 3.465439e-03
28 12/150 3.465439e-03
29 5/21 3.465439e-03
30 14/200 3.465439e-03
31 9/86 3.599549e-03
32 13/179 4.132635e-03
33 14/206 4.138333e-03
34 4/12 4.138333e-03
35 6/36 4.138333e-03
36 6/36 4.138333e-03
37 4/13 5.534832e-03
38 10/116 5.996412e-03
39 8/75 6.522020e-03
40 15/246 6.747256e-03
41 8/76 6.815715e-03
42 5/26 7.188531e-03
43 6/42 8.442499e-03
44 4/15 8.610638e-03
45 6/43 9.020169e-03
46 6/43 9.020169e-03
47 4/16 1.036024e-02
48 4/16 1.036024e-02
49 11/155 1.203458e-02
50 9/110 1.486776e-02
51 6/48 1.506933e-02
52 3/8 1.587997e-02
53 3/8 1.587997e-02
54 3/8 1.587997e-02
55 3/8 1.587997e-02
56 4/19 1.742104e-02
57 4/19 1.742104e-02
58 4/19 1.742104e-02
59 4/20 2.100328e-02
60 3/9 2.100328e-02
61 6/53 2.100328e-02
62 6/53 2.100328e-02
63 6/53 2.100328e-02
64 13/230 2.410747e-02
65 3/10 2.756818e-02
66 4/22 2.756818e-02
67 5/39 3.188841e-02
68 4/23 3.188841e-02
69 7/81 3.532415e-02
70 4/24 3.659696e-02
71 5/41 3.798370e-02
72 3/12 4.430934e-02
73 3/12 4.430934e-02
74 5/43 4.532399e-02
75 4/26 4.658082e-02
Genes
1 CSF3;TNFRSF6B;RORC;TNF;IL18RAP;PSMD3;TRIM26;HLA-DPA1;FCER1G;IL4R;IL1R1;IL1R2;IL13;HLA-B;HLA-C;HLA-A;HLA-F;IRF1;CSNK2B;LTA;LTB;HLA-DQB2;HLA-DQB1;GATA3;IL1RL2;SOCS1;IL21R;STAT6;HLA-DQA2;HLA-DQA1;STAT5A;TNFSF18;IL33;HLA-DRB5;TSLP;CCL20;PSMB8;IL2;PSMB9;IL4;TNFSF4;HLA-DPB1;HLA-DRA;TRIM38;TRIM31;IL7R;MAP3K14;HLA-DRB1;IL18R1
2 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;IRF1;HLA-DPB1;HLA-DRA;TRIM26;TRIM38;TRIM31;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
3 GATA3;AGPAT1;AIF1;TNF;AGER;FLOT1;STAT6;TRIM27;HLA-DPA1;IL33;XRCC6;FCER1G;TSLP;IL1R1;IL13;HLA-A;BTN3A2;IL2;TLR1;IL4;LACC1;PTPRC;TNFSF4;IRF1;HLA-DPB1;CHI3L1;TLR6;LTB;HSPA1B;IL18R1;HSPA1A
4 HLA-DRB5;FCER1G;HLA-A;KIF11;HLA-F;HLA-DMA;HLA-DMB;KIF3A;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
5 HLA-DRB5;CCL20;HLA-B;HLA-C;HLA-A;HLA-F;AIF1;IRF1;HLA-DPB1;HLA-DRA;TRIM26;TRIM38;TRIM31;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
6 HLA-DRB5;FCER1G;KIF11;HLA-DMA;HLA-DMB;KIF3A;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
7 HLA-DRB5;FCER1G;KIF11;HLA-DMA;HLA-DMB;KIF3A;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
8 HLA-DRB5;BTN2A2;HLA-A;GATA3;BTNL2;BTN3A2;SPPL3;PSMB8;PSMB9;PTPRC;PSMD3;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
9 HLA-DRB5;BTN2A2;HLA-A;GATA3;BTNL2;BTN3A2;SPPL3;PSMB8;PSMB9;PTPRC;PSMD3;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
10 TAP2;TAP1;HLA-DRA;HLA-A;HLA-F;HLA-DRB1
11 PTPRC;HLA-B;HLA-DRA;HLA-A;HLA-F;AGER;HLA-DRB1
12 TNFSF18;TNFRSF6B;CCL20;GATA3;NR1D1;NPNT;TNF;PSMB8;PSMB9;ZFP36L1;TNFSF4;PSMD3;LTA;CHI3L1;LTB;MAP3K14
13 HLA-DMA;HLA-DMB;HLA-DRA;HLA-DRB1
14 FCER1G;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F
15 FCER1G;PSMD3;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;PSMB8;PSMB9
16 HLA-B;HLA-C;HLA-A;HLA-F
17 HLA-B;HLA-C;HLA-A;HLA-F
18 IL4;HLA-DMB;PTPRC;TNFSF4;HLA-DPB1;AGER;AIF1;IL2;HLA-DPA1
19 HLA-B;HLA-C;HLA-A;HLA-F
20 SOCS1;TNFSF4;HLA-DRA;HLA-DRB1
21 PSMD3;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;PSMB8;PSMB9
22 IL1R1;TNFSF4;HLA-DPB1;TRIM27;BTN3A2;HLA-A;IL18R1;HLA-DPA1
23 IL4;HLA-DMB;PTPRC;TNFSF4;HLA-DPB1;HLA-A;GATA3;AIF1;HLA-DPA1
24 TNFSF18;IL4;HLA-DMB;PTPRC;TNFSF4;HLA-DPB1;AIF1;HLA-DRB1;HLA-DPA1
25 HLA-DMB;PTPRC;TNFSF4;HLA-DRA;IL2;HLA-DRB1
26 STAT5A;CSF3;IL4R;SMAD3;CCL20;IL1R1;IL1R2;IL13;RORC;GATA3;NR1D1;NPNT;TNF;AIF1;IL2;ZFP36L1;IL4;IL1RL2;SOCS1;IRF1;CHI3L1;STAT6;IL18R1;HLA-DPA1
27 IL4;TSLP;TNFSF4;GATA3
28 IL33;PTPRC;BTN2A2;TNFSF4;HLA-B;FLOT1;GATA3;BTNL2;BTN3A2;AGPAT1;TNF;HLA-DRB1
29 IL4;IL1R1;TNFSF4;HLA-A;IL18R1
30 TCF19;AFF3;TOB2;AIF1;POU5F1;ZFP36L1;PTPRC;FLOT1;RNF39;TRIM26;TRIM27;TRIM38;TRIM31;TRIM10
31 IL1R1;TNFSF4;HLA-DPB1;TRIM27;BTN3A2;HLA-A;HLA-DRB1;IL18R1;HLA-DPA1
32 HLA-B;HLA-C;HLA-A;HLA-F;IL4;NCR3;TNFSF4;IRF1;HLA-DRA;PDCD1;HLA-DRB1;MICA;MICB
33 IL33;TSLP;IL1R1;IL13;GATA3;NR1D1;TNF;IL2;IL4;LACC1;IL1RL2;SHARPIN;TNFSF4;GPSM3
34 SOCS1;TNFSF4;HLA-DRA;HLA-DRB1
35 IL4;TLR1;IL13;AGER;AIF1;TNF
36 TNFSF4;HLA-B;HLA-DRA;HLA-A;HLA-F;HLA-DRB1
37 SOCS1;TNFSF4;HLA-DRA;HLA-DRB1
38 TNFSF18;TNFRSF6B;TNFSF4;PSMD3;LTA;LTB;TNF;MAP3K14;PSMB8;PSMB9
39 IL4;HLA-DMB;PTPRC;TNFSF4;HLA-DPB1;AIF1;IL2;HLA-DPA1
40 TNFSF18;CSF3;SMAD3;TNF;AGER;IL18RAP;FLOT1;TRIM26;TRIM27;TLR6;TRIM38;TRIM31;HSPA1B;IL18R1;HSPA1A
41 TLR1;IL33;TSLP;TNFSF4;TLR6;TNF;AGER;AIF1
42 HLA-B;HLA-DRA;HLA-A;HLA-F;HLA-DRB1
43 IL33;TSLP;TNFSF4;AGER;AIF1;TNF
44 DDR1;EFEMP2;RIC1;LRP1
45 NCR3;HLA-B;HLA-DRA;HLA-A;HLA-F;HLA-DRB1
46 IL4;SOCS1;TNFSF4;HLA-DRA;GATA3;HLA-DRB1
47 IL4;TSLP;TNFSF4;GATA3
48 HLA-B;HLA-A;HLA-F;MICA
49 TNFSF18;IL18RAP;FLOT1;TRIM27;TLR6;TRIM38;TNF;AGER;HSPA1B;IL18R1;HSPA1A
50 TLR1;IL33;TSLP;TNFSF4;HLA-B;TLR6;TNF;AGER;AIF1
51 IL4;TSLP;TNFSF4;IL13;AGER;HLA-DRB1
52 TAP2;TAP1;HLA-A
53 C4B;C4A;C2
54 HLA-DRA;AGER;HLA-DRB1
55 C4B;C4A;C2
56 STAT5A;SOCS1;TSLP;IL7R
57 IL4;IL33;NR1D1;TNF
58 STAT5A;SOCS1;TSLP;IL7R
59 TNFSF4;HLA-DRA;HLA-A;HLA-DRB1
60 TNFSF4;HLA-DRA;HLA-DRB1
61 C4B;C4A;HLA-DRA;CFB;HLA-DRB1;C2
62 IL33;TSLP;TNFSF4;TNF;AGER;AIF1
63 C4B;C4A;FCER1G;PTPRC;TNF;C2
64 NRROS;CCL20;AIF1;TNF;TLR1;IL4;NCR3;TNFSF4;ADORA1;CHI3L1;TLR10;TLR6;ABCF1
65 TNFSF4;HLA-DRA;HLA-DRB1
66 IL4;AGER;AIF1;TNF
67 TRIM26;TRIM27;TRIM38;TRIM31;HLA-DRB1
68 FCER1G;TNFSF4;GATA3;HLA-DRB1
69 TLR1;PTPRC;CHI3L1;TLR6;TNF;HSPA1B;HSPA1A
70 BAG6;NCR3;FCER1G;MICB
71 FCER1G;PTPRC;GATA3;IL2;ZFP36L1
72 IL4;MYOG;FLOT1
73 ZC3H10;TNF;POU5F1
74 BAG6;NCR3;PTPRC;IL21R;IL2
75 SOCS1;IRF1;TNFSF4;IL2
[1] "GO_Cellular_Component_2021"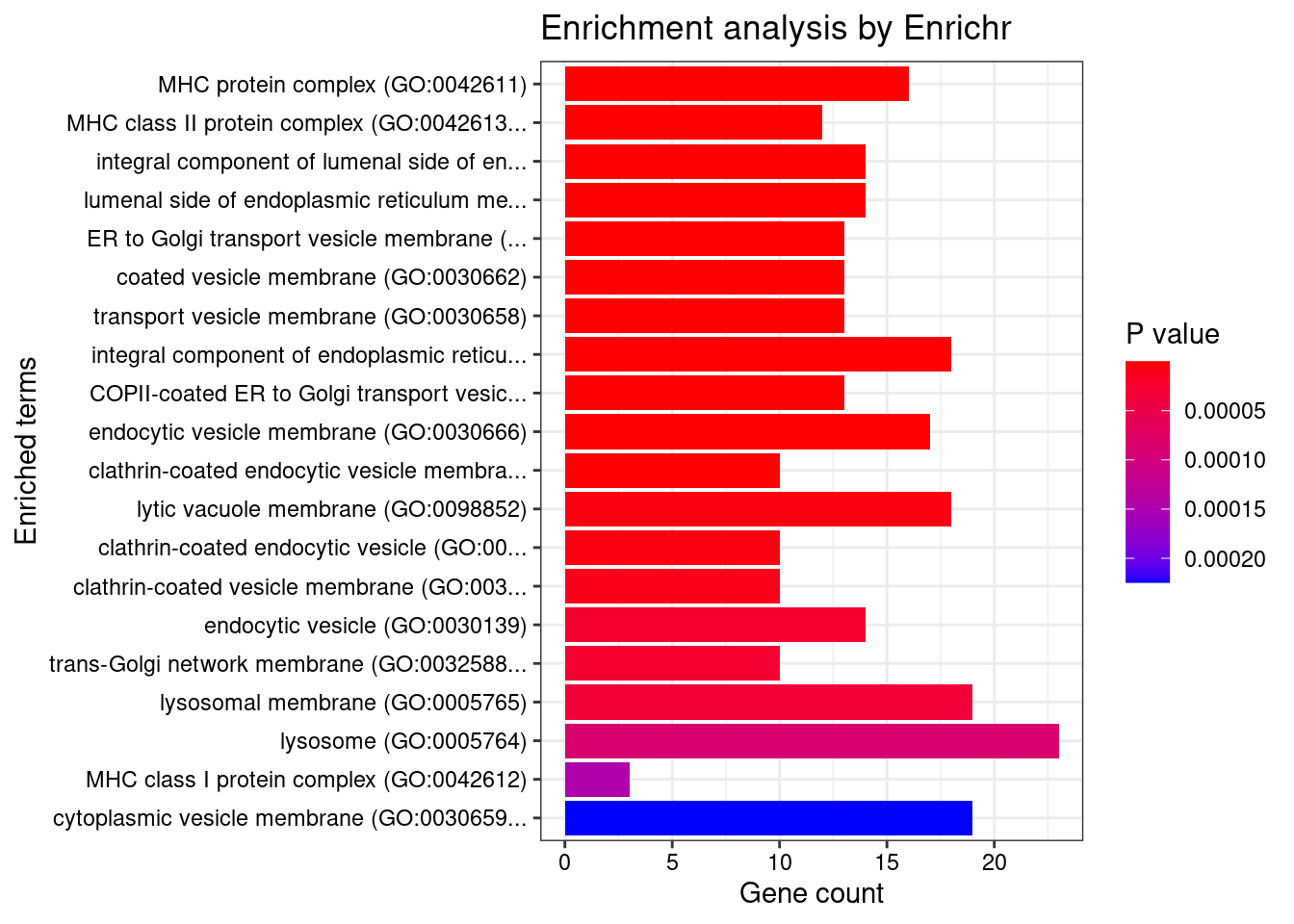
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term
1 MHC protein complex (GO:0042611)
2 MHC class II protein complex (GO:0042613)
3 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556)
4 lumenal side of endoplasmic reticulum membrane (GO:0098553)
5 ER to Golgi transport vesicle membrane (GO:0012507)
6 coated vesicle membrane (GO:0030662)
7 transport vesicle membrane (GO:0030658)
8 integral component of endoplasmic reticulum membrane (GO:0030176)
9 COPII-coated ER to Golgi transport vesicle (GO:0030134)
10 endocytic vesicle membrane (GO:0030666)
11 clathrin-coated endocytic vesicle membrane (GO:0030669)
12 lytic vacuole membrane (GO:0098852)
13 clathrin-coated endocytic vesicle (GO:0045334)
14 clathrin-coated vesicle membrane (GO:0030665)
15 endocytic vesicle (GO:0030139)
16 trans-Golgi network membrane (GO:0032588)
17 lysosomal membrane (GO:0005765)
18 lysosome (GO:0005764)
19 MHC class I protein complex (GO:0042612)
20 cytoplasmic vesicle membrane (GO:0030659)
21 phagocytic vesicle membrane (GO:0030670)
22 phagocytic vesicle (GO:0045335)
23 bounding membrane of organelle (GO:0098588)
24 late endosome membrane (GO:0031902)
25 trans-Golgi network (GO:0005802)
26 spermatoproteasome complex (GO:1990111)
27 Golgi membrane (GO:0000139)
Overlap Adjusted.P.value
1 16/20 3.865995e-22
2 12/13 4.084271e-18
3 14/28 1.836309e-15
4 14/28 1.836309e-15
5 13/54 1.306444e-09
6 13/55 1.400376e-09
7 13/60 3.907652e-09
8 18/142 1.150352e-08
9 13/79 1.104969e-07
10 17/158 3.514375e-07
11 10/69 1.878058e-05
12 18/267 1.104033e-04
13 10/85 1.104033e-04
14 10/90 1.716790e-04
15 14/189 3.493980e-04
16 10/99 3.493980e-04
17 19/330 4.314108e-04
18 23/477 1.005313e-03
19 3/6 1.613461e-03
20 19/380 2.336214e-03
21 6/45 2.436225e-03
22 8/100 7.931024e-03
23 28/767 1.325767e-02
24 6/68 1.956829e-02
25 12/239 2.511754e-02
26 2/5 3.007040e-02
27 18/472 4.945907e-02
Genes
1 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
2 HLA-DRB5;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
3 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
4 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
5 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
6 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
7 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
8 HLA-DRB5;ATF6B;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;SPPL3;HLA-DPB1;HLA-DRA;ESYT1;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
9 HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-F;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
10 HLA-DRB5;LRP1;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;HLA-DPB1;HLA-DRA;IL7R;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
11 HLA-DRB5;HLA-DPB1;HLA-DRA;IL7R;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
12 STARD3;HLA-DRB5;LRP1;HLA-F;AP5B1;HLA-DMA;HLA-DMB;HLA-DPB1;FLOT1;HLA-DRA;HLA-DOA;HLA-DQA2;HLA-DOB;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
13 HLA-DRB5;HLA-DPB1;HLA-DRA;IL7R;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
14 HLA-DRB5;HLA-DPB1;HLA-DRA;IL7R;HLA-DQA2;HLA-DQB2;HLA-DQA1;HLA-DRB1;HLA-DPA1;HLA-DQB1
15 RAB5B;HLA-DRB5;LRP1;FMNL1;HLA-DPB1;HLA-DRA;RIN3;SRGAP2;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
16 HLA-DRB5;RIC1;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
17 STARD3;HLA-DRB5;LRP1;CLEC16A;HLA-F;AP5B1;HLA-DMA;HLA-DMB;HLA-DPB1;FLOT1;HLA-DRA;HLA-DOA;HLA-DQA2;HLA-DOB;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
18 STARD3;HLA-DRB5;LRP1;HLA-F;AP5B1;HLA-DMA;NEU4;HLA-DMB;NAGLU;GLB1;NEU1;HLA-DPB1;FLOT1;HLA-DRA;PPT2;HLA-DOA;HLA-DQA2;HLA-DOB;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
19 HLA-B;HLA-C;HLA-A
20 HLA-DRB5;RAB5B;LRP1;HLA-B;HLA-C;ITPR3;SPPL3;GJA1;PTPRC;ORMDL3;ERBB2;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
21 HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F
22 FMNL1;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-F;SRGAP2
23 B4GALT3;RAB5B;LRP1;NOTCH4;ITPR3;SPPL3;GJA1;ORMDL3;ERBB2;HLA-DQA2;HLA-DQA1;HLA-DPA1;HLA-DRB5;RIC1;DMPK;TAP2;B3GALT4;HLA-B;TAP1;HLA-C;HLA-A;HLA-F;PTPRC;HLA-DPB1;HLA-DRA;HLA-DRB1;HLA-DQB2;HLA-DQB1
24 STARD3;HLA-DMA;HLA-DRB5;HLA-DMB;HLA-DRA;HLA-DRB1
25 HLA-DRB5;RIC1;ARL5C;MLANA;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
26 PSMB8;PSMB9
27 B4GALT3;HLA-DRB5;RIC1;NOTCH4;B3GALT4;HLA-B;HLA-C;HLA-A;HLA-F;GJA1;HLA-DPB1;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DRB1;HLA-DQB2;HLA-DPA1;HLA-DQB1
[1] "GO_Molecular_Function_2021"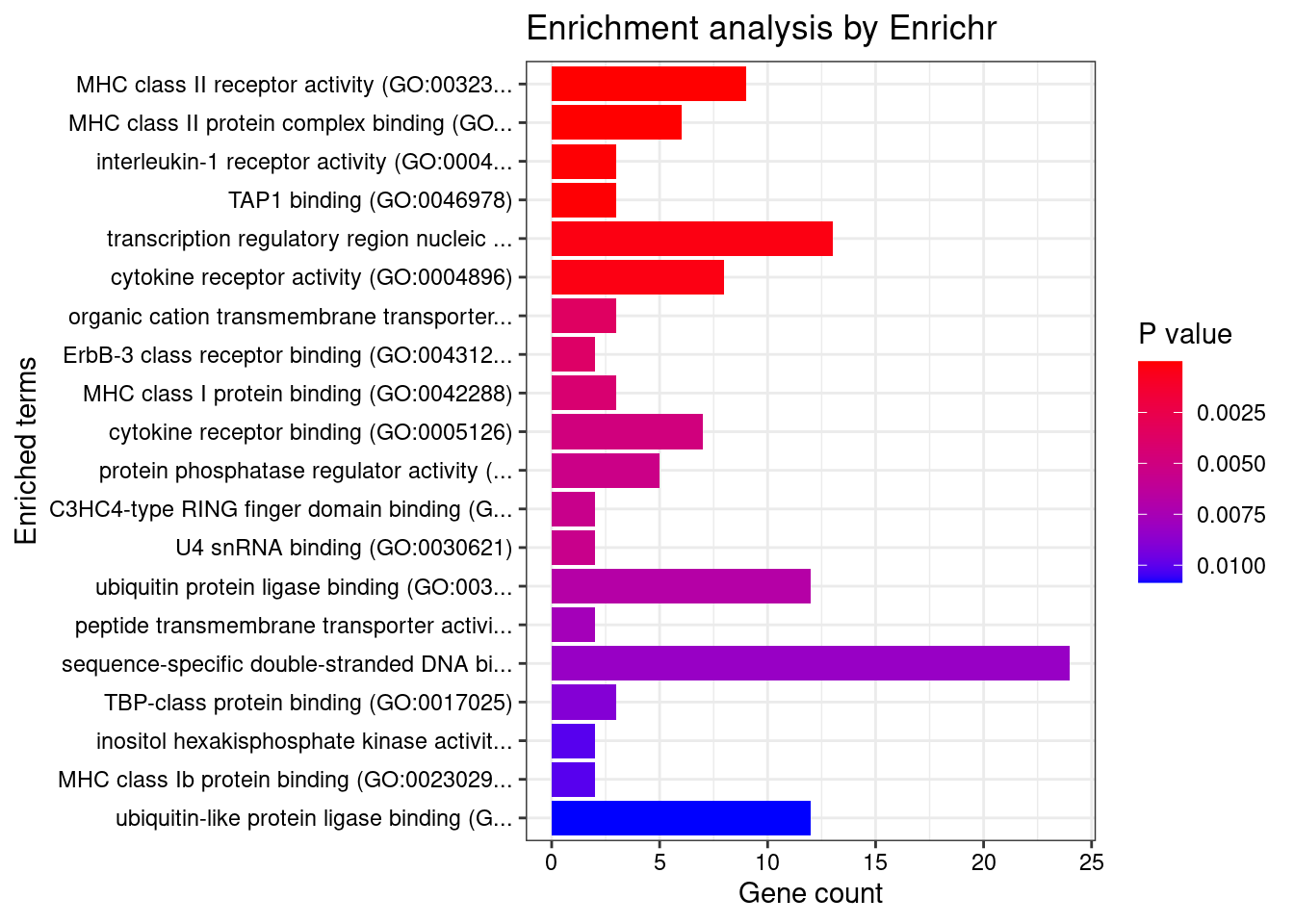
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term
1 MHC class II receptor activity (GO:0032395)
2 MHC class II protein complex binding (GO:0023026)
3 interleukin-1 receptor activity (GO:0004908)
4 TAP1 binding (GO:0046978)
5 transcription regulatory region nucleic acid binding (GO:0001067)
6 cytokine receptor activity (GO:0004896)
Overlap Adjusted.P.value
1 9/10 1.473833e-12
2 6/17 1.047572e-04
3 3/5 6.562964e-03
4 3/5 6.562964e-03
5 13/212 2.080962e-02
6 8/88 2.080962e-02
Genes
1 HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
2 HLA-DMA;HLA-DMB;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DRB1
3 IL1RL2;IL1R1;IL1R2
4 TAP2;TAP1;HLA-F
5 XRCC6;SMAD3;ZBTB14;THRA;ATF6B;GATA3;NR1D1;TNF;POU5F1;HHEX;TBL1XR1;IRF1;FOXA3
6 IL1RL2;IL18RAP;IL4R;IL1R1;IL1R2;IL21R;IL7R;IL18R1Enrichment analysis for cTWAS genes in top tissues separately
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
ctwas_genes_tissue <- df[[tissue]]$ctwas
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}
}Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"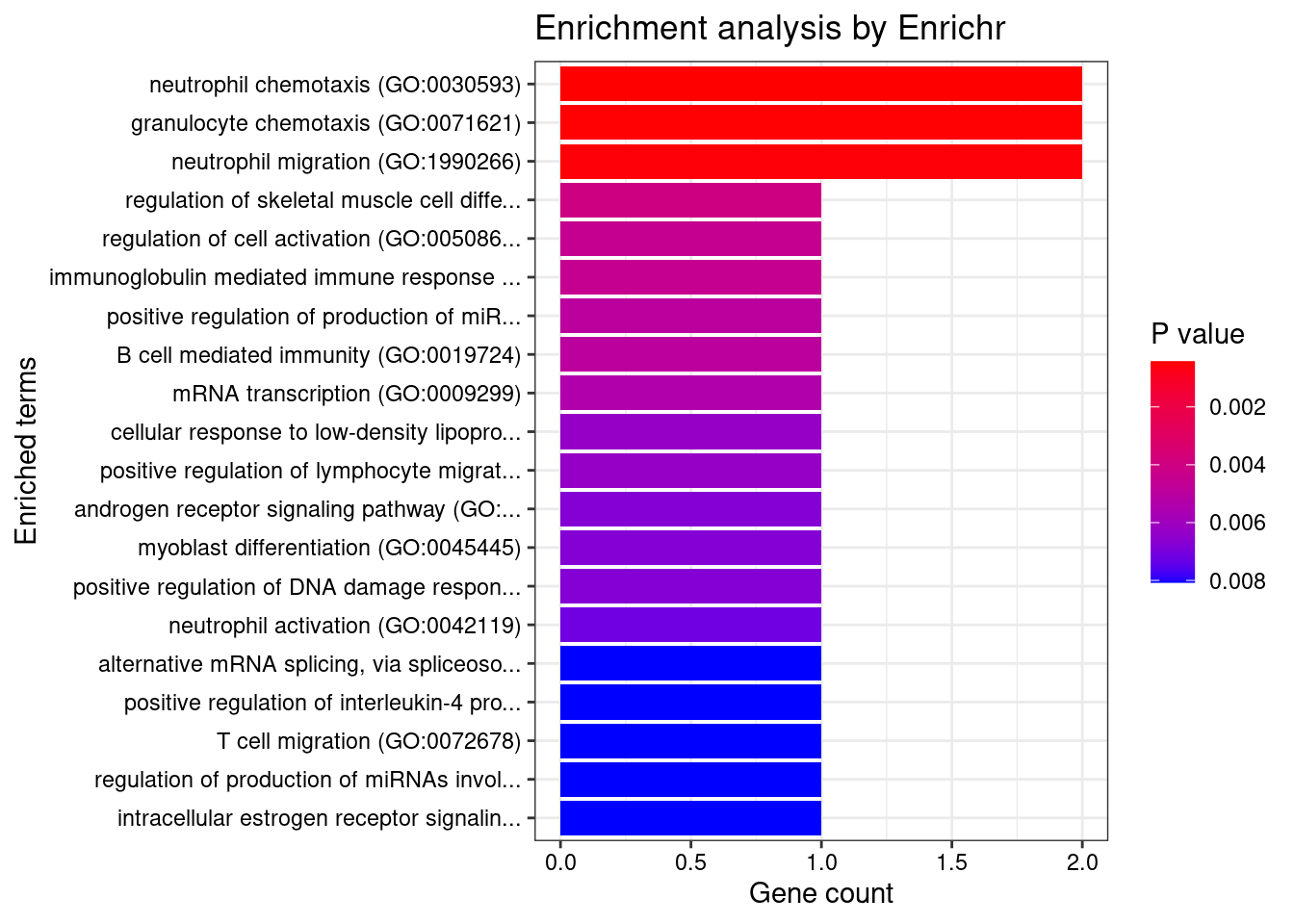
Term
1 neutrophil chemotaxis (GO:0030593)
2 granulocyte chemotaxis (GO:0071621)
3 neutrophil migration (GO:1990266)
4 regulation of skeletal muscle cell differentiation (GO:2001014)
5 regulation of cell activation (GO:0050865)
6 immunoglobulin mediated immune response (GO:0016064)
7 positive regulation of production of miRNAs involved in gene silencing by miRNA (GO:1903800)
8 B cell mediated immunity (GO:0019724)
9 mRNA transcription (GO:0009299)
10 cellular response to low-density lipoprotein particle stimulus (GO:0071404)
11 positive regulation of lymphocyte migration (GO:2000403)
12 androgen receptor signaling pathway (GO:0030521)
13 myoblast differentiation (GO:0045445)
14 positive regulation of DNA damage response, signal transduction by p53 class mediator (GO:0043517)
15 neutrophil activation (GO:0042119)
16 alternative mRNA splicing, via spliceosome (GO:0000380)
17 positive regulation of interleukin-4 production (GO:0032753)
18 T cell migration (GO:0072678)
19 regulation of production of miRNAs involved in gene silencing by miRNA (GO:1903798)
20 intracellular estrogen receptor signaling pathway (GO:0030520)
21 myeloid cell activation involved in immune response (GO:0002275)
22 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (GO:0002460)
23 regulation of T cell migration (GO:2000404)
24 calcium-mediated signaling using intracellular calcium source (GO:0035584)
25 regulation of platelet activation (GO:0010543)
26 positive regulation of signal transduction by p53 class mediator (GO:1901798)
27 regulation of interleukin-4 production (GO:0032673)
28 neuromuscular junction development (GO:0007528)
29 immune response-activating cell surface receptor signaling pathway (GO:0002429)
30 positive regulation of T cell migration (GO:2000406)
31 positive regulation of gene silencing by miRNA (GO:2000637)
32 T cell activation involved in immune response (GO:0002286)
33 regulation of DNA damage response, signal transduction by p53 class mediator (GO:0043516)
34 signal transduction by p53 class mediator (GO:0072331)
35 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474)
36 intrinsic apoptotic signaling pathway by p53 class mediator (GO:0072332)
37 regulation of androgen receptor signaling pathway (GO:0060765)
38 regulation of intracellular steroid hormone receptor signaling pathway (GO:0033143)
39 regulation of protein tyrosine kinase activity (GO:0061097)
40 lymphocyte migration (GO:0072676)
41 nucleocytoplasmic transport (GO:0006913)
42 T cell differentiation (GO:0030217)
43 monocyte chemotaxis (GO:0002548)
44 positive regulation of protein tyrosine kinase activity (GO:0061098)
45 lymphocyte chemotaxis (GO:0048247)
46 mRNA catabolic process (GO:0006402)
47 intracellular steroid hormone receptor signaling pathway (GO:0030518)
48 positive regulation of RNA metabolic process (GO:0051254)
49 regulation of pri-miRNA transcription by RNA polymerase II (GO:1902893)
Overlap Adjusted.P.value Genes
1 2/70 0.02104704 FCER1G;CCL20
2 2/73 0.02104704 FCER1G;CCL20
3 2/77 0.02104704 FCER1G;CCL20
4 1/9 0.04522556 DDX5
5 1/10 0.04522556 FCER1G
6 1/10 0.04522556 FCER1G
7 1/11 0.04522556 DDX5
8 1/11 0.04522556 FCER1G
9 1/12 0.04522556 DDX5
10 1/14 0.04522556 FCER1G
11 1/14 0.04522556 CCL20
12 1/15 0.04522556 DDX5
13 1/15 0.04522556 DDX5
14 1/15 0.04522556 DDX5
15 1/16 0.04522556 FCER1G
16 1/18 0.04522556 DDX5
17 1/18 0.04522556 FCER1G
18 1/18 0.04522556 CCL20
19 1/18 0.04522556 DDX5
20 1/18 0.04522556 DDX5
21 1/19 0.04522556 FCER1G
22 1/20 0.04522556 BACH2
23 1/20 0.04522556 CCL20
24 1/21 0.04522556 CCL20
25 1/22 0.04522556 FCER1G
26 1/22 0.04522556 DDX5
27 1/23 0.04522556 FCER1G
28 1/24 0.04522556 DOK7
29 1/24 0.04522556 FCER1G
30 1/25 0.04553053 CCL20
31 1/27 0.04633059 DDX5
32 1/28 0.04633059 FCER1G
33 1/28 0.04633059 DDX5
34 1/33 0.04997650 DDX5
35 1/33 0.04997650 FCER1G
36 1/36 0.04997650 DDX5
37 1/36 0.04997650 DDX5
38 1/37 0.04997650 DDX5
39 1/39 0.04997650 DOK7
40 1/40 0.04997650 CCL20
41 1/40 0.04997650 BACH2
42 1/41 0.04997650 FCER1G
43 1/42 0.04997650 CCL20
44 1/42 0.04997650 DOK7
45 1/44 0.04997650 CCL20
46 1/44 0.04997650 DDX5
47 1/44 0.04997650 DDX5
48 1/45 0.04997650 DDX5
49 1/45 0.04997650 DDX5
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"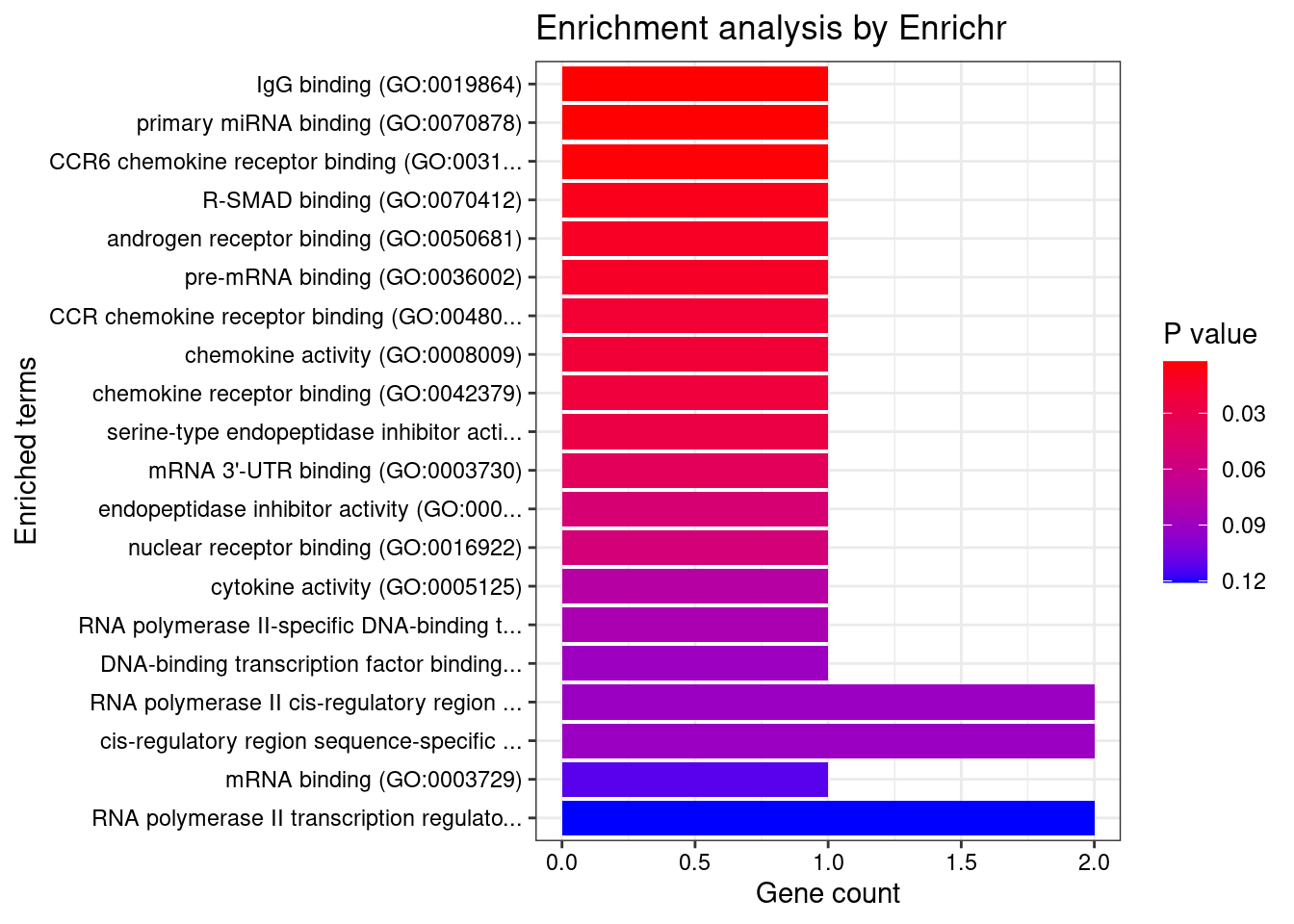
Term Overlap Adjusted.P.value
1 IgG binding (GO:0019864) 1/5 0.02726682
2 primary miRNA binding (GO:0070878) 1/6 0.02726682
3 CCR6 chemokine receptor binding (GO:0031731) 1/7 0.02726682
Genes
1 FCER1G
2 DDX5
3 CCL20
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"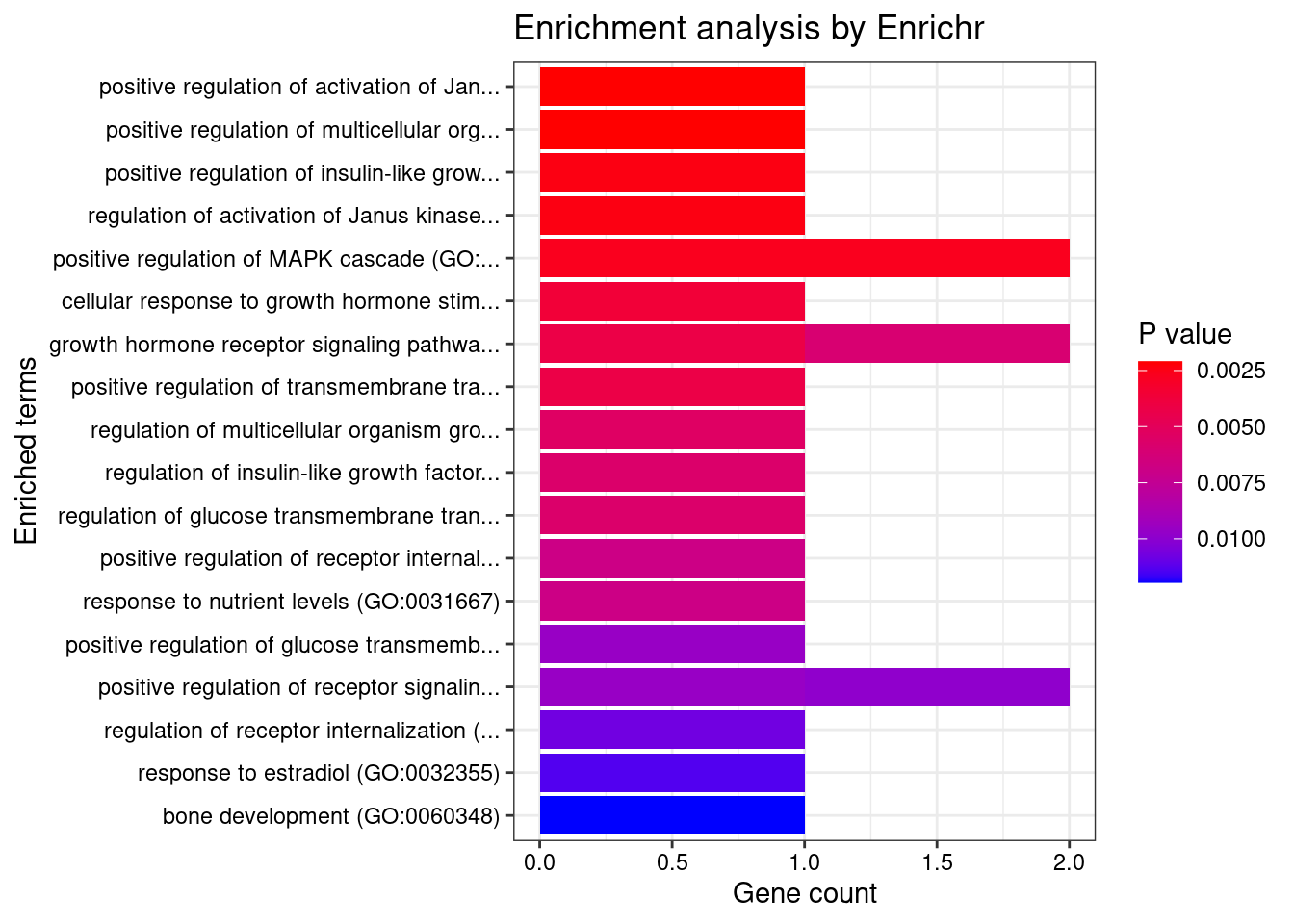
Term
1 positive regulation of activation of Janus kinase activity (GO:0010536)
2 positive regulation of multicellular organism growth (GO:0040018)
3 positive regulation of insulin-like growth factor receptor signaling pathway (GO:0043568)
4 regulation of activation of Janus kinase activity (GO:0010533)
5 positive regulation of MAPK cascade (GO:0043410)
6 cellular response to growth hormone stimulus (GO:0071378)
7 growth hormone receptor signaling pathway via JAK-STAT (GO:0060397)
8 positive regulation of transmembrane transport (GO:0034764)
9 regulation of multicellular organism growth (GO:0040014)
10 regulation of insulin-like growth factor receptor signaling pathway (GO:0043567)
11 regulation of glucose transmembrane transport (GO:0010827)
12 growth hormone receptor signaling pathway (GO:0060396)
13 positive regulation of receptor internalization (GO:0002092)
14 response to nutrient levels (GO:0031667)
15 positive regulation of glucose transmembrane transport (GO:0010828)
16 positive regulation of receptor signaling pathway via STAT (GO:1904894)
17 positive regulation of receptor signaling pathway via JAK-STAT (GO:0046427)
18 regulation of receptor internalization (GO:0002090)
19 response to estradiol (GO:0032355)
20 bone development (GO:0060348)
21 positive regulation of receptor-mediated endocytosis (GO:0048260)
22 regulation of receptor signaling pathway via JAK-STAT (GO:0046425)
23 positive regulation of developmental growth (GO:0048639)
24 positive regulation of tyrosine phosphorylation of STAT protein (GO:0042531)
25 response to organic cyclic compound (GO:0014070)
26 regulation of tyrosine phosphorylation of STAT protein (GO:0042509)
27 positive regulation of MAP kinase activity (GO:0043406)
28 positive regulation of growth (GO:0045927)
29 positive regulation of phosphatidylinositol 3-kinase signaling (GO:0014068)
30 regulation of peptidyl-tyrosine phosphorylation (GO:0050730)
31 regulation of MAP kinase activity (GO:0043405)
32 positive regulation of cellular metabolic process (GO:0031325)
33 positive regulation of protein serine/threonine kinase activity (GO:0071902)
34 regulation of phosphatidylinositol 3-kinase signaling (GO:0014066)
35 response to lipid (GO:0033993)
Overlap Adjusted.P.value Genes
1 1/7 0.02310039 GH1
2 1/7 0.02310039 GH1
3 1/8 0.02310039 GH1
4 1/8 0.02310039 GH1
5 2/274 0.02310039 GH1;ARHGAP8
6 1/12 0.02310039 GH1
7 1/14 0.02310039 GH1
8 1/14 0.02310039 GH1
9 1/18 0.02310039 GH1
10 1/19 0.02310039 GH1
11 1/19 0.02310039 GH1
12 1/20 0.02310039 GH1
13 1/23 0.02310039 GH1
14 1/23 0.02310039 GH1
15 1/32 0.02726104 GH1
16 1/32 0.02726104 GH1
17 1/33 0.02726104 GH1
18 1/36 0.02806260 GH1
19 1/38 0.02806260 GH1
20 1/40 0.02806260 GH1
21 1/44 0.02868247 GH1
22 1/45 0.02868247 GH1
23 1/52 0.03167543 GH1
24 1/59 0.03359111 GH1
25 1/60 0.03359111 GH1
26 1/68 0.03572813 GH1
27 1/69 0.03572813 GH1
28 1/76 0.03708376 GH1
29 1/77 0.03708376 GH1
30 1/92 0.04275078 GH1
31 1/97 0.04338556 GH1
32 1/105 0.04338556 GH1
33 1/106 0.04338556 GH1
34 1/106 0.04338556 GH1
35 1/114 0.04528154 GH1
[1] "GO_Cellular_Component_2021"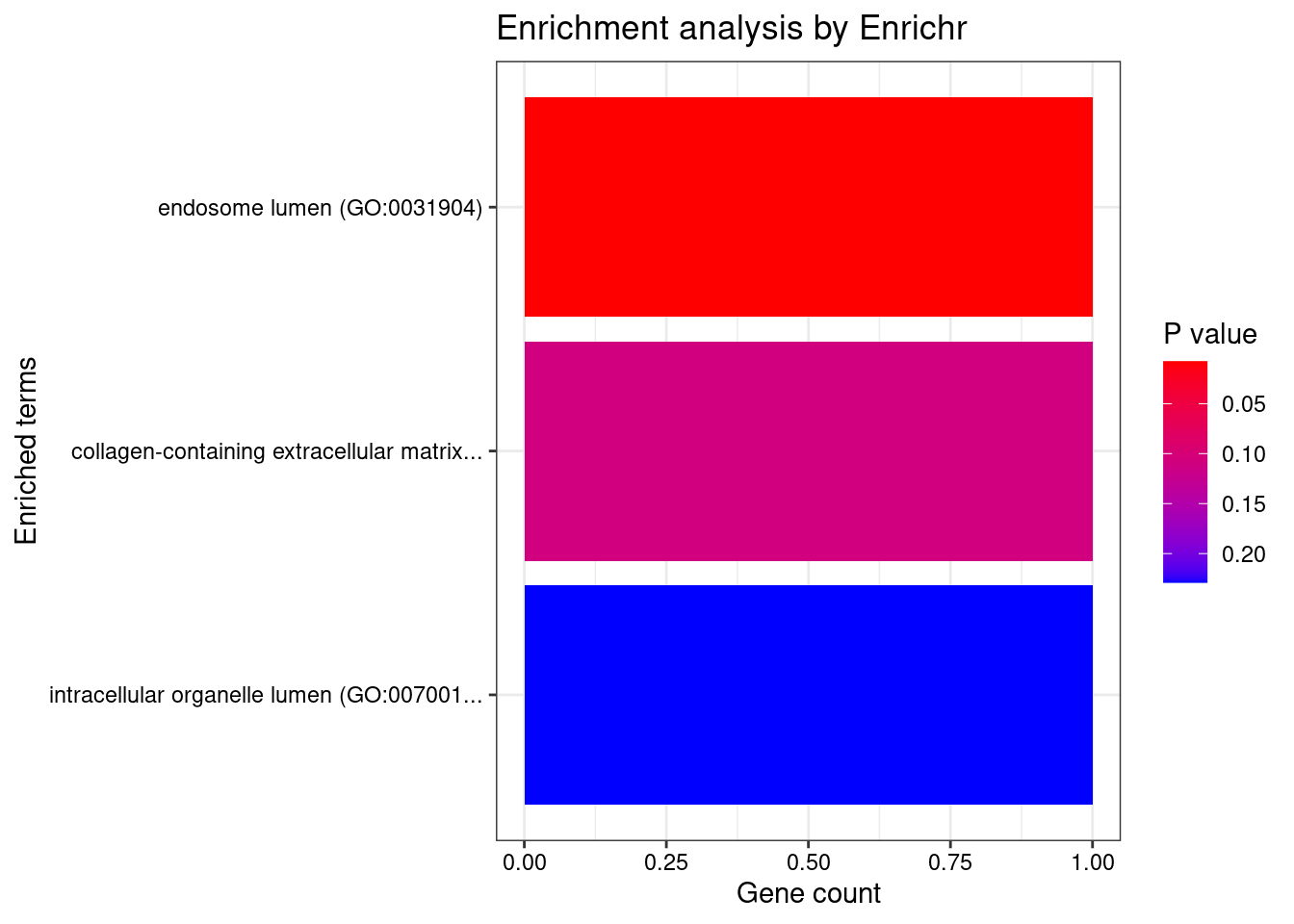
Term Overlap Adjusted.P.value Genes
1 endosome lumen (GO:0031904) 1/26 0.02332672 GH1
[1] "GO_Molecular_Function_2021"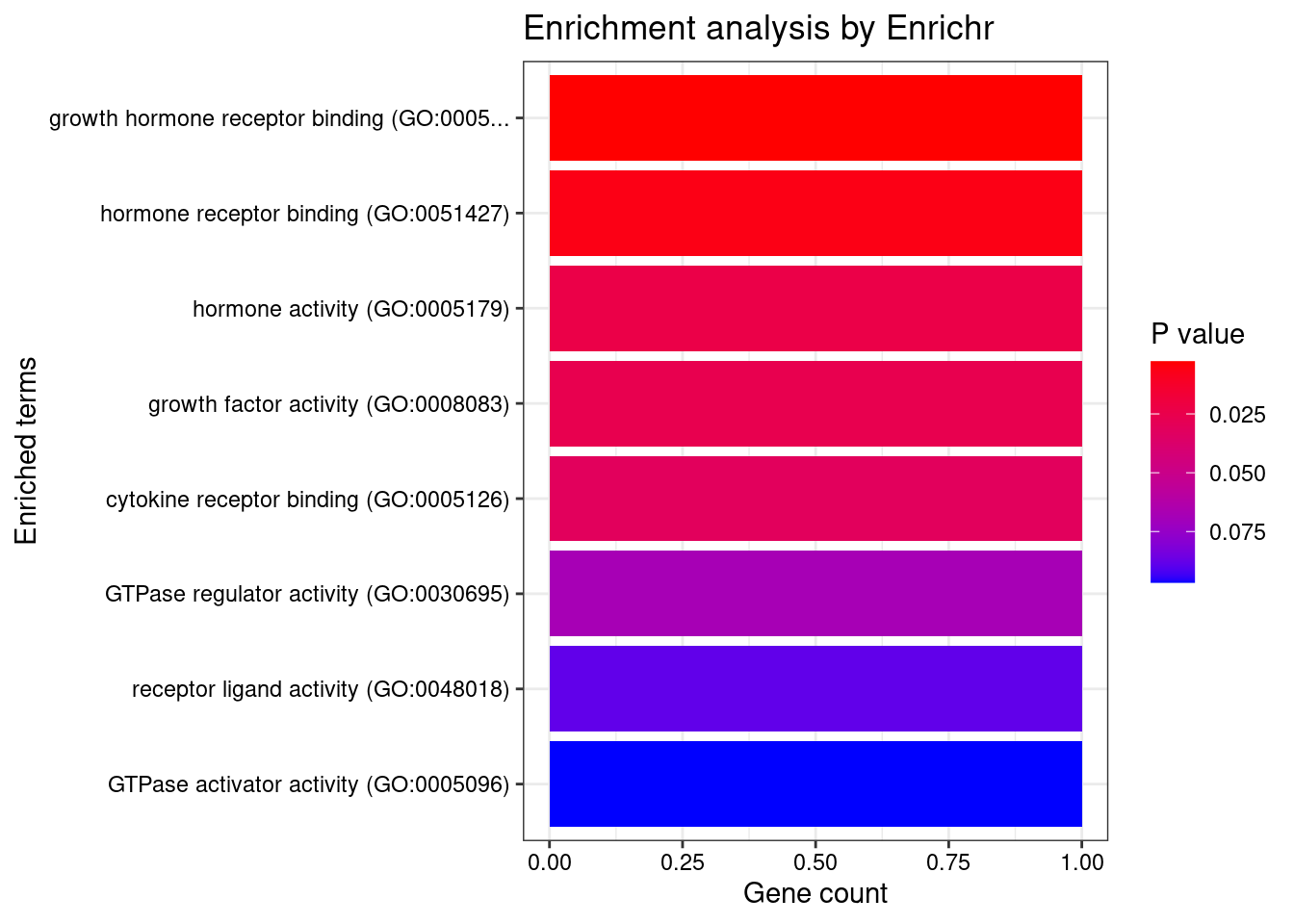
Term Overlap Adjusted.P.value
1 growth hormone receptor binding (GO:0005131) 1/9 0.02157805
2 hormone receptor binding (GO:0051427) 1/21 0.02513677
3 hormone activity (GO:0005179) 1/78 0.04974895
4 growth factor activity (GO:0008083) 1/87 0.04974895
5 cytokine receptor binding (GO:0005126) 1/105 0.04974895
Genes
1 GH1
2 GH1
3 GH1
4 GH1
5 GH1
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"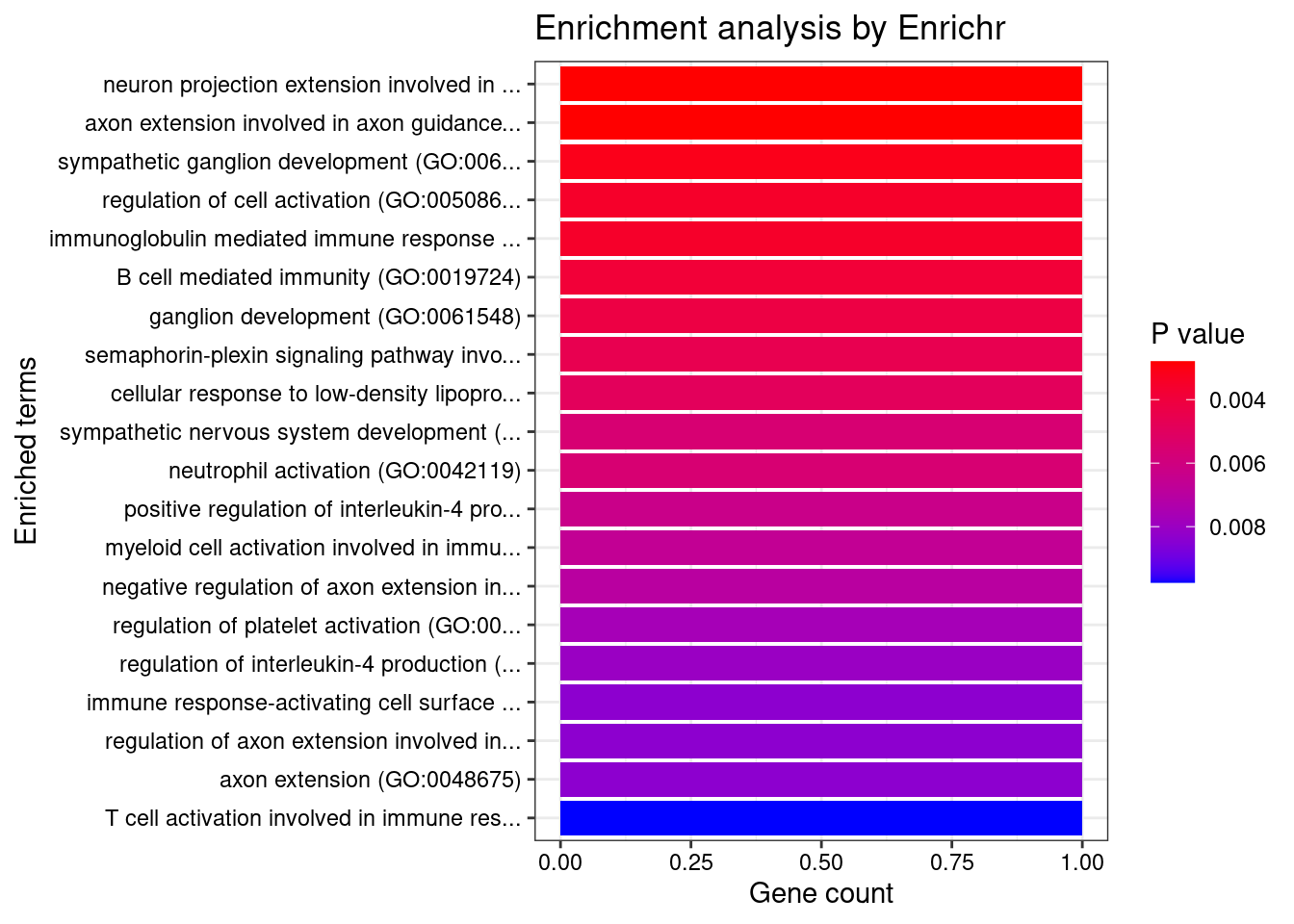
Term
1 neuron projection extension involved in neuron projection guidance (GO:1902284)
2 axon extension involved in axon guidance (GO:0048846)
3 sympathetic ganglion development (GO:0061549)
4 regulation of cell activation (GO:0050865)
5 immunoglobulin mediated immune response (GO:0016064)
6 B cell mediated immunity (GO:0019724)
7 ganglion development (GO:0061548)
8 semaphorin-plexin signaling pathway involved in neuron projection guidance (GO:1902285)
9 cellular response to low-density lipoprotein particle stimulus (GO:0071404)
10 sympathetic nervous system development (GO:0048485)
11 neutrophil activation (GO:0042119)
12 positive regulation of interleukin-4 production (GO:0032753)
13 myeloid cell activation involved in immune response (GO:0002275)
14 negative regulation of axon extension involved in axon guidance (GO:0048843)
15 regulation of platelet activation (GO:0010543)
16 regulation of interleukin-4 production (GO:0032673)
17 immune response-activating cell surface receptor signaling pathway (GO:0002429)
18 regulation of axon extension involved in axon guidance (GO:0048841)
19 axon extension (GO:0048675)
20 T cell activation involved in immune response (GO:0002286)
21 negative regulation of gene expression, epigenetic (GO:0045814)
22 negative regulation of axon extension (GO:0030517)
23 negative regulation of chemotaxis (GO:0050922)
24 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474)
25 neuron projection extension (GO:1990138)
26 negative chemotaxis (GO:0050919)
27 semaphorin-plexin signaling pathway (GO:0071526)
28 neural crest cell migration (GO:0001755)
29 negative regulation of G0 to G1 transition (GO:0070317)
30 regulation of G0 to G1 transition (GO:0070316)
31 T cell differentiation (GO:0030217)
32 neural crest cell development (GO:0014032)
33 receptor internalization (GO:0031623)
34 ameboidal-type cell migration (GO:0001667)
35 positive regulation of phagocytosis (GO:0050766)
36 receptor metabolic process (GO:0043112)
37 regulation of phagocytosis (GO:0050764)
38 negative regulation of cell cycle process (GO:0010948)
39 neutrophil chemotaxis (GO:0030593)
40 Fc-gamma receptor signaling pathway (GO:0038094)
41 granulocyte chemotaxis (GO:0071621)
42 Fc receptor mediated stimulatory signaling pathway (GO:0002431)
43 negative regulation of developmental process (GO:0051093)
44 neutrophil migration (GO:1990266)
45 antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590)
46 regulation of gene expression, epigenetic (GO:0040029)
47 positive regulation of transport (GO:0051050)
Overlap Adjusted.P.value Genes
1 1/8 0.03216218 SEMA3F
2 1/8 0.03216218 SEMA3F
3 1/9 0.03216218 SEMA3F
4 1/10 0.03216218 FCER1G
5 1/10 0.03216218 FCER1G
6 1/11 0.03216218 FCER1G
7 1/12 0.03216218 SEMA3F
8 1/13 0.03216218 SEMA3F
9 1/14 0.03216218 FCER1G
10 1/16 0.03216218 SEMA3F
11 1/16 0.03216218 FCER1G
12 1/18 0.03216218 FCER1G
13 1/19 0.03216218 FCER1G
14 1/20 0.03216218 SEMA3F
15 1/22 0.03216218 FCER1G
16 1/23 0.03216218 FCER1G
17 1/24 0.03216218 FCER1G
18 1/24 0.03216218 SEMA3F
19 1/24 0.03216218 SEMA3F
20 1/28 0.03227327 FCER1G
21 1/30 0.03227327 SUZ12
22 1/31 0.03227327 SEMA3F
23 1/32 0.03227327 SEMA3F
24 1/33 0.03227327 FCER1G
25 1/33 0.03227327 SEMA3F
26 1/33 0.03227327 SEMA3F
27 1/35 0.03295160 SEMA3F
28 1/37 0.03329374 SEMA3F
29 1/38 0.03329374 SUZ12
30 1/41 0.03358953 SUZ12
31 1/41 0.03358953 FCER1G
32 1/45 0.03569309 SEMA3F
33 1/49 0.03766547 FCER1G
34 1/52 0.03838918 SEMA3F
35 1/53 0.03838918 FCER1G
36 1/58 0.03971019 FCER1G
37 1/58 0.03971019 FCER1G
38 1/64 0.04262670 SUZ12
39 1/70 0.04377807 FCER1G
40 1/72 0.04377807 FCER1G
41 1/73 0.04377807 FCER1G
42 1/74 0.04377807 FCER1G
43 1/77 0.04377807 SUZ12
44 1/77 0.04377807 FCER1G
45 1/78 0.04377807 FCER1G
46 1/82 0.04499563 SUZ12
47 1/91 0.04880592 FCER1G
[1] "GO_Cellular_Component_2021"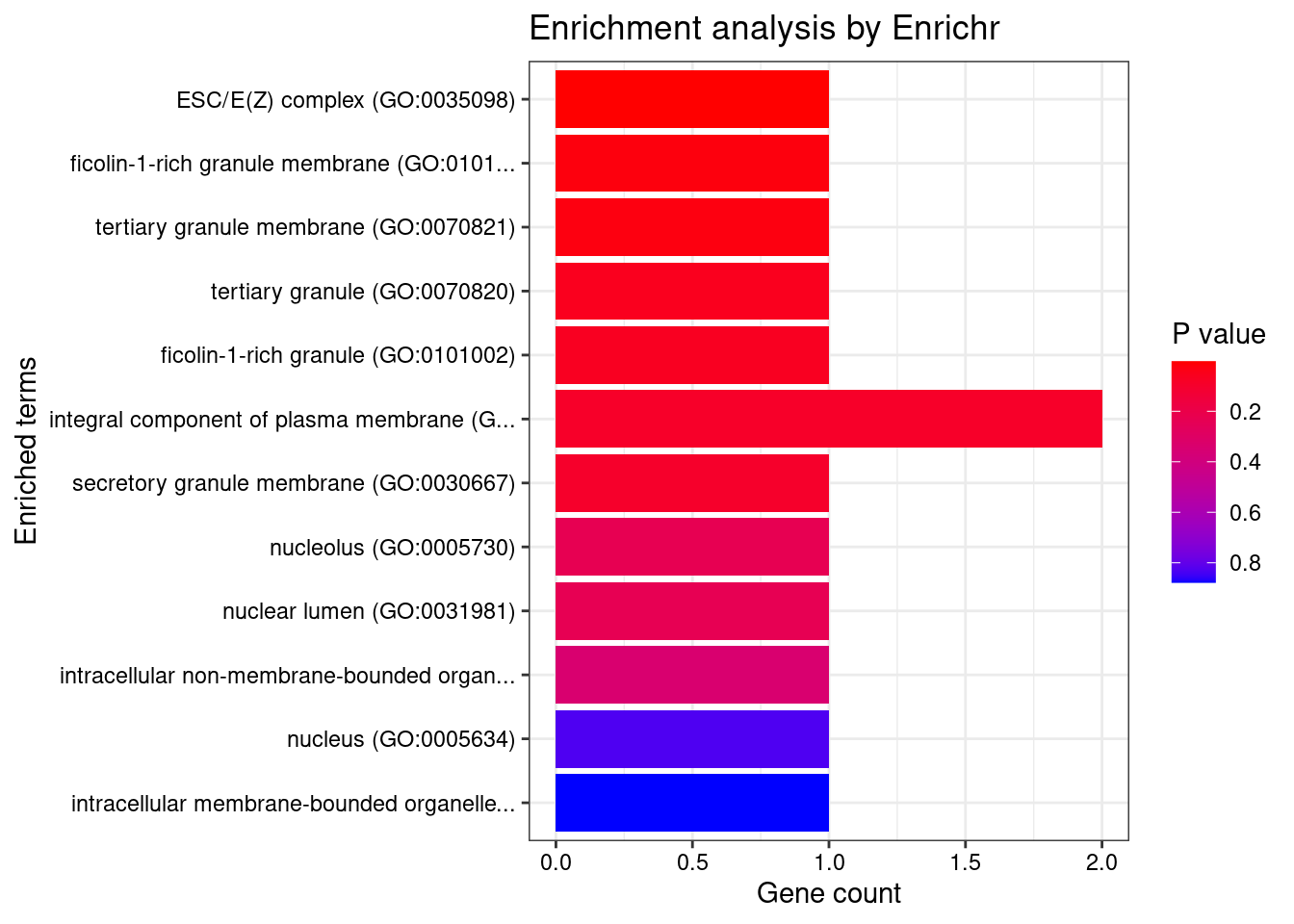
Term Overlap Adjusted.P.value Genes
1 ESC/E(Z) complex (GO:0035098) 1/8 0.03356418 SUZ12
[1] "GO_Molecular_Function_2021"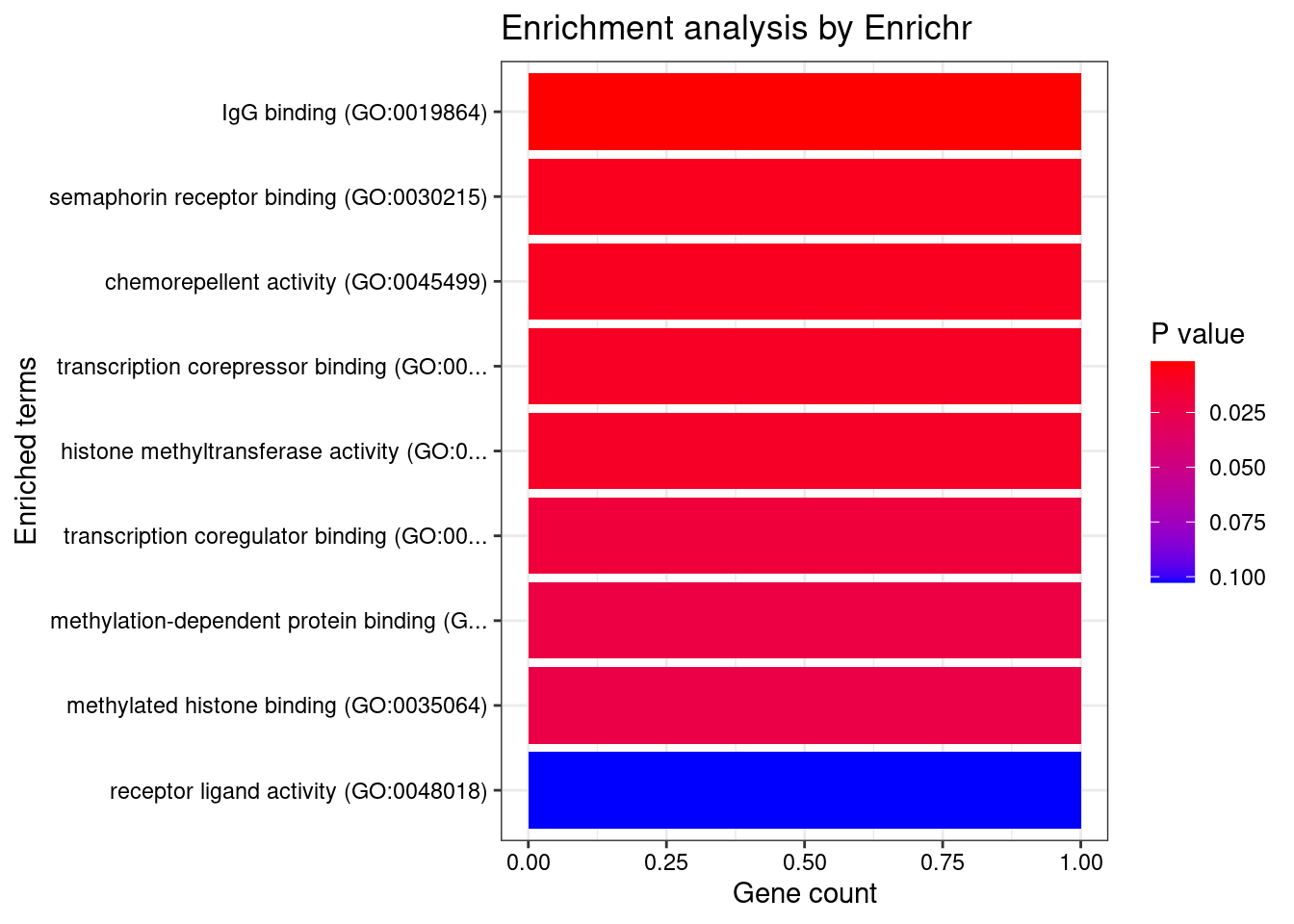
Term Overlap
1 IgG binding (GO:0019864) 1/5
2 semaphorin receptor binding (GO:0030215) 1/23
3 chemorepellent activity (GO:0045499) 1/25
4 transcription corepressor binding (GO:0001222) 1/29
5 histone methyltransferase activity (GO:0042054) 1/32
6 transcription coregulator binding (GO:0001221) 1/53
7 methylation-dependent protein binding (GO:0140034) 1/63
8 methylated histone binding (GO:0035064) 1/68
Adjusted.P.value Genes
1 0.01574025 FCER1G
2 0.02006629 SEMA3F
3 0.02006629 SEMA3F
4 0.02006629 SUZ12
5 0.02006629 SUZ12
6 0.02650719 SUZ12
7 0.02650719 SUZ12
8 0.02650719 SUZ12
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"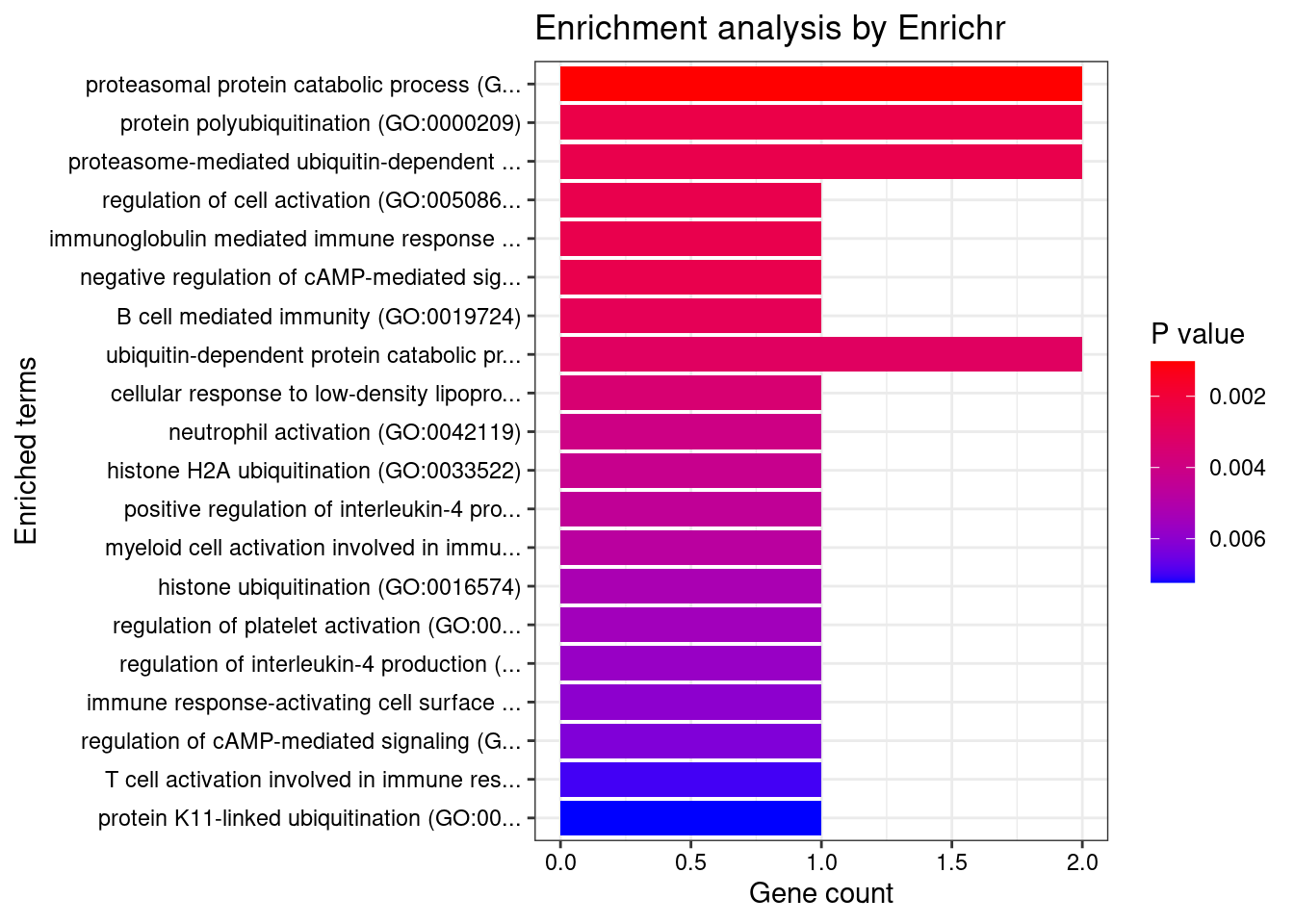
Term
1 proteasomal protein catabolic process (GO:0010498)
2 protein polyubiquitination (GO:0000209)
3 proteasome-mediated ubiquitin-dependent protein catabolic process (GO:0043161)
4 regulation of cell activation (GO:0050865)
5 immunoglobulin mediated immune response (GO:0016064)
6 negative regulation of cAMP-mediated signaling (GO:0043951)
7 B cell mediated immunity (GO:0019724)
8 ubiquitin-dependent protein catabolic process (GO:0006511)
9 cellular response to low-density lipoprotein particle stimulus (GO:0071404)
10 neutrophil activation (GO:0042119)
11 histone H2A ubiquitination (GO:0033522)
12 positive regulation of interleukin-4 production (GO:0032753)
13 myeloid cell activation involved in immune response (GO:0002275)
14 histone ubiquitination (GO:0016574)
15 regulation of platelet activation (GO:0010543)
16 regulation of interleukin-4 production (GO:0032673)
17 immune response-activating cell surface receptor signaling pathway (GO:0002429)
18 regulation of cAMP-mediated signaling (GO:0043949)
19 T cell activation involved in immune response (GO:0002286)
20 protein K11-linked ubiquitination (GO:0070979)
21 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474)
22 protein monoubiquitination (GO:0006513)
23 T cell differentiation (GO:0030217)
24 protein K63-linked ubiquitination (GO:0070534)
25 postreplication repair (GO:0006301)
26 receptor internalization (GO:0031623)
27 positive regulation of phagocytosis (GO:0050766)
28 protein K48-linked ubiquitination (GO:0070936)
29 receptor metabolic process (GO:0043112)
30 regulation of phagocytosis (GO:0050764)
31 regulation of tumor necrosis factor-mediated signaling pathway (GO:0010803)
32 response to light stimulus (GO:0009416)
33 I-kappaB kinase/NF-kappaB signaling (GO:0007249)
34 protein autoubiquitination (GO:0051865)
35 neutrophil chemotaxis (GO:0030593)
36 Fc-gamma receptor signaling pathway (GO:0038094)
37 granulocyte chemotaxis (GO:0071621)
38 regulation of cytokine-mediated signaling pathway (GO:0001959)
39 Fc receptor mediated stimulatory signaling pathway (GO:0002431)
40 neutrophil migration (GO:1990266)
41 antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590)
42 negative regulation of defense response (GO:0031348)
43 positive regulation of transport (GO:0051050)
44 antigen processing and presentation of exogenous peptide antigen via MHC class II (GO:0019886)
45 response to UV (GO:0009411)
46 antigen processing and presentation of peptide antigen via MHC class II (GO:0002495)
47 antigen processing and presentation of exogenous peptide antigen (GO:0002478)
48 Fc-epsilon receptor signaling pathway (GO:0038095)
49 Fc receptor signaling pathway (GO:0038093)
50 histone modification (GO:0016570)
51 cellular response to lectin (GO:1990858)
52 stimulatory C-type lectin receptor signaling pathway (GO:0002223)
53 negative regulation of response to external stimulus (GO:0032102)
54 innate immune response activating cell surface receptor signaling pathway (GO:0002220)
55 positive regulation of canonical Wnt signaling pathway (GO:0090263)
Overlap Adjusted.P.value Genes
1 2/205 0.02771084 SHARPIN;UBE2B
2 2/314 0.02771084 SHARPIN;UBE2B
3 2/321 0.02771084 SHARPIN;UBE2B
4 1/10 0.02771084 FCER1G
5 1/10 0.02771084 FCER1G
6 1/10 0.02771084 UBE2B
7 1/11 0.02771084 FCER1G
8 2/354 0.02771084 SHARPIN;UBE2B
9 1/14 0.02771084 FCER1G
10 1/16 0.02771084 FCER1G
11 1/17 0.02771084 UBE2B
12 1/18 0.02771084 FCER1G
13 1/19 0.02771084 FCER1G
14 1/21 0.02771084 UBE2B
15 1/22 0.02771084 FCER1G
16 1/23 0.02771084 FCER1G
17 1/24 0.02771084 FCER1G
18 1/25 0.02771084 UBE2B
19 1/28 0.02891857 FCER1G
20 1/29 0.02891857 UBE2B
21 1/33 0.03132780 FCER1G
22 1/39 0.03531969 UBE2B
23 1/41 0.03550947 FCER1G
24 1/44 0.03650896 UBE2B
25 1/46 0.03663440 UBE2B
26 1/49 0.03720632 FCER1G
27 1/53 0.03720632 FCER1G
28 1/57 0.03720632 UBE2B
29 1/58 0.03720632 FCER1G
30 1/58 0.03720632 FCER1G
31 1/58 0.03720632 SHARPIN
32 1/61 0.03734690 UBE2B
33 1/62 0.03734690 SHARPIN
34 1/65 0.03767238 UBE2B
35 1/70 0.03767238 FCER1G
36 1/72 0.03767238 FCER1G
37 1/73 0.03767238 FCER1G
38 1/74 0.03767238 SHARPIN
39 1/74 0.03767238 FCER1G
40 1/77 0.03775661 FCER1G
41 1/78 0.03775661 FCER1G
42 1/85 0.04013729 SHARPIN
43 1/91 0.04194602 FCER1G
44 1/98 0.04304962 FCER1G
45 1/99 0.04304962 UBE2B
46 1/100 0.04304962 FCER1G
47 1/103 0.04338466 FCER1G
48 1/111 0.04355674 FCER1G
49 1/112 0.04355674 FCER1G
50 1/114 0.04355674 UBE2B
51 1/115 0.04355674 FCER1G
52 1/115 0.04355674 FCER1G
53 1/118 0.04355674 SHARPIN
54 1/119 0.04355674 FCER1G
55 1/130 0.04666649 UBE2B
[1] "GO_Cellular_Component_2021"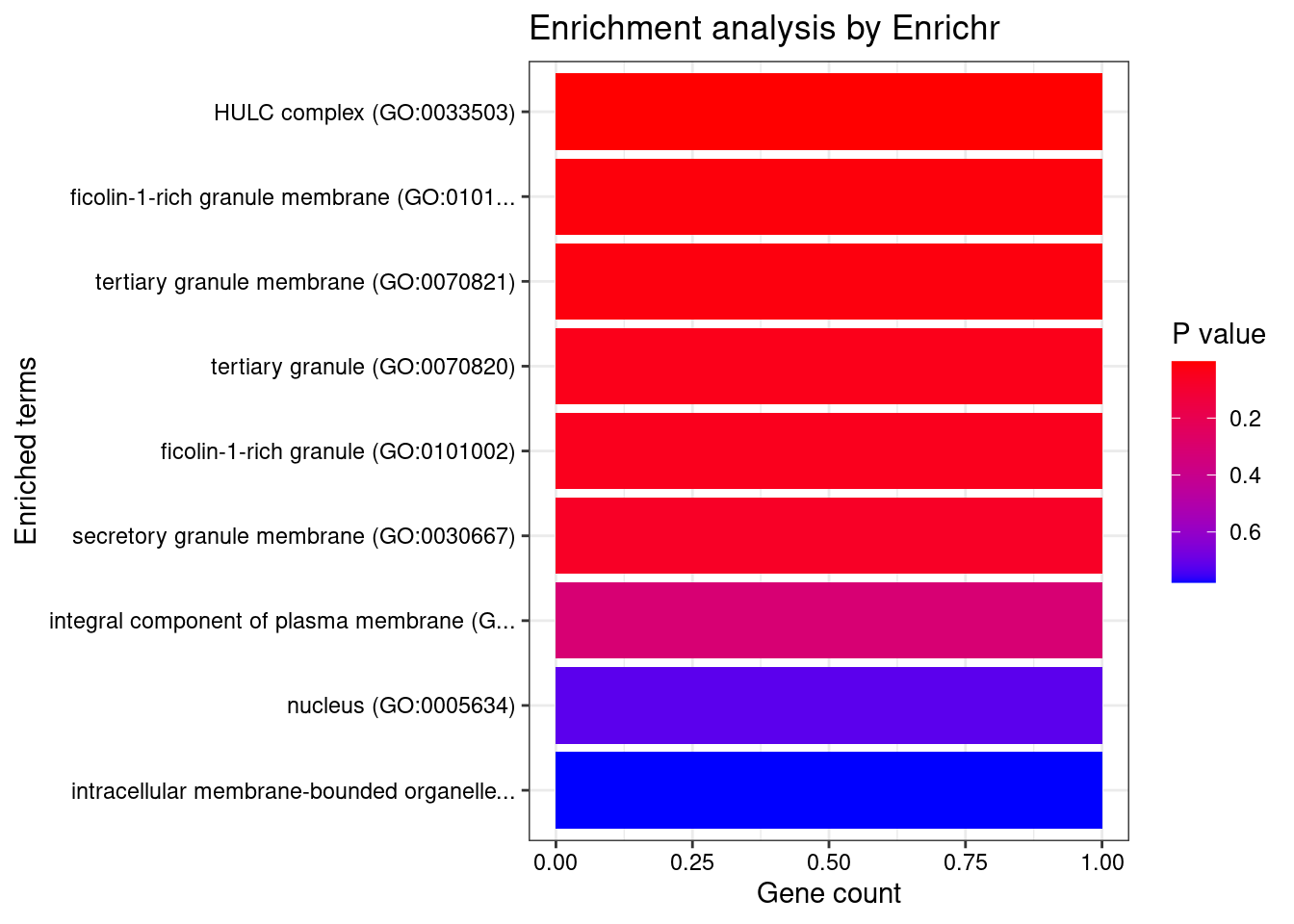
Term Overlap Adjusted.P.value Genes
1 HULC complex (GO:0033503) 1/5 0.01124526 UBE2B
[1] "GO_Molecular_Function_2021"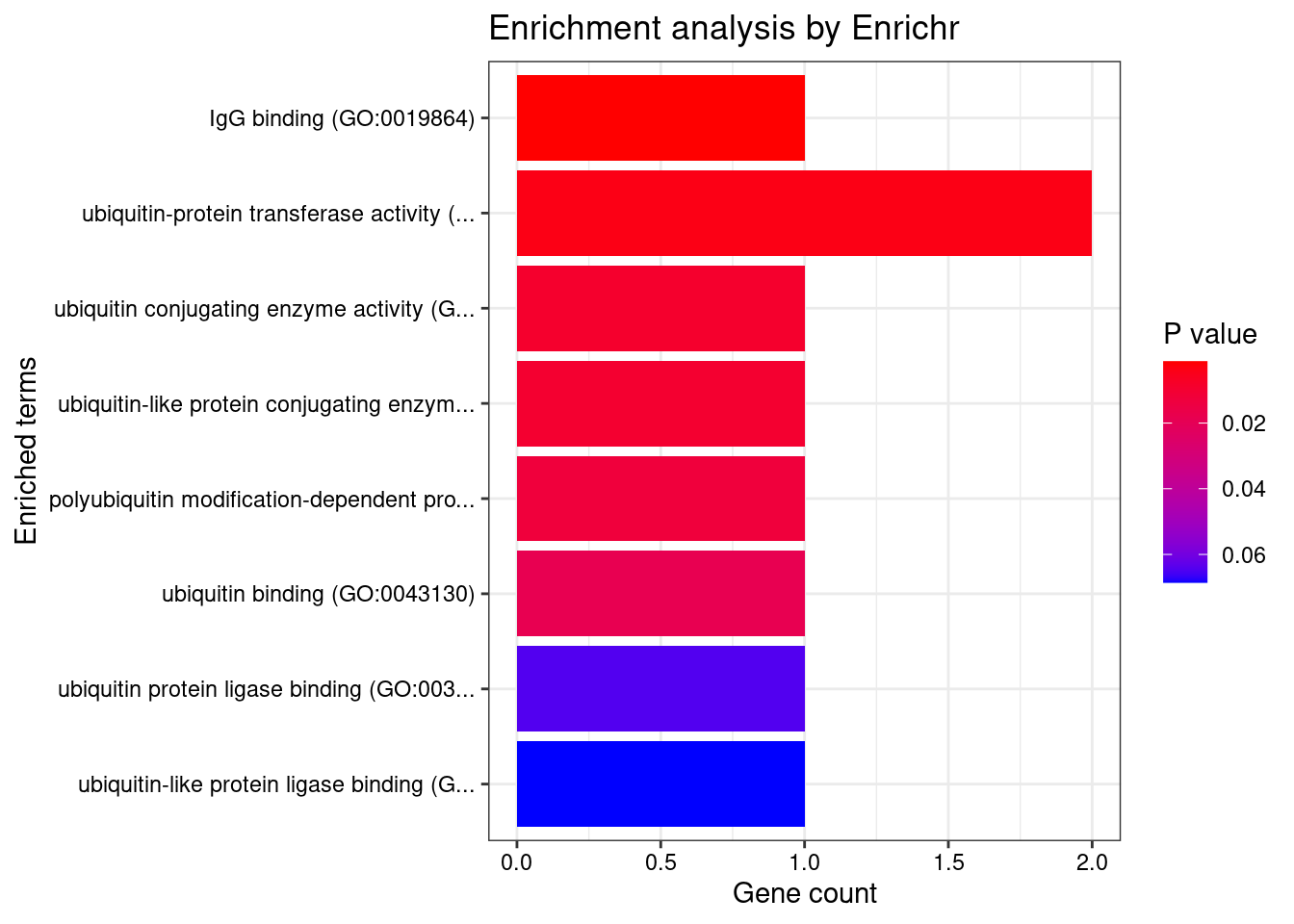
Term
1 IgG binding (GO:0019864)
2 ubiquitin-protein transferase activity (GO:0004842)
3 ubiquitin conjugating enzyme activity (GO:0061631)
4 ubiquitin-like protein conjugating enzyme activity (GO:0061650)
5 polyubiquitin modification-dependent protein binding (GO:0031593)
6 ubiquitin binding (GO:0043130)
Overlap Adjusted.P.value Genes
1 1/5 0.009995787 FCER1G
2 2/392 0.014738563 SHARPIN;UBE2B
3 1/35 0.018929624 UBE2B
4 1/38 0.018929624 UBE2B
5 1/51 0.020298056 SHARPIN
6 1/74 0.024487045 SHARPIN
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"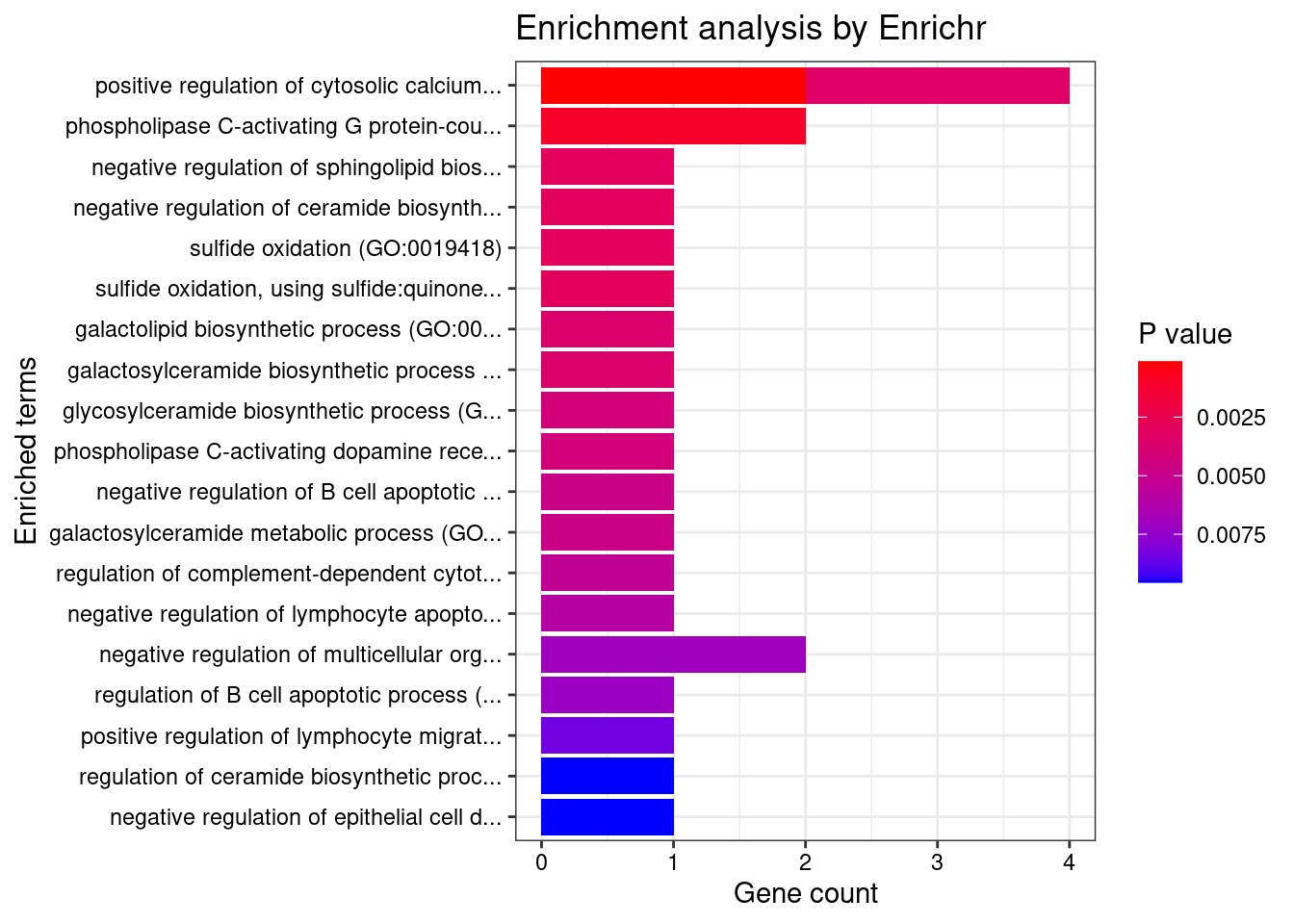
Term
1 positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway (GO:0051482)
2 phospholipase C-activating G protein-coupled receptor signaling pathway (GO:0007200)
3 negative regulation of sphingolipid biosynthetic process (GO:0090155)
4 negative regulation of ceramide biosynthetic process (GO:1900060)
5 sulfide oxidation (GO:0019418)
6 sulfide oxidation, using sulfide:quinone oxidoreductase (GO:0070221)
7 positive regulation of cytosolic calcium ion concentration (GO:0007204)
8 galactolipid biosynthetic process (GO:0019375)
9 galactosylceramide biosynthetic process (GO:0006682)
10 glycosylceramide biosynthetic process (GO:0046476)
11 phospholipase C-activating dopamine receptor signaling pathway (GO:0060158)
12 negative regulation of B cell apoptotic process (GO:0002903)
13 galactosylceramide metabolic process (GO:0006681)
14 regulation of complement-dependent cytotoxicity (GO:1903659)
15 negative regulation of lymphocyte apoptotic process (GO:0070229)
16 negative regulation of multicellular organismal process (GO:0051241)
17 regulation of B cell apoptotic process (GO:0002902)
Overlap Adjusted.P.value Genes
1 2/28 0.01434511 GNA15;GPR18
2 2/81 0.04274781 GNA15;GPR18
3 1/5 0.04274781 ORMDL3
4 1/5 0.04274781 ORMDL3
5 1/5 0.04274781 SUOX
6 1/5 0.04274781 SUOX
7 2/147 0.04274781 GNA15;GPR18
8 1/6 0.04274781 B4GALT3
9 1/6 0.04274781 B4GALT3
10 1/7 0.04274781 B4GALT3
11 1/7 0.04274781 GNA15
12 1/8 0.04274781 ORMDL3
13 1/8 0.04274781 B4GALT3
14 1/9 0.04464395 IL13
15 1/10 0.04628472 ORMDL3
16 2/214 0.04898046 PLCL1;IL13
17 1/12 0.04898046 ORMDL3
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"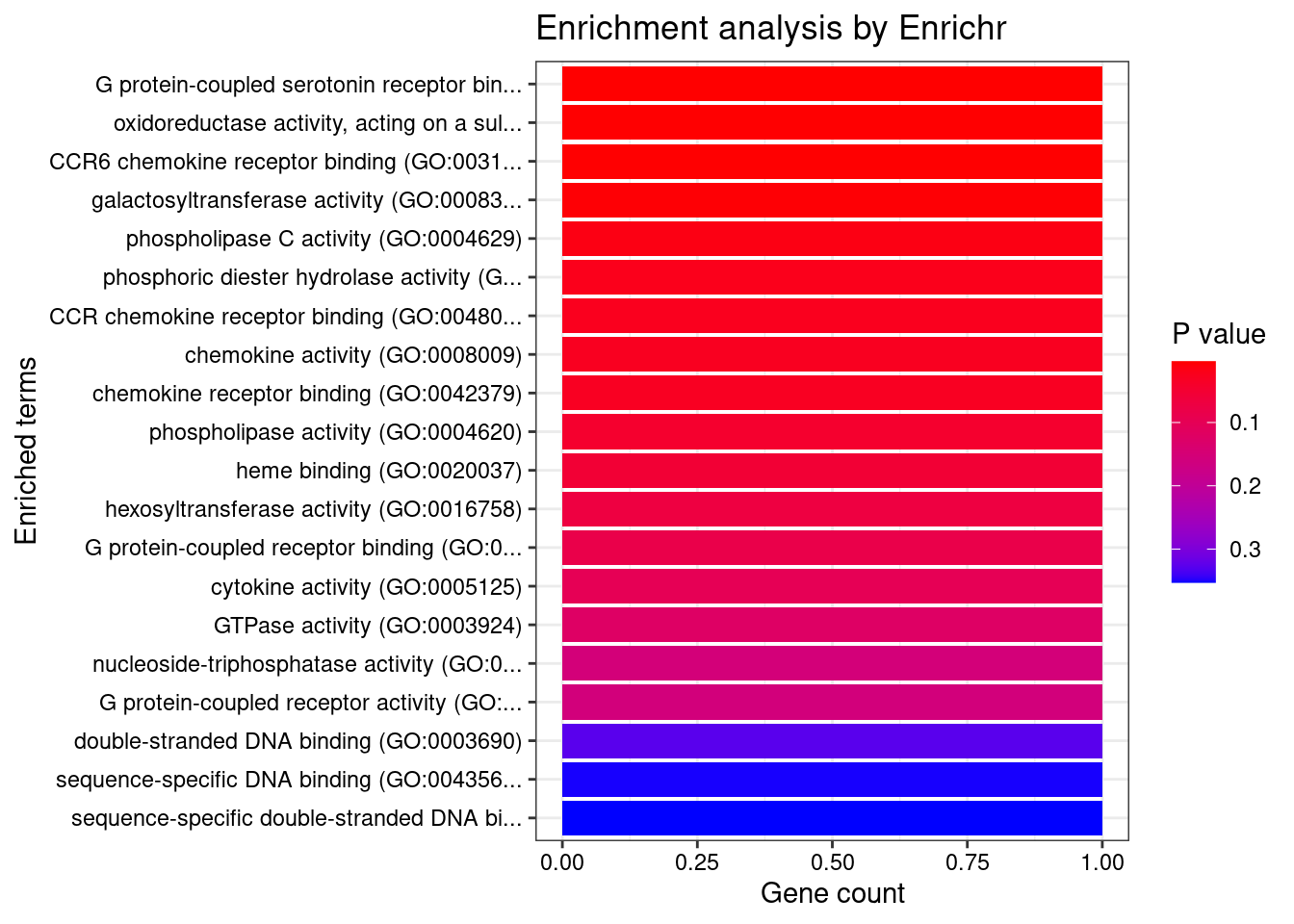
Term
1 G protein-coupled serotonin receptor binding (GO:0031821)
2 oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor (GO:0016670)
3 CCR6 chemokine receptor binding (GO:0031731)
4 galactosyltransferase activity (GO:0008378)
Overlap Adjusted.P.value Genes
1 1/6 0.03214645 GNA15
2 1/7 0.03214645 SUOX
3 1/7 0.03214645 CCL20
4 1/11 0.03784533 B4GALT3
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] enrichR_3.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 compiler_3.6.1 pillar_1.6.1 later_0.8.0
[5] git2r_0.26.1 workflowr_1.6.2 tools_3.6.1 digest_0.6.20
[9] evaluate_0.14 lifecycle_1.0.0 tibble_3.1.2 gtable_0.3.0
[13] pkgconfig_2.0.3 rlang_0.4.11 DBI_1.1.1 curl_3.3
[17] yaml_2.2.0 xfun_0.8 httr_1.4.1 stringr_1.4.0
[21] dplyr_1.0.7 knitr_1.23 generics_0.0.2 fs_1.3.1
[25] vctrs_0.3.8 tidyselect_1.1.0 rprojroot_2.0.2 grid_3.6.1
[29] glue_1.4.2 R6_2.5.0 fansi_0.5.0 rmarkdown_1.13
[33] farver_2.1.0 purrr_0.3.4 ggplot2_3.3.3 magrittr_2.0.1
[37] whisker_0.3-2 scales_1.1.0 promises_1.0.1 htmltools_0.3.6
[41] ellipsis_0.3.2 colorspace_1.4-1 httpuv_1.5.1 labeling_0.3
[45] utf8_1.2.1 stringi_1.4.3 munsell_0.5.0 rjson_0.2.20
[49] crayon_1.4.1