Standing height - all weights
wesleycrouse
2022-02-28
Last updated: 2022-04-12
Checks: 7 0
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 107bb6d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
working directory clean
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-a-389_allweights.Rmd) and HTML (docs/ukb-a-389_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b67c150 | wesleycrouse | 2022-04-11 | additional enrichment analysis |
| html | b67c150 | wesleycrouse | 2022-04-11 | additional enrichment analysis |
| html | 95e0f8e | wesleycrouse | 2022-04-07 | scroll bar |
| Rmd | a4575d7 | wesleycrouse | 2022-04-07 | formating |
| Rmd | 4f7234f | wesleycrouse | 2022-04-07 | scroll bars on reports |
| html | 4f7234f | wesleycrouse | 2022-04-07 | scroll bars on reports |
| Rmd | 4c816ef | wesleycrouse | 2022-04-07 | format testing |
| html | 4c816ef | wesleycrouse | 2022-04-07 | format testing |
| Rmd | b3b8825 | wesleycrouse | 2022-04-07 | cleaning up disgenet |
| html | b3b8825 | wesleycrouse | 2022-04-07 | cleaning up disgenet |
| Rmd | 25e8811 | wesleycrouse | 2022-04-07 | disgenet by tissue group |
| html | 25e8811 | wesleycrouse | 2022-04-07 | disgenet by tissue group |
| Rmd | 7fff562 | wesleycrouse | 2022-04-07 | disgenet by tissue |
| html | 7fff562 | wesleycrouse | 2022-04-07 | disgenet by tissue |
| Rmd | aa2135e | wesleycrouse | 2022-04-07 | disgenet by tissue |
| html | aa2135e | wesleycrouse | 2022-04-07 | disgenet by tissue |
| Rmd | 0d8109c | wesleycrouse | 2022-04-07 | disgenet |
| html | 0d8109c | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | f0ff987 | wesleycrouse | 2022-04-07 | disgenet |
| html | f0ff987 | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | 69ea5e1 | wesleycrouse | 2022-04-07 | disgenet |
| html | 69ea5e1 | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | 90a4ded | wesleycrouse | 2022-04-07 | disgenet |
| html | 90a4ded | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | c3ea700 | wesleycrouse | 2022-04-07 | fixing disgenet results |
| html | c3ea700 | wesleycrouse | 2022-04-07 | fixing disgenet results |
| Rmd | bfaad60 | wesleycrouse | 2022-04-07 | more disgenet tinkering |
| html | bfaad60 | wesleycrouse | 2022-04-07 | more disgenet tinkering |
| Rmd | 1932c04 | wesleycrouse | 2022-04-07 | disgenet |
| html | 1932c04 | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | 67267e0 | wesleycrouse | 2022-04-07 | exploring disgenet |
| html | 67267e0 | wesleycrouse | 2022-04-07 | exploring disgenet |
| Rmd | 9a8dc01 | wesleycrouse | 2022-04-07 | disgenet |
| html | 9a8dc01 | wesleycrouse | 2022-04-07 | disgenet |
| Rmd | 696b089 | wesleycrouse | 2022-04-07 | adding disgenet |
| html | 696b089 | wesleycrouse | 2022-04-07 | adding disgenet |
| Rmd | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| Rmd | f7e9822 | wesleycrouse | 2022-04-06 | testing subsections |
| html | f7e9822 | wesleycrouse | 2022-04-06 | testing subsections |
| Rmd | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | d0f6e53 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| Rmd | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| html | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| Rmd | e5cfa54 | wesleycrouse | 2022-04-04 | more kegg results |
| html | e5cfa54 | wesleycrouse | 2022-04-04 | more kegg results |
| Rmd | 36ef7d6 | wesleycrouse | 2022-04-04 | kegg results |
| html | 36ef7d6 | wesleycrouse | 2022-04-04 | kegg results |
| Rmd | aec6dd2 | wesleycrouse | 2022-04-04 | printing kegg results |
| html | aec6dd2 | wesleycrouse | 2022-04-04 | printing kegg results |
| Rmd | 74cdb65 | wesleycrouse | 2022-04-04 | layout |
| html | 74cdb65 | wesleycrouse | 2022-04-04 | layout |
| Rmd | 23f0259 | wesleycrouse | 2022-04-04 | layout |
| html | 23f0259 | wesleycrouse | 2022-04-04 | layout |
| Rmd | b999e70 | wesleycrouse | 2022-04-04 | fixing display of results |
| html | b999e70 | wesleycrouse | 2022-04-04 | fixing display of results |
| Rmd | dd02af5 | wesleycrouse | 2022-04-04 | kegg |
| html | dd02af5 | wesleycrouse | 2022-04-04 | kegg |
| Rmd | 34ca036 | wesleycrouse | 2022-04-04 | kegg for individual tissues |
| html | 34ca036 | wesleycrouse | 2022-04-04 | kegg for individual tissues |
| Rmd | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | 364b716 | wesleycrouse | 2022-04-02 | TWAS FP based on confidence sets |
| Rmd | c9809d4 | wesleycrouse | 2022-04-01 | additional TWAS FP analyses |
| Rmd | a9dcb4d | wesleycrouse | 2022-04-01 | alternative TWAS FP figures based on confidence sets |
| html | a9dcb4d | wesleycrouse | 2022-04-01 | alternative TWAS FP figures based on confidence sets |
| Rmd | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| html | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| html | 1c45bb5 | wesleycrouse | 2022-03-30 | fixing formatting |
| html | ad4604a | wesleycrouse | 2022-03-30 | Adding numbers to results |
| html | 18f0b45 | wesleycrouse | 2022-03-24 | format |
| Rmd | 16289f6 | wesleycrouse | 2022-03-24 | improving layout |
| Rmd | 8397beb | wesleycrouse | 2022-03-24 | format |
| html | 8397beb | wesleycrouse | 2022-03-24 | format |
| Rmd | 27e1022 | wesleycrouse | 2022-03-24 | layout |
| html | 27e1022 | wesleycrouse | 2022-03-24 | layout |
| Rmd | 85641ef | wesleycrouse | 2022-03-24 | layout |
| html | 85641ef | wesleycrouse | 2022-03-24 | layout |
| html | 717e77e | wesleycrouse | 2022-03-23 | adjusting heatmaps |
| Rmd | ae26765 | wesleycrouse | 2022-03-23 | plots |
| html | ae26765 | wesleycrouse | 2022-03-23 | plots |
| Rmd | aefd338 | wesleycrouse | 2022-03-23 | adjusting heatmap |
| html | aefd338 | wesleycrouse | 2022-03-23 | adjusting heatmap |
| Rmd | 64ee362 | wesleycrouse | 2022-03-23 | adjusting gene-tissue heatmap |
| html | 64ee362 | wesleycrouse | 2022-03-23 | adjusting gene-tissue heatmap |
| html | b5e392d | wesleycrouse | 2022-03-23 | tables |
| Rmd | 1b1fcaf | wesleycrouse | 2022-03-23 | chart for tissue specificity |
| Rmd | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | bbf031d | wesleycrouse | 2022-03-23 | adjusting sections |
| Rmd | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | 60f39e6 | wesleycrouse | 2022-03-22 | additional traits |
| Rmd | 073f2a3 | wesleycrouse | 2022-03-22 | enrichment analysis for all weights |
| html | 073f2a3 | wesleycrouse | 2022-03-22 | enrichment analysis for all weights |
| Rmd | ba908fe | wesleycrouse | 2022-03-21 | more traits for all weight analysis |
| Rmd | 42bb4a7 | wesleycrouse | 2022-03-18 | adjusting plot |
| html | 42bb4a7 | wesleycrouse | 2022-03-18 | adjusting plot |
| html | 3643de3 | wesleycrouse | 2022-03-18 | enrichment by tissue group |
| Rmd | 63a5188 | wesleycrouse | 2022-03-18 | adding tissue groups |
| Rmd | 09644c4 | wesleycrouse | 2022-03-18 | format |
| html | 09644c4 | wesleycrouse | 2022-03-18 | format |
| Rmd | 1cce45c | wesleycrouse | 2022-03-18 | more formatting |
| html | 1cce45c | wesleycrouse | 2022-03-18 | more formatting |
| Rmd | c96b149 | wesleycrouse | 2022-03-18 | formatting height report |
| html | c96b149 | wesleycrouse | 2022-03-18 | formatting height report |
| Rmd | d0b131e | wesleycrouse | 2022-03-18 | cleaning up height analysis |
| html | d0b131e | wesleycrouse | 2022-03-18 | cleaning up height analysis |
| Rmd | ecfacf7 | wesleycrouse | 2022-03-03 | tissue-specific enrichment analysis |
| html | ecfacf7 | wesleycrouse | 2022-03-03 | tissue-specific enrichment analysis |
| html | 380982d | wesleycrouse | 2022-03-01 | fixing typo in all weight reports |
| html | 76fa2cd | wesleycrouse | 2022-03-01 | cleaning up all weight reports |
| html | 2509c32 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
| Rmd | 962fd16 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
| Rmd | b4f3106 | wesleycrouse | 2022-03-01 | cleaning up height report |
| html | b4f3106 | wesleycrouse | 2022-03-01 | cleaning up height report |
| Rmd | ddcbd9b | wesleycrouse | 2022-02-28 | all weight analysis |
| html | ddcbd9b | wesleycrouse | 2022-02-28 | all weight analysis |
| Rmd | c73888b | wesleycrouse | 2022-02-28 | adding all weight analysis for height |
| html | c73888b | wesleycrouse | 2022-02-28 | adding all weight analysis for height |
options(width=1000)trait_id <- "ukb-a-389"
trait_name <- "Standing height"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)
results_dirs <- results_dirs[-grep("All",results_dirs)]Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions and effects
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
# ctwas_res$region_cs_tag <- paste(ctwas_res$region_tag, ctwas_res$cs_index, sep="_")
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes_index <- ctwas_gene_res$susie_pip>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_genes_index]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#
# twas_genes_index <- abs(ctwas_gene_res$z) > sig_thresh
# twas_genes <- ctwas_gene_res$genename[twas_genes_index]
#
# #gene PIPs and z scores
# gene_pips <- ctwas_gene_res[,c("genename", "region_tag", "susie_pip", "z", "region_cs_tag")]
#
# #total PIPs by region
# regions <- unique(ctwas_gene_res$region_tag)
# region_pips <- data.frame(region=regions, stringsAsFactors=F)
# region_pips$gene_pip <- sapply(regions, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_tag==x])})
# region_pips$snp_pip <- sapply(regions, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_tag==x])})
# region_pips$snp_maxz <- sapply(regions, function(x){max(abs(ctwas_snp_res$z[ctwas_snp_res$region_tag==x]))})
#
# #total PIPs by causal set
# regions_cs <- unique(ctwas_gene_res$region_cs_tag)
# region_cs_pips <- data.frame(region_cs=regions_cs, stringsAsFactors=F)
# region_cs_pips$gene_pip <- sapply(regions_cs, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_cs_tag==x])})
# region_cs_pips$snp_pip <- sapply(regions_cs, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_cs_tag==x])})
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes,
# gene_pips=gene_pips,
# region_pips=region_pips,
# sig_thresh=sig_thresh,
# region_cs_pips=region_cs_pips)
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL,
stringsAsFactors=F)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)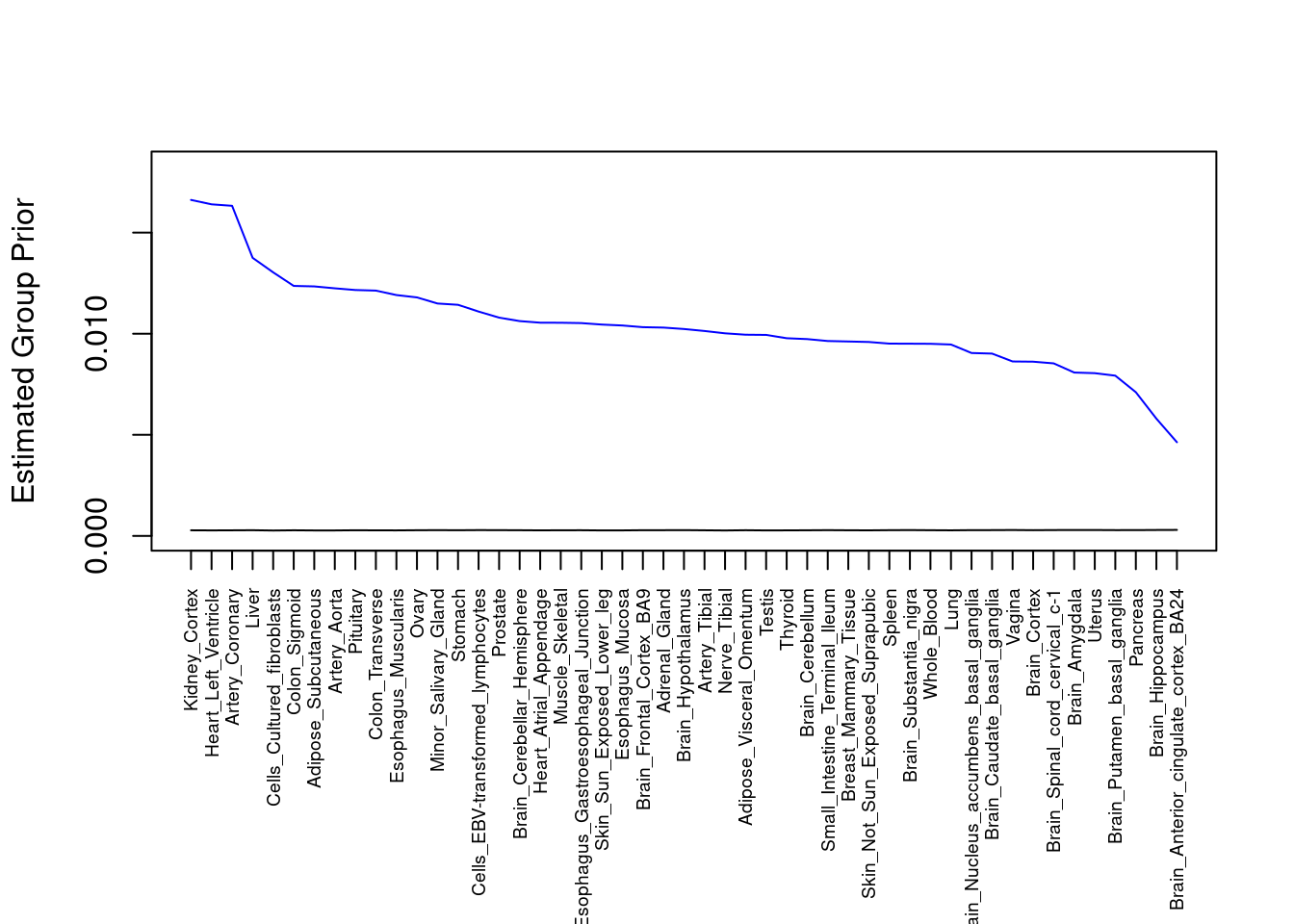
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)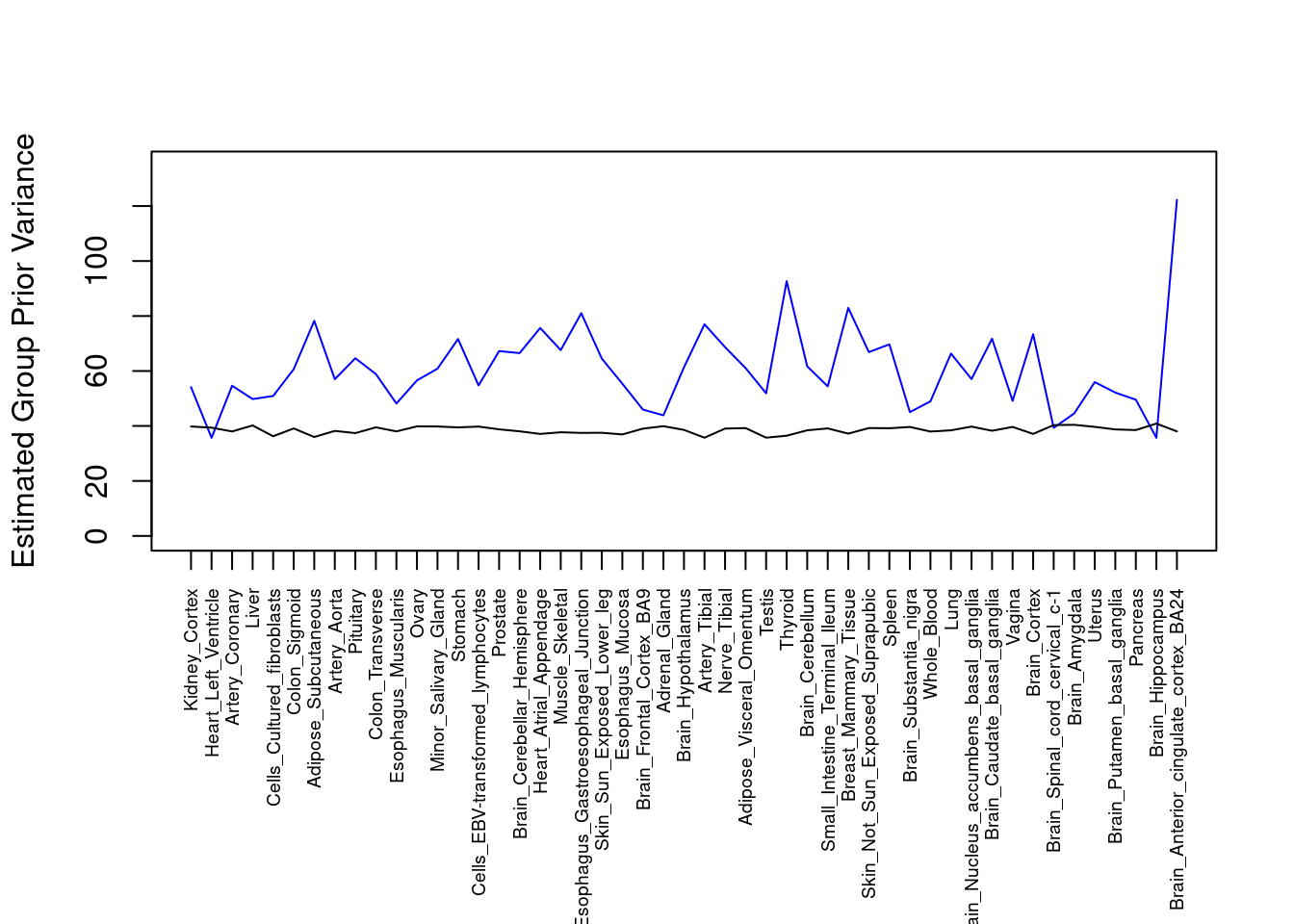
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)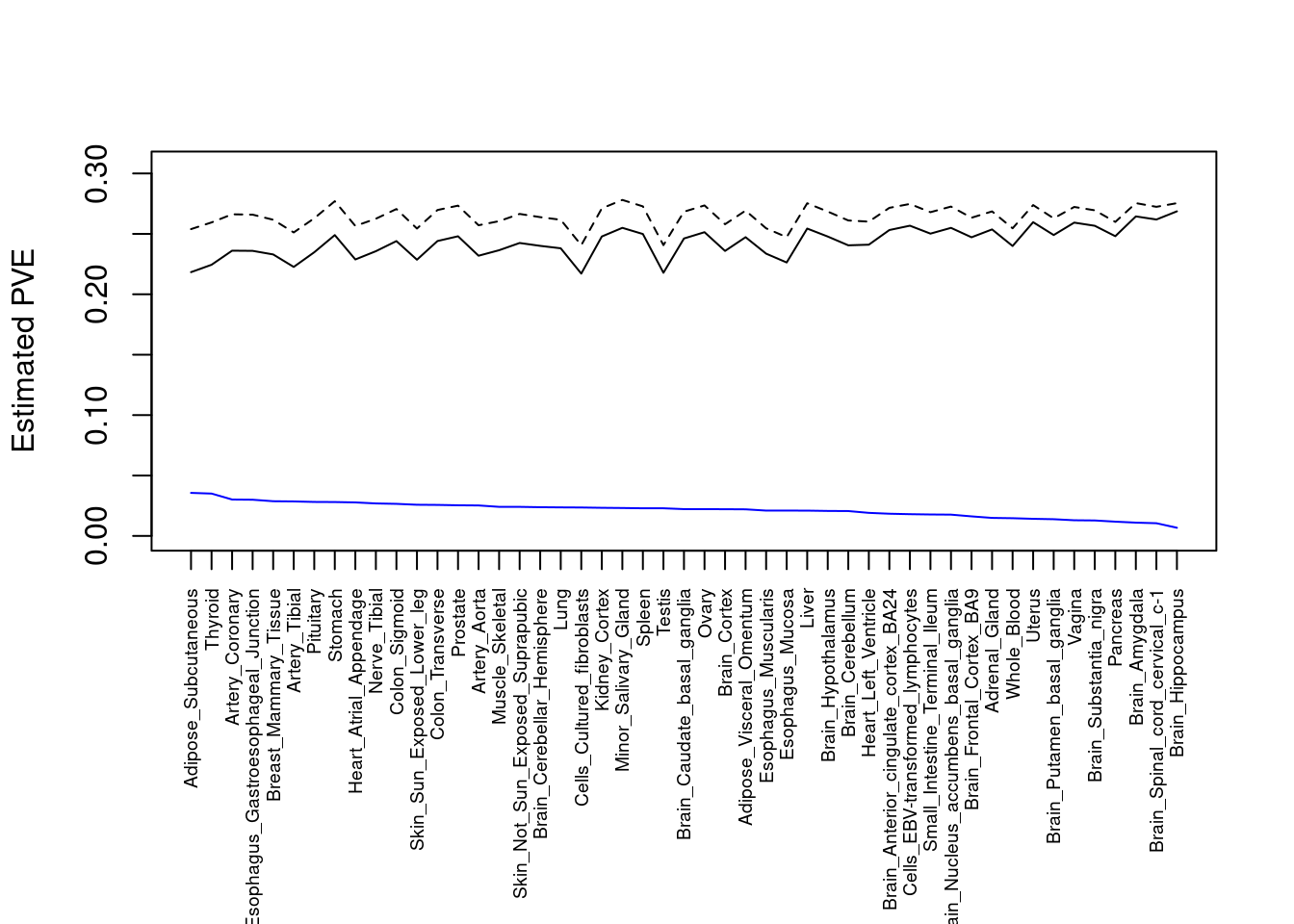
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")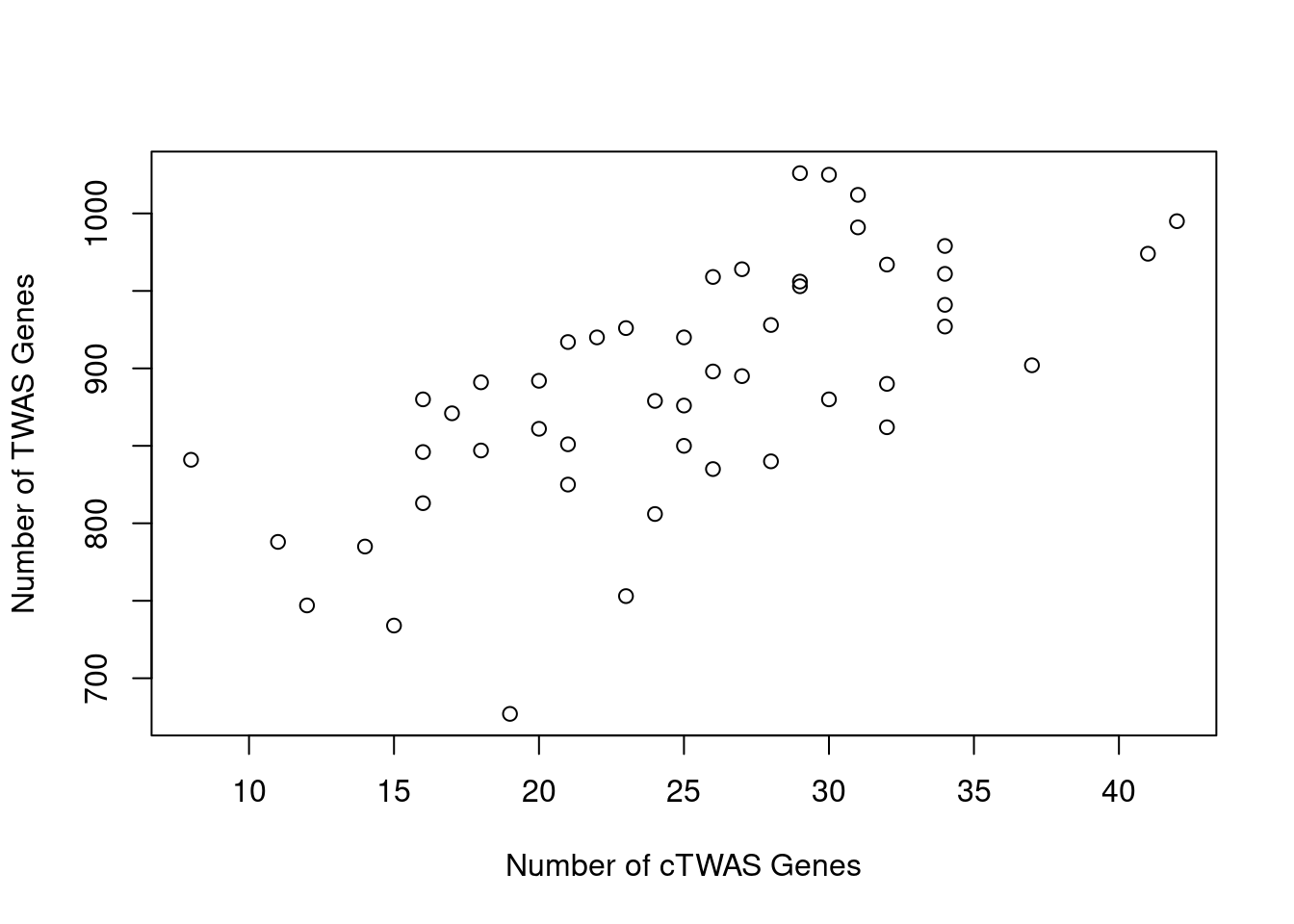
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 319#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 3781Enrichment analysis for cTWAS genes
GO
#enrichment for cTWAS genes using enrichR
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Cellular_Component_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Molecular_Function_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
KEGG
#enrichment for cTWAS genes using KEGG
library(WebGestaltR)******************************************* ** Welcome to WebGestaltR ! ** *******************************************background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#listGeneSet()
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!enrichResult[,c("description", "size", "overlap", "FDR", "userId")]NULLDisGeNET
#enrichment for cTWAS genes using DisGeNET
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <- disease_enrichment(entities=ctwas_genes, vocabulary = "HGNC", database = "CURATED")RP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDMUCL1 gene(s) from the input list not found in DisGeNET CURATEDZNF3 gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDHOOK2 gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDZBTB38 gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDZYG11B gene(s) from the input list not found in DisGeNET CURATEDSHE gene(s) from the input list not found in DisGeNET CURATEDBET1L gene(s) from the input list not found in DisGeNET CURATEDRGP1 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDNATD1 gene(s) from the input list not found in DisGeNET CURATEDNUPR2 gene(s) from the input list not found in DisGeNET CURATEDRP4-798A10.4 gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDRP11-140I16.3 gene(s) from the input list not found in DisGeNET CURATEDMETTL21B gene(s) from the input list not found in DisGeNET CURATEDSPCS2 gene(s) from the input list not found in DisGeNET CURATEDCELF6 gene(s) from the input list not found in DisGeNET CURATEDDCST2 gene(s) from the input list not found in DisGeNET CURATEDDYRK4 gene(s) from the input list not found in DisGeNET CURATEDC3orf18 gene(s) from the input list not found in DisGeNET CURATEDPARPBP gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDEFCAB8 gene(s) from the input list not found in DisGeNET CURATEDRFTN1 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDC14orf80 gene(s) from the input list not found in DisGeNET CURATEDSAV1 gene(s) from the input list not found in DisGeNET CURATEDC17orf74 gene(s) from the input list not found in DisGeNET CURATEDC17orf82 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDSLC2A12 gene(s) from the input list not found in DisGeNET CURATEDMETTL8 gene(s) from the input list not found in DisGeNET CURATEDC2orf49 gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDPLCD4 gene(s) from the input list not found in DisGeNET CURATEDTRAPPC2B gene(s) from the input list not found in DisGeNET CURATEDLPCAT1 gene(s) from the input list not found in DisGeNET CURATEDTMCC1 gene(s) from the input list not found in DisGeNET CURATEDRP5-1085F17.3 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDSYTL2 gene(s) from the input list not found in DisGeNET CURATEDSH3D21 gene(s) from the input list not found in DisGeNET CURATEDRNF181 gene(s) from the input list not found in DisGeNET CURATEDC2orf40 gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDRAD18 gene(s) from the input list not found in DisGeNET CURATEDCOPZ2 gene(s) from the input list not found in DisGeNET CURATEDFAM180B gene(s) from the input list not found in DisGeNET CURATEDNGDN gene(s) from the input list not found in DisGeNET CURATEDSPAG8 gene(s) from the input list not found in DisGeNET CURATEDUBE2Q1 gene(s) from the input list not found in DisGeNET CURATEDJMJD4 gene(s) from the input list not found in DisGeNET CURATEDZNF219 gene(s) from the input list not found in DisGeNET CURATEDTC2N gene(s) from the input list not found in DisGeNET CURATEDTM2D1 gene(s) from the input list not found in DisGeNET CURATEDRP11-1277A3.3 gene(s) from the input list not found in DisGeNET CURATEDPSRC1 gene(s) from the input list not found in DisGeNET CURATEDEZH1 gene(s) from the input list not found in DisGeNET CURATEDOVAAL gene(s) from the input list not found in DisGeNET CURATEDTULP4 gene(s) from the input list not found in DisGeNET CURATEDUGGT2 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDMRPS9 gene(s) from the input list not found in DisGeNET CURATEDUNC50 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDMIR193BHG gene(s) from the input list not found in DisGeNET CURATEDC1QTNF4 gene(s) from the input list not found in DisGeNET CURATEDRP11-78O7.2 gene(s) from the input list not found in DisGeNET CURATEDSPRYD3 gene(s) from the input list not found in DisGeNET CURATEDARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDUPK1A gene(s) from the input list not found in DisGeNET CURATEDGRIN3B gene(s) from the input list not found in DisGeNET CURATEDEIF4EBP3 gene(s) from the input list not found in DisGeNET CURATEDCCDC127 gene(s) from the input list not found in DisGeNET CURATEDUSP37 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATEDFAM208A gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDSLC45A4 gene(s) from the input list not found in DisGeNET CURATEDAL132709.1 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDFOXN2 gene(s) from the input list not found in DisGeNET CURATEDSYCE2 gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDRNF167 gene(s) from the input list not found in DisGeNET CURATEDPRRT3 gene(s) from the input list not found in DisGeNET CURATEDCCL4L2 gene(s) from the input list not found in DisGeNET CURATEDATP5G1 gene(s) from the input list not found in DisGeNET CURATEDLINC01124 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDTATDN2 gene(s) from the input list not found in DisGeNET CURATEDTBC1D3H gene(s) from the input list not found in DisGeNET CURATEDUSP49 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDTPCN2 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDCTD-3224K15.3 gene(s) from the input list not found in DisGeNET CURATEDZNF827 gene(s) from the input list not found in DisGeNET CURATEDRFX3-AS1 gene(s) from the input list not found in DisGeNET CURATEDTRIM34 gene(s) from the input list not found in DisGeNET CURATEDPAGR1 gene(s) from the input list not found in DisGeNET CURATEDAPOBEC3G gene(s) from the input list not found in DisGeNET CURATEDSYNGR2 gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDTMEM150A gene(s) from the input list not found in DisGeNET CURATEDCNOT9 gene(s) from the input list not found in DisGeNET CURATEDARRDC2 gene(s) from the input list not found in DisGeNET CURATEDAF067845.3 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDPRSS27 gene(s) from the input list not found in DisGeNET CURATEDSLC16A9 gene(s) from the input list not found in DisGeNET CURATEDDDX42 gene(s) from the input list not found in DisGeNET CURATEDTMEM129 gene(s) from the input list not found in DisGeNET CURATEDCFAP54 gene(s) from the input list not found in DisGeNET CURATEDC1QTNF1 gene(s) from the input list not found in DisGeNET CURATEDCCDC187 gene(s) from the input list not found in DisGeNET CURATEDKIAA1644 gene(s) from the input list not found in DisGeNET CURATEDUBE2Z gene(s) from the input list not found in DisGeNET CURATEDRP11-509I21.2 gene(s) from the input list not found in DisGeNET CURATEDRNF169 gene(s) from the input list not found in DisGeNET CURATEDHEMK1 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDTHUMPD1 gene(s) from the input list not found in DisGeNET CURATEDRP11-503L19.1 gene(s) from the input list not found in DisGeNET CURATEDPODN gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATEDPLCD3 gene(s) from the input list not found in DisGeNET CURATEDNFE2L1 gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDPSMA1 gene(s) from the input list not found in DisGeNET CURATEDIL17RE gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDLINC01001 gene(s) from the input list not found in DisGeNET CURATEDCRIP1 gene(s) from the input list not found in DisGeNET CURATEDAPOLD1 gene(s) from the input list not found in DisGeNET CURATEDNEU3 gene(s) from the input list not found in DisGeNET CURATEDCCDC169 gene(s) from the input list not found in DisGeNET CURATEDZNF213 gene(s) from the input list not found in DisGeNET CURATEDPAPD4 gene(s) from the input list not found in DisGeNET CURATEDFAM129A gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDCCDC57 gene(s) from the input list not found in DisGeNET CURATEDSMIM24 gene(s) from the input list not found in DisGeNET CURATEDif (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes=ctwas_genes)
#background is union of genes analyzed in all tissue
background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background]})
####################
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
hyp_score_padj gene_set nset ngenes percent padj
1 gwascatalog 5959 141 0.02366169 4.799366e-10
2 mgi_essential 2299 54 0.02348847 9.283940e-04
3 clinvar_path_likelypath 2766 58 0.02096891 5.576615e-03
4 fda_approved_drug_targets 350 7 0.02000000 3.235696e-01
5 core_essentials_hart 265 4 0.01509434 5.498685e-01Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Cellular_Component_2021
Term Overlap Adjusted.P.value
1 MHC protein complex (GO:0042611) 18/20 5.229887e-09
2 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556) 19/28 2.443073e-06
3 lumenal side of endoplasmic reticulum membrane (GO:0098553) 19/28 2.443073e-06
4 MHC class II protein complex (GO:0042613) 12/13 2.443073e-06
5 ER to Golgi transport vesicle membrane (GO:0012507) 24/54 1.340537e-03
6 coated vesicle membrane (GO:0030662) 23/55 5.414556e-03
7 COPII-coated ER to Golgi transport vesicle (GO:0030134) 29/79 9.674325e-03
8 lysosome (GO:0005764) 119/477 3.156488e-02
9 endocytic vesicle membrane (GO:0030666) 47/158 3.156488e-02
10 lytic vacuole membrane (GO:0098852) 71/267 4.906135e-02
11 MHC class I protein complex (GO:0042612) 5/6 4.906135e-02
12 nucleus (GO:0005634) 918/4484 4.930597e-02
Genes
1 HLA-DRB5;HFE;HLA-B;HLA-C;HLA-A;HLA-F;HLA-E;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
2 HLA-DRB5;SPPL2C;SPPL2B;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;CALR;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
3 HLA-DRB5;SPPL2C;SPPL2B;HLA-B;HLA-C;HLA-A;HLA-F;SPPL3;HLA-G;HLA-E;HLA-DPB1;HLA-DRA;CALR;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
4 HLA-DRB5;HLA-DMA;HLA-DMB;HLA-DPB1;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1
5 HLA-DRB5;PEF1;SAR1B;PDCD6;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;HLA-DPB1;TMED2;HLA-DRA;TMED7;CNIH2;HLA-DQA2;CNIH3;HLA-DQB2;HLA-DQA1;SEC31B;HLA-DRB1;HLA-DPA1;HLA-DQB1
6 HLA-DRB5;SAR1B;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;TMEM199;AP1G1;HLA-DPB1;TMED2;HLA-DRA;CNIH2;HLA-DQA2;CNIH3;HLA-DQB2;HLA-DQA1;SEC31B;HLA-DRB1;HLA-DPA1;HLA-DQB1
7 SAR1B;CTSZ;LMAN2;TMED2;TMED7;TMED6;HLA-DQA2;HLA-DQA1;HLA-DPA1;HLA-DRB5;TICAM2;SEC16A;HLA-B;HLA-C;HLA-A;HLA-F;HLA-G;HLA-E;SEC23IP;HLA-DPB1;HLA-DRA;ERGIC3;CNIH2;ERGIC2;CNIH3;HLA-DQB2;CNIH4;HLA-DRB1;HLA-DQB1
8 SPPL2C;SPPL2B;TMEM97;LAPTM4A;AP1S1;SLC39A8;HLA-DOA;HLA-DOB;NSF;CUBN;USP4;SLC30A2;VPS33A;PRKCD;OMD;SLC11A2;EPDR1;TSC2;HLA-F;AP5B1;PLA2G15;WDR81;ATP6V0D1;SEH1L;KCNE2;ABCB6;PSEN2;ABCB9;GNS;NEU3;MYO6;NEU1;ATP6V0A2;SLC38A9;HLA-DQA2;HLA-DQA1;AP1M2;ARSB;ATP6V0A1;CD164;HLA-DRB5;CYBRD1;CP;VASN;GFAP;MFHAS1;DAB2;TCN2;GNB2;OGN;PLEKHM1;HLA-DRA;SDC1;MAN2B1;PPT2;HLA-DRB1;TINAGL1;SLC44A2;CTSZ;ECE1;CTSW;TCIRG1;ABCC10;AP1G1;FLOT1;CTSH;ACP2;CTSF;UBXN6;HLA-DPA1;SPHK2;AP3B1;SLC39A14;TMEM175;VAMP8;NPC1;NPC2;TMBIM1;COL6A1;TLR9;TMEM106B;VAMP4;RAB7B;ATP6V0C;HLA-DQB2;HLA-DQB1;PCYOX1;RAMP2;LRP1;SRC;VPS4A;WDR24;TMEM165;ACAN;CLN3;HLA-DMA;HLA-DMB;DRAM1;CXCR2;GPC5;FNIP1;LDLR;SLC17A2;SLC17A3;SLC17A4;SLC12A4;AP3D1;MTOR;TPCN2;CLCN7;GALNS;GLB1;HLA-DPB1;OCIAD1;GOPC;ATP13A2;OCIAD2;AGRN;CD68
9 LRP1;CAMK2A;AP2A1;TCIRG1;WNT6;ATP6V0A2;APOE;HLA-DQA2;KIAA0319;LDLR;HLA-DQA1;WNT3;WLS;ATP6V0A1;WNT4;HLA-DPA1;HLA-DRB5;WNT3A;NOS3;PTCH1;WNT5A;HLA-B;TAP2;HLA-C;TAP1;WNT7A;CYBA;HLA-A;HLA-F;HLA-G;IGF2R;HLA-E;DNM2;VAMP8;RAB35;NOSTRIN;HLA-DPB1;HLA-DRA;CALR;ATP6V0D1;VAMP4;ATP6V0C;HLA-DRB1;HLA-DQB2;VAMP2;HLA-DQB1;VAMP3
10 SLC44A2;SPPL2C;SPPL2B;ECE1;TCIRG1;ABCC10;LAPTM4A;AP1G1;FLOT1;AP1S1;SLC39A8;HLA-DOA;HLA-DOB;UBXN6;HLA-DPA1;NSF;SPHK2;SLC30A2;VPS33A;SLC11A2;AP3B1;HLA-F;AP5B1;SLC39A14;TMEM175;VAMP8;WDR81;NPC1;COL6A1;TMBIM1;TMEM106B;ATP6V0D1;ATP6V0C;HLA-DQB2;HLA-DQB1;SEH1L;ABCB6;LRP1;PSEN2;ABCB9;WDR24;TMEM165;NEU3;CLN3;HLA-DMA;HLA-DMB;MYO6;ATP6V0A2;SLC38A9;FNIP1;HLA-DQA2;HLA-DQA1;AP1M2;ATP6V0A1;HLA-DRB5;SLC12A4;AP3D1;CYBRD1;CP;VASN;MTOR;TPCN2;CLCN7;DAB2;GNB2;HLA-DPB1;HLA-DRA;GOPC;ATP13A2;CD68;HLA-DRB1
11 HFE;HLA-B;HLA-C;HLA-A;HLA-E
12 ATF1;MDC1;JRK;SPI1;EHMT2;TESK2;PSMD9;ALKBH5;AKT1;PRKACA;HES7;DICER1;ZNF14;DDX39B;DDIT3;CLNS1A;PSME4;PADI2;ATF7;CLOCK;SF1;SLBP;URI1;SHMT1;FLII;AGAP2;NCAPG;IQGAP1;MYO6;ZBTB7A;HEXIM1;CTC1;HEXIM2;HACE1;BTBD18;TIPARP;PBX2;PBX4;SNF8;FLI1;NR2F6;CS;MOV10;EIF6;TDP2;PSMG1;EZH2;ZNF493;ZNF44;CTCF;PRKAG3;MYPN;MED13;CHAF1A;HEY2;NELFE;ZNF484;SS18;ZNF483;DFFA;DUSP3;RING1;RIPK3;SPHK2;SIRT6;DYNLL1;POU5F1;SIRT3;DDB2;MRPL44;PSMA6;PPARG;ERGIC2;PPARD;ZNF471;ZNF470;ZNF518B;DLST;ZNF518A;HTT;CENPB;FOXS1;EXOSC10;PSMB3;SAMD1;MEF2D;SPDEF;ZNF462;TRMT10A;SLF2;YLPM1;GATAD2A;SKI;PSMC5;ZNF71;PSMC3;GID8;ZNF219;CRLF3;CENPP;ZNF696;FRK;MYLK2;MTCH2;PDCD6;CSE1L;NAB2;MAP3K7CL;RORA;CASC3;RUVBL2;SCMH1;KPNA6;PABPC4L;KPNA5;SLC25A42;KPNA2;PITX1;ZNF443;ZNF442;PIAS4;BNIP3L;IK;KHNYN;FBXW7;KMT5C;ZBTB38;UBE2E1;UBE2E2;TC2N;CDC25A;MAPKAPK3;MSH3;RAD54L2;KPNB1;BLM;MAX;ARL3;FOXO6;FOXO3;GLIS1;FOXO1;GLIS2;SAV1;STK31;EIF1AD;ATXN3;STK36;AMDHD2;ZKSCAN3;ZKSCAN5;SMYD4;IP6K3;IP6K2;ZNF420;TIA1;MYPOP;RFXANK;ARPC5;EEF2;MCC;FOXN2;DRAP1;ZC3HC1;MLXIPL;STK24;ARPC2;SP3;MDM4;ZNF655;ZNF652;NFE2L1;CELF1;FHL2;FHL3;CELF6;FASLG;FOXM1;ZNF408;PPP6R3;HMG20A;NKX3-2;ZNF641;BOK;DST;RAD23A;FOXL1;MSL1;PPM1B;SLC25A19;ZNF638;ZNF878;LCOR;BIRC6;TOP1;UHRF1;MGRN1;PPM1M;ZBTB42;BARX1;CXXC5;MLLT6;PPM1G;PARD6A;CCDC85B;TP53INP2;ATXN1L;E2F1;IGF2BP1;ZNF623;IGF2BP3;BEND6;PRRX1;GZMA;DONSON;TBX3;TBX2;TRIM39;EIF3G;STUB1;PABPC1L;TRIM32;EIF4A1;CCNJ;STH;CPNE7;CCNF;NUDT5;NUDT3;UBE2L3;CCAR1;MCIDAS;SPATA24;CPNE1;FAM53C;FAM53B;FAM53A;TRIM27;BSN;CCNL1;ZFP1;SFMBT1;IMP3;SFMBT2;ACTN4;SFRP4;CSNK2B;CD2BP2;DXO;ALPK3;LAP3;TRIB1;TP53;CCNO;PXT1;L3MBTL3;ZBTB1;ZBTB4;ZNF709;MIER3;XRRA1;MIER1;DHX36;SYMPK;ZNF700;HIVEP3;APOE;ZBED6;APOBEC3C;EGR2;APOBEC3F;APOBEC3G;CRADD;APOBEC3H;XRCC3;SMARCA5;XRCC1;PTK6;NTAN1;SMARCA4;ZBTB6;TH;ID4;NDUFAF3;INTS4;GRB2;ARMC12;MKX;CALR;FBXL6;FOXE3;FOXE1;PPP1R11;PPP1R10;PTPRN;N4BP2L2;RSRC1;SUFU;UIMC1;ARIH2;DHX16;ACADS;NCOA1;ZHX2;NCOA6;SREBF2;SUPT3H;TOX4;PDIK1L;G2E3;NCOA7;PTMA;FOXC1;PCNA;DOT1L;PRICKLE4;SERTAD3;RXRA;EXOSC4;EXOSC9;HSF1;SCAND1;ZNF585B;EXOSC2;RIBC2;ZNF585A;NPM1;ERCC6L2;AKR1C3;PEX14;PPP1CA;TFAP4;PER3;TARDBP;FOXA3;ZFP28;CRTC3;PRDM4;UBXN2A;UBE3D;WWC1;FHOD1;UBE2Z;NR3C1;ACTG1;CCND3;CCND2;RGS3;CCND1;BANF1;TEKT4;PCID2;BORA;UBE4B;WBP11;POU3F3;ADAM19;SPOCD1;BRMS1;CCNE1;SBDS;HOMEZ;DDX6;UBIAD1;DDX4;PSMD13;PSEN2;PDS5B;PDS5A;NKAPL;IRAK2;CAMTA1;CYC1;PIBF1;VDR;PHF11;HMGA1;HMGA2;CLK3;ISG20;NASP;UBE2N;NABP2;TAB1;SERGEF;UBE2K;FGF11;PICALM;PHF19;RERE;DYRK4;GSK3A;DDX46;CHD8;HP1BP3;DDX42;ADAR;RERG;UBE2Q1;GATAD1;MYBL2;CNPPD1;ETV1;IRAK3;DDX51;ETV4;UTP18;ETV6;ETV7;CCNA2;ELF2;IRF3;IRF1;CRY1;FBP1;LAMA5;RRN3;DDX25;SAMHD1;PSMB10;ADD1;AURKB;ATAD2B;AURKA;CAND2;ATOH7;CLN3;TNKS2;PABPN1;TDG;TBX20;TRA2A;DROSHA;RALY;VHL;PDLIM4;ATF7IP;EYA4;MAPK14;CDK11B;MAPK15;TBX18;MAPK13;MAPK10;CCS;REST;ERCC3;SPATA33;CARM1;TAF8;QRICH1;RBM45;NUP37;RPL4;RPL5;DCAF7;ELK4;NYNRIN;HOXA9;CLP1;ZMIZ2;SOX15;CREB3L2;CDK20;PIP4K2B;DIP2B;HOXA7;CDK5RAP3;HOXA4;IER3;TSPYL1;ZNF160;PRKCD;SOX11;HOXB3;SRFBP1;TSPYL6;VPS25;HOXB7;HOXB6;TSPYL4;ZNF398;HOXB5;CTBP1;XPC;LDHA;WDR70;NTHL1;PRDX1;PTPMT1;MCMBP;NKX2-5;NKX2-4;RREB1;NHEJ1;PHC2;ZNF142;RAB3IP;NFATC3;NR1D2;UPF3A;NR1D1;INO80;FOSL2;NFATC4;FOSL1;TADA2B;FIBP;HNRNPK;TRIP6;SNAI1;CDK10;RAD18;PHF2;SETD2;CDKN1B;TCF25;PHF23;CATIP;TRIM7;SPIN1;CIC;ZNF367;TIGD3;TCF19;BRD2;TESC;TET2;TET1;GNL2;NUDT16L1;TERF2;GNL3;ITPKC;RNF169;MTF1;ITPKA;RARA;AHRR;SF3B2;WDR26;SF3B6;GPS2;NSD1;UBN1;NSD2;RPS3;RPL13;RPL18;RPL17;ZNF345;ZNF101;CDKN2D;SF3A3;HIPK4;ZNF341;TIGAR;CDKN2C;GSN;SF3A2;HSPA1L;MGMT;PCGF2;SAAL1;NR1H3;WWP2;ELP5;GTF2H4;DEF6;RYBP;NFIB;NFIC;HYLS1;ZNF576;HSPA1B;ZNF574;HSPA1A;N6AMT1;ZNF573;IRS1;GTF2B;CTNND1;UBP1;PARK7;RPL10A;CLU;FGF2;EDC4;HMGN4;MAEA;SALL2;TUBB3;CHP1;OIP5;ZNF568;MAF1;MEN1;HMGN1;CSNK1G3;TLE3;USP7;MAP2K2;MAMSTR;USP4;USP3;IGFBP3;PGD;RNF40;CLIP1;ILF3;KIAA1614;NCL;ERF;SNRPG;TSSK6;ZNF799;SNRPE;ZNF311;IPPK;DNMT1;TNKS;ANKRD11;CUL3;NOTCH4;PHB;AK9;BAG6;BAG5;NACC1;DVL2;SAFB2;ZNF789;ZNF304;FANCA;FANCC;FANCE;LSM5;LSM3;COPRS;CDK6;CDK4;CNOT2;MAFF;BCL2;NF1;MAFK;ZNF774;RAD9A;CALCOCO2;SYNE2;PAGR1;SART1;SIN3A;DNMT3B;WDR5;TLK2;TLK1;HIF1AN;TNPO1;ZNF521;RFC5;RFC1;DNMT3A;SMC1B;PTBP3;PTP4A2;NPEPL1;PKM;ESPL1;CTDSP1;ESRP2;ZNF514;ZNF513;TESMIN;LCORL;NSRP1;SMARCD1;NFAT5;SMARCD2;VPS4A;TULP3;TTK;PDHB;DPCD;DNAJB2;NFS1;HIRIP3;POLR2A;ZNF507;POLR2B;NOCT;RSBN1L;RBBP8;ASCC2;STOX1;JAZF1;MLXIP;SPAG8;RNF25;PTPN12;PTPN13;PTPN14;MTOR;PML;KLF1;APC;ZGLP1;INSM1;SYCP3;PFKM;TRMT61A;CCDC62;GFI1;HDAC11;NFU1;ZNF608;ZNF606;HABP4;DPF2;ZNF844;ARL6IP4;KAT8;SOX8;SKP2;ABT1;TIPIN;KRT4;METTL1;NFKBIL1;METTL3;SLC11A2;OVOL1;JMJD8;RUNX2;INPP4A;XRN1;KATNAL2;XRN2;DNA2;NEIL1;TELO2;TEX264;DCUN1D4;TWIST2;C2CD4A;TWIST1;TXN;RPAP2;GMCL1;DGUOK;ZNF827;INPP5E;CDT1;ZRANB1;INSR;PDE2A;MANF;GADD45G;PARP10;TPX2;STAG1;FUBP3;RNPS1;DET1;FEN1;FAF1;CITED4;SRA1;RSF1;MEOX2;LIMD1;HINT1;XPO1;HINT3;CLIC1;IZUMO4;STRADA;TUBB;TOP3A;PROX2;CBFA2T2;CBFA2T3;MLF1;MLF2;UGDH;RAD51C;WAC;HNRNPH3;MAPT;UMPS;DNASE1L2;DTYMK;NOL9;AIF1;ARNTL;TLDC2;GPBP1L1;DCPS;SUZ12;RYK;CDX1;XPO7;GRHL1;MLH1;CTNNB1;MEGF8;ZNF354C;MPHOSPH6;KDM1A;PRAG1;DSCC1;HDLBP;MYRFL;NUCKS1;ARID4B;CWC25;EFTUD2;SCGN;CHCHD2;MEIOC;KNTC1;LMO1;TRAPPC2L;TOMM34;TSC2;NOSIP;TRAPPC2B;KCTD1;LGALS9C;MAU2;FANCD2;CMPK1;CHMP4B;MATR3;NUPR2;FIZ1;ZCCHC8;STK4;ZMAT2;SOHLH2;BPTF;CBX7;CBX6;HELQ;TGFB1;SMAD3;SMAD9;EIF2S2;POU2F2;SMAD6;ESR1;ESR2;SMAD7;DIAPH1;FER;KIF18B;BLMH;BRWD1;FGFR2;USP36;USP37;NVL;USP34;SIX1;USP39;NR2E3;PKD1;SENP6;SNAPC5;SDR16C5;SIX6;SIX4;SIX5;PPP2R5E;TRPS1;SNAPC4;POLL;KLF11;USP47;DCXR;AKTIP;AXIN1;PPP2R5A;PPP2R5D;PPP2R5C;AXIN2;ARID1A;SENP2;SENP3;TGFBR1;SENP1;DDX19B;LATS2;KDM7A;POLDIP2;RGS14;ATF6B;USP10;LEF1;CAMK2A;MAPK9;MCRIP2;FAM120A;MAPK1;GBP4;MAPK3;KDM4B;STN1;KDM4C;NOS3;ARID3A;ARID3B;TICRR;KANSL1;RPS6KB2;CDK2AP1;RGS12;SSBP4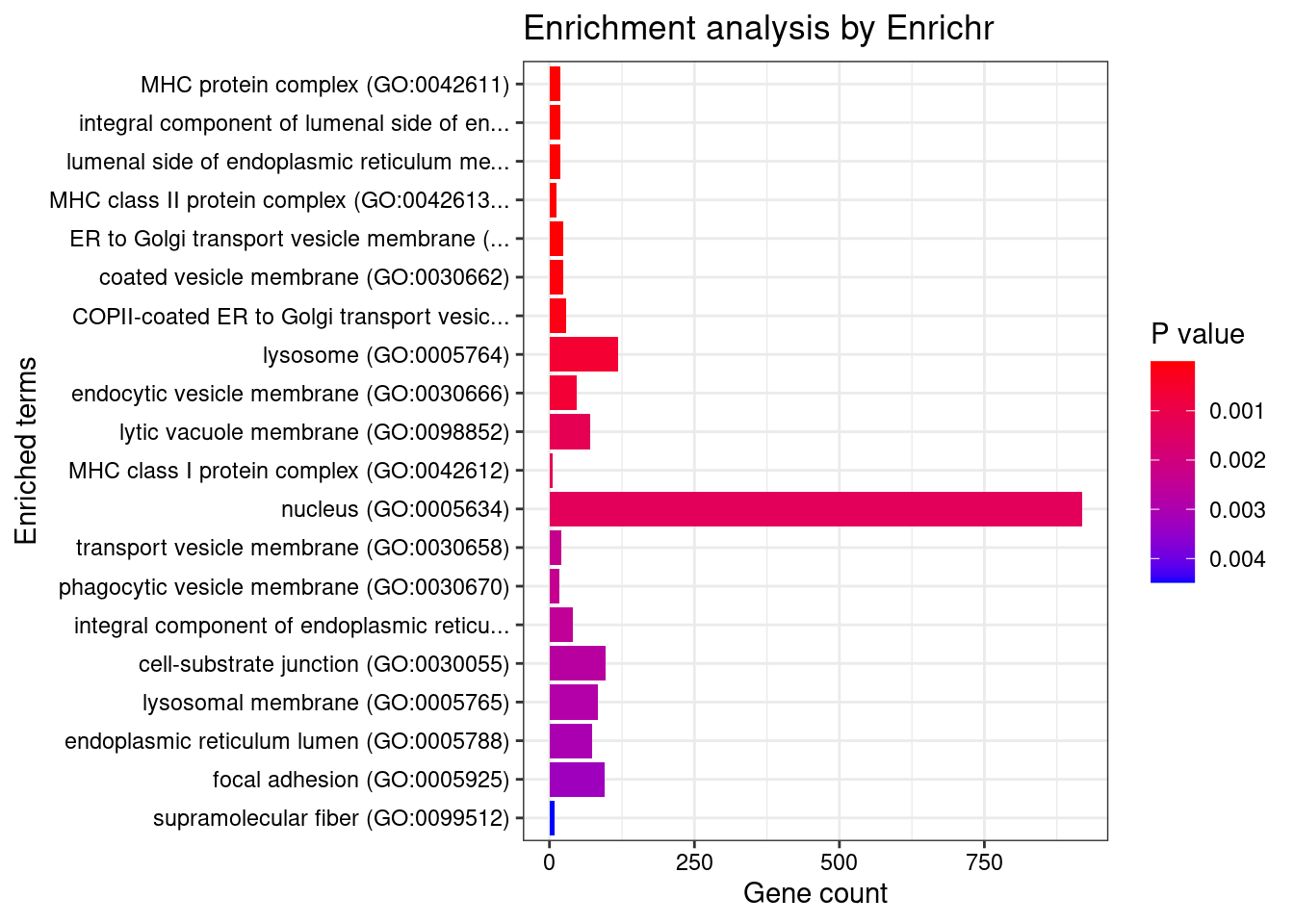
GO_Molecular_Function_2021
Term Overlap Adjusted.P.value Genes
1 MHC class II receptor activity (GO:0032395) 9/10 0.002759325 HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1;HLA-DPA1;HLA-DQB1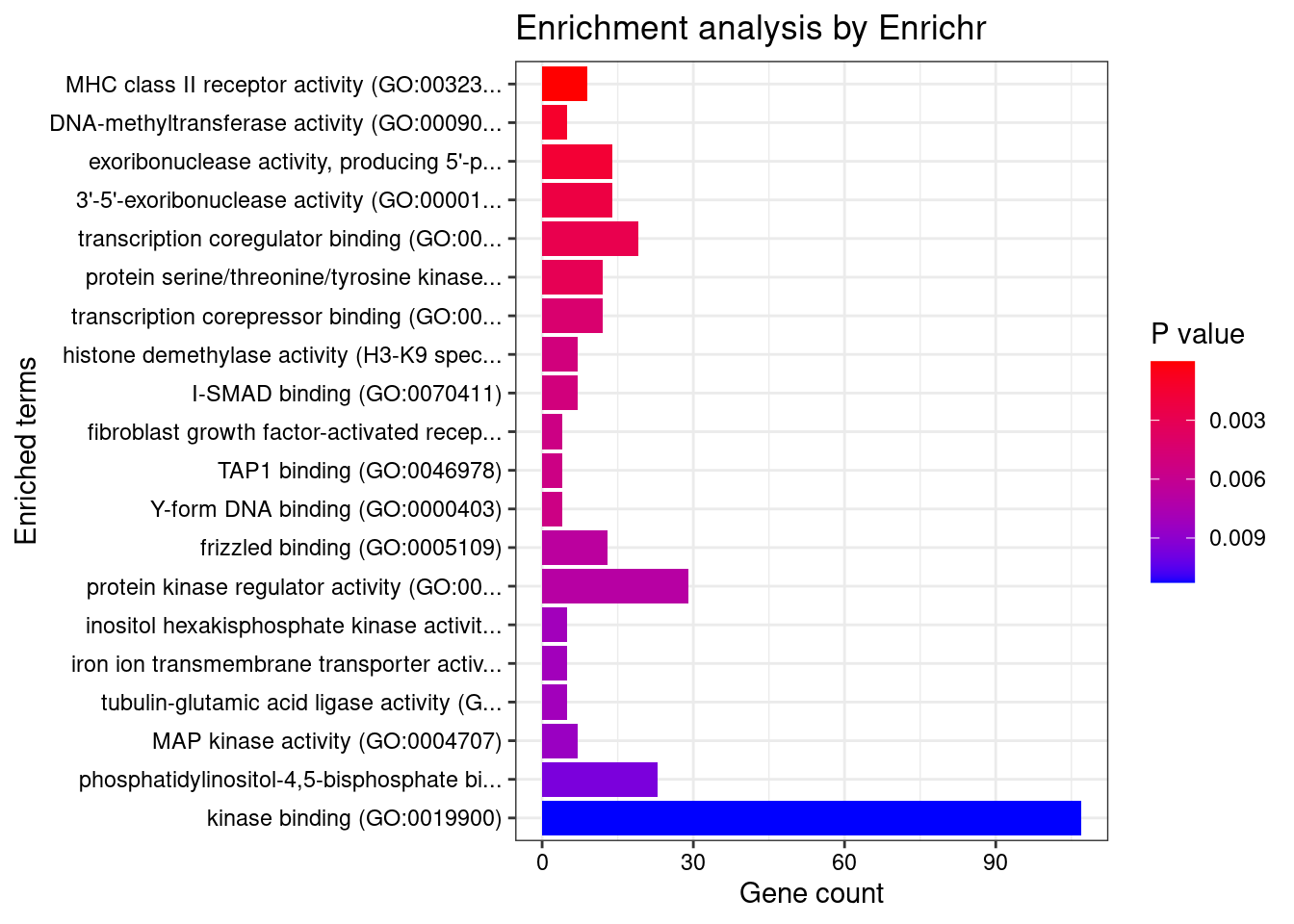
Enrichment analysis for cTWAS genes in top tissues separately
GO
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 41
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Thyroid
Number of cTWAS Genes in Tissue: 31
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Artery_Coronary
Number of cTWAS Genes in Tissue: 37
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 response to vitamin D (GO:0033280) 2/9 0.03959705 CYP27B1;SFRP1
2 response to organic cyclic compound (GO:0014070) 3/60 0.03959705 CYP27B1;SFRP1;ABCB4
3 ceramide transport (GO:0035627) 2/14 0.04253236 ABCB4;PLEKHA3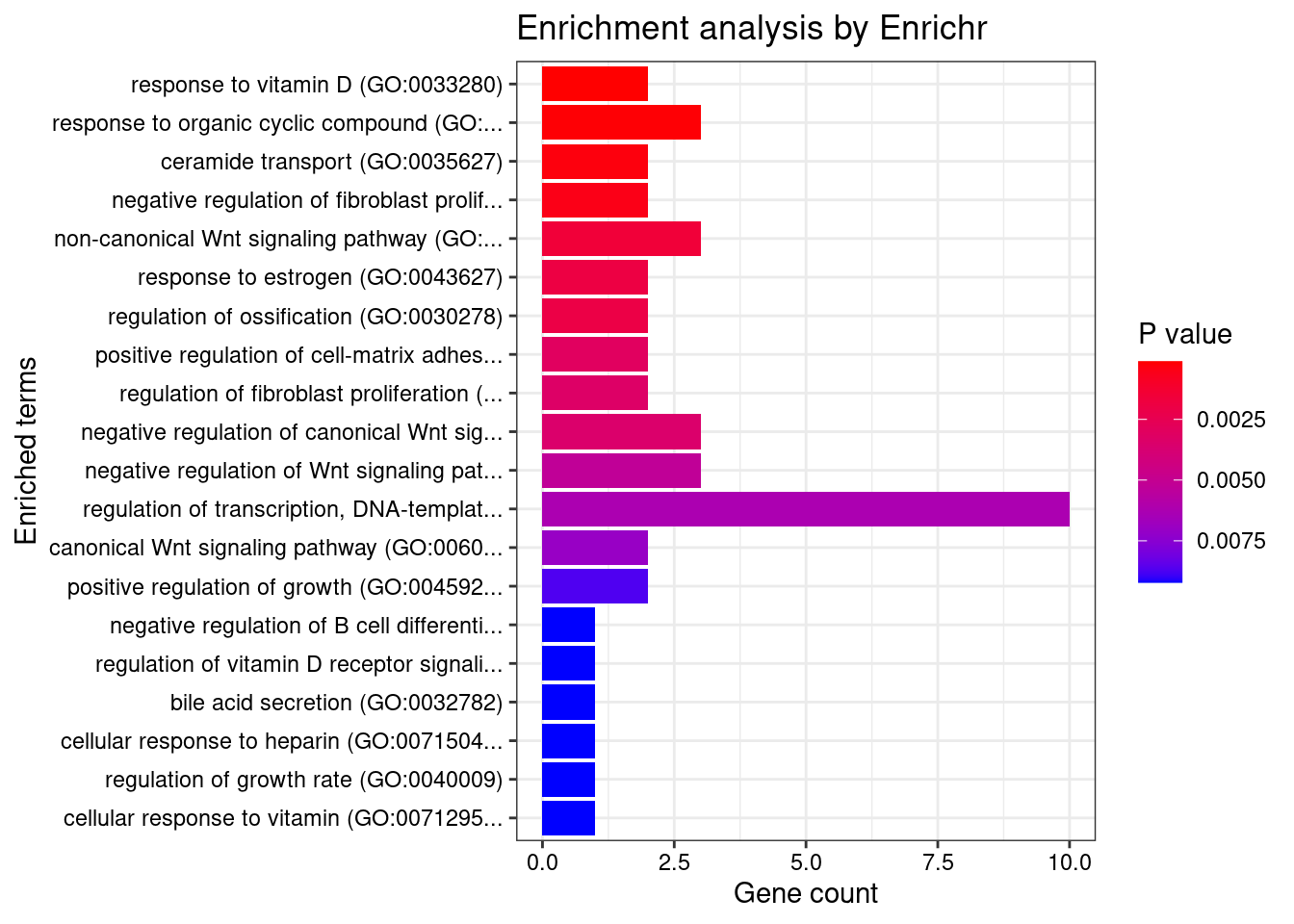
Esophagus_Gastroesophageal_Junction
Number of cTWAS Genes in Tissue: 34
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Breast_Mammary_Tissue
Number of cTWAS Genes in Tissue: 23
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of pri-miRNA transcription by RNA polymerase II (GO:1902893) 3/45 0.005662126 REST;SMAD3;NFATC4
2 negative regulation of alcohol biosynthetic process (GO:1902931) 2/13 0.011795821 CYP27B1;REST
3 negative regulation of pri-miRNA transcription by RNA polymerase II (GO:1902894) 2/14 0.011795821 REST;NFATC4
4 positive regulation of cysteine-type endopeptidase activity involved in apoptotic process (GO:0043280) 3/119 0.025841403 REST;DAP;SMAD3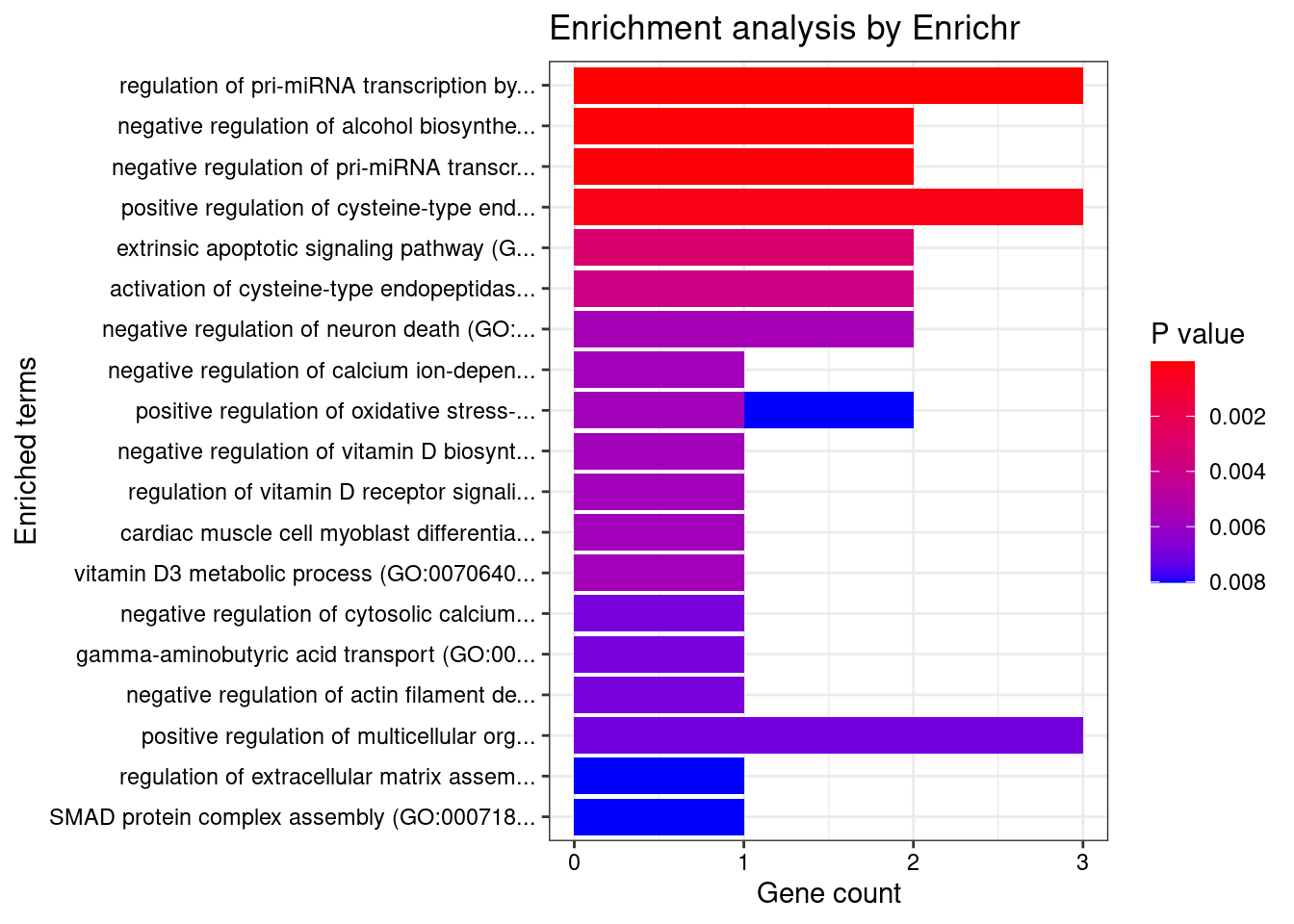
KEGG
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- NULL
try(enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_tissue, referenceGene=background_tissue,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F))
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
} Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 41
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Thyroid
Number of cTWAS Genes in Tissue: 31
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Artery_Coronary
Number of cTWAS Genes in Tissue: 37
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...
description size overlap FDR userId
1 Wnt signaling pathway 75 4 0.02038269 DVL3;SFRP1;CCND2;NFATC4
Esophagus_Gastroesophageal_Junction
Number of cTWAS Genes in Tissue: 34
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...
description size overlap FDR userId
1 Arginine and proline metabolism 32 3 0.0307298 P4HA2;CKB;SMOX
Breast_Mammary_Tissue
Number of cTWAS Genes in Tissue: 23
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_tissue, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
} Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 41ARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDSLC16A9 gene(s) from the input list not found in DisGeNET CURATEDRP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDHOOK2 gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDC2orf40 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDCCL4L2 gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
71 Rickets 0.006143348 2/21 4/9703
5 Sertoli-Leydig cell tumor of ovary 0.014642221 1/21 1/9703
27 Gynandroblastoma 0.014642221 1/21 1/9703
64 Noonan Syndrome 0.014642221 2/21 24/9703
74 Sertoli Cell Tumor 0.014642221 1/21 1/9703
95 Sertoli-Leydig Cell Tumor 0.014642221 1/21 1/9703
98 Hereditary orotic aciduria 0.014642221 1/21 1/9703
106 Jansen type metaphyseal chondrodysplasia 0.014642221 1/21 1/9703
107 Orotic aciduria 0.014642221 1/21 1/9703
108 Hereditary orotic aciduria, type 1 0.014642221 1/21 1/9703
125 Epidermolysis bullosa simplex, Ogna type 0.014642221 1/21 1/9703
139 Myasthenic Syndromes, Congenital 0.014642221 2/21 24/9703
153 Pleuropulmonary blastoma 0.014642221 1/21 1/9703
167 ABCD syndrome 0.014642221 1/21 1/9703
168 HIRSCHSPRUNG DISEASE, SUSCEPTIBILITY TO, 2 0.014642221 1/21 1/9703
169 Eiken Skeletal Dysplasia 0.014642221 1/21 1/9703
173 Failure of Tooth Eruption, Primary 0.014642221 1/21 1/9703
175 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.014642221 1/21 1/9703
176 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.014642221 1/21 1/9703
179 Chondrodysplasia, blomstrand type 0.014642221 1/21 1/9703
181 Combined Oxidative Phosphorylation Deficiency 3 0.014642221 1/21 1/9703
182 Rhabdomyosarcoma, Embryonal, 2 0.014642221 1/21 1/9703
195 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.014642221 1/21 1/9703
198 AGAMMAGLOBULINEMIA 6, AUTOSOMAL RECESSIVE 0.014642221 1/21 1/9703
199 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.014642221 1/21 1/9703
209 DICER1 syndrome 0.014642221 1/21 1/9703
212 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.014642221 1/21 1/9703
213 AGAMMAGLOBULINEMIA 6 0.014642221 1/21 1/9703
217 ROBINOW SYNDROME, AUTOSOMAL DOMINANT 3 0.014642221 1/21 1/9703
218 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.014642221 1/21 1/9703
219 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.014642221 1/21 1/9703
220 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.014642221 1/21 1/9703
226 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.014642221 1/21 1/9703
230 GLOBAL DEVELOPMENTAL DELAY, LUNG CYSTS, OVERGROWTH, AND WILMS TUMOR 0.014642221 1/21 1/9703
13 Colonic Neoplasms 0.021162652 3/21 152/9703
43 Pulmonary Hypertension 0.021162652 2/21 40/9703
44 Lambert-Eaton Myasthenic Syndrome 0.021162652 1/21 2/9703
96 Sex Cord-Stromal Tumor 0.021162652 1/21 2/9703
109 Vitamin D-dependent rickets, type 1 0.021162652 1/21 2/9703
118 Euthyroid Goiter 0.021162652 1/21 2/9703
119 Malignant Granulosa Cell Tumor 0.021162652 1/21 2/9703
129 Myasthenic Syndrome 0.021162652 1/21 2/9703
131 Thyroid carcinoma 0.021162652 2/21 44/9703
148 Congenital hypoplastic anemia 0.021162652 1/21 2/9703
162 NUT midline carcinoma 0.021162652 1/21 2/9703
185 Alcohol Toxicity 0.021162652 1/21 2/9703
190 Epidermolysis Bullosa Simplex With Pyloric Atresia 0.021162652 1/21 2/9703
11 Malignant tumor of colon 0.022172885 3/21 159/9703
41 Hyperparathyroidism 0.028121310 1/21 3/9703
178 Epidermolysis bullosa with pyloric atresia 0.028121310 1/21 3/9703
183 POSTAXIAL POLYDACTYLY, TYPE B 0.028121310 1/21 3/9703
211 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.028121310 1/21 3/9703
224 Polydactyly, Postaxial, Type A1 0.028121310 1/21 3/9703
20 Enchondromatosis 0.030541413 1/21 4/9703
46 Acute Erythroblastic Leukemia 0.030541413 1/21 4/9703
97 POLYDACTYLY, POSTAXIAL 0.030541413 1/21 4/9703
105 Robinow Syndrome 0.030541413 1/21 4/9703
121 Partial atrioventricular canal 0.030541413 1/21 4/9703
166 Myasthenic Syndrome, Congenital, with Facial Dysmorphism, associated with Acetylcholine Receptor Deficiency 0.030541413 1/21 4/9703
171 Perisylvian syndrome 0.030541413 1/21 4/9703
172 WAARDENBURG SYNDROME, TYPE 4A 0.030541413 1/21 4/9703
180 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.030541413 1/21 4/9703
204 Male Germ Cell Tumor 0.030541413 1/21 4/9703
216 MYASTHENIC SYNDROME, CONGENITAL, 1A, SLOW-CHANNEL 0.030541413 1/21 4/9703
225 Robinow Syndrome, Autosomal Dominant 0.030541413 1/21 4/9703
82 Polymyositis 0.033958000 1/21 5/9703
104 Polymyositis Ossificans 0.033958000 1/21 5/9703
132 Polymyositis, Idiopathic 0.033958000 1/21 5/9703
159 Granulosa Cell Cancer 0.033958000 1/21 5/9703
164 Congenital muscular hypertrophy-cerebral syndrome 0.033958000 1/21 5/9703
174 Cornelia de Lange Syndrome 3 0.033958000 1/21 5/9703
191 Waardenburg Syndrome Type 2 0.033958000 1/21 5/9703
227 Cornelia de Lange Syndrome 1 0.033958000 1/21 5/9703
17 Diabetes Mellitus, Non-Insulin-Dependent 0.035623279 3/21 221/9703
117 Aplasia Cutis Congenita 0.038592958 1/21 6/9703
201 Waardenburg Syndrome 0.038592958 1/21 6/9703
210 Postaxial polydactyly type A 0.038592958 1/21 6/9703
16 Communicating Hydrocephalus 0.040745473 1/21 7/9703
112 Hydrocephalus Ex-Vacuo 0.040745473 1/21 7/9703
127 Post-Traumatic Hydrocephalus 0.040745473 1/21 7/9703
130 Obstructive Hydrocephalus 0.040745473 1/21 7/9703
158 Cerebral ventriculomegaly 0.040745473 1/21 7/9703
177 Hyperphosphatasia with Mental Retardation 0.040745473 1/21 7/9703
193 Polycystic kidney disease, type 2 0.040745473 1/21 7/9703
196 Fetal Cerebral Ventriculomegaly 0.040745473 1/21 7/9703
39 Congenital Hydrocephalus 0.041188107 1/21 8/9703
75 Hereditary spherocytosis 0.041188107 1/21 8/9703
111 Perinatal Subarachnoid Hemorrhage 0.041188107 1/21 8/9703
113 Cornelia De Lange Syndrome 0.041188107 1/21 8/9703
126 Subarachnoid Hemorrhage, Spontaneous 0.041188107 1/21 8/9703
138 Subarachnoid Hemorrhage, Aneurysmal 0.041188107 1/21 8/9703
143 Subarachnoid Hemorrhage, Intracranial 0.041188107 1/21 8/9703
146 Polycystic Kidney, Type 1 Autosomal Dominant Disease 0.041188107 1/21 8/9703
165 Agammaglobulinemia, non-Bruton type 0.041188107 1/21 8/9703
197 Aqueductal Stenosis 0.041188107 1/21 8/9703
200 Heterotaxy Syndrome 0.041188107 1/21 8/9703
38 Hydrocephalus 0.043997415 1/21 9/9703
68 Pituitary Adenoma 0.043997415 1/21 9/9703
77 Subarachnoid Hemorrhage 0.043997415 1/21 9/9703
80 Polycystic Kidney, Autosomal Dominant 0.043997415 1/21 9/9703
214 Abnormality of the face 0.043997415 1/21 9/9703
4 Anemia, Sickle Cell 0.048356954 1/21 10/9703
Thyroid
Number of cTWAS Genes in Tissue: 31RP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDMRPS9 gene(s) from the input list not found in DisGeNET CURATEDSH3D21 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDFAM208A gene(s) from the input list not found in DisGeNET CURATEDPODN gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDCCDC57 gene(s) from the input list not found in DisGeNET CURATEDPLCD4 gene(s) from the input list not found in DisGeNET CURATEDSPRYD3 gene(s) from the input list not found in DisGeNET CURATEDSPAG8 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
8 Malignant Neoplasms 0.03058413 3/16 128/9703
30 Hepatosplenomegaly 0.03058413 1/16 1/9703
125 Preterm premature rupture of membranes (disorder) 0.03058413 1/16 1/9703
143 Acute Coronary Syndrome 0.03058413 2/16 33/9703
149 DEVELOPMENTAL DYSPLASIA OF THE HIP 1 0.03058413 1/16 1/9703
158 Angel shaped phalangoepiphyseal dysplasia 0.03058413 1/16 1/9703
180 OSTEOGENESIS IMPERFECTA, TYPE X 0.03058413 1/16 1/9703
185 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.03058413 1/16 1/9703
194 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.03058413 1/16 1/9703
196 Abnormality of brain morphology 0.03058413 1/16 1/9703
202 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.03058413 1/16 1/9703
79 Multiple synostosis syndrome 0.03149114 1/16 3/9703
101 Chondrodysplasia, Grebe type 0.03149114 1/16 3/9703
104 Vitamin D-dependent rickets, type 1 0.03149114 1/16 2/9703
109 Multiple synostoses syndrome 1 0.03149114 1/16 3/9703
136 Acute schizophrenia 0.03149114 1/16 3/9703
148 Brachydactyly syndrome type C 0.03149114 1/16 2/9703
160 MULTIPLE SYNOSTOSES SYNDROME 2 0.03149114 1/16 2/9703
163 Fibular hypoplasia and complex brachydactyly 0.03149114 1/16 3/9703
164 SYMPHALANGISM, PROXIMAL 0.03149114 1/16 2/9703
165 Brachydactyly type C 0.03149114 1/16 3/9703
166 BRACHYDACTYLY, TYPE A1 (disorder) 0.03149114 1/16 3/9703
168 POSTAXIAL POLYDACTYLY, TYPE B 0.03149114 1/16 3/9703
170 Alcohol Toxicity 0.03149114 1/16 2/9703
176 Acromesomelic dysplasia Hunter-Thompson type 0.03149114 1/16 3/9703
184 Ovarian clear cell carcinoma 0.03149114 1/16 3/9703
186 BRACHYDACTYLY, TYPE A1, C 0.03149114 1/16 2/9703
189 SYMPHALANGISM, PROXIMAL, 1A 0.03149114 1/16 2/9703
191 SYMPHALANGISM, PROXIMAL, 1B 0.03149114 1/16 2/9703
193 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.03149114 1/16 3/9703
195 Abnormality of the respiratory system 0.03149114 1/16 2/9703
200 Polydactyly, Postaxial, Type A1 0.03149114 1/16 3/9703
64 Rickets 0.03442522 1/16 4/9703
90 Liposarcoma, Pleomorphic 0.03442522 1/16 4/9703
93 POLYDACTYLY, POSTAXIAL 0.03442522 1/16 4/9703
114 Osteoarthritis, Knee 0.03442522 1/16 4/9703
159 BRACHYDACTYLY, TYPE A2 0.03442522 1/16 4/9703
161 Perisylvian syndrome 0.03442522 1/16 4/9703
167 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.03442522 1/16 4/9703
70 Polymyositis 0.03899844 1/16 5/9703
100 Polymyositis Ossificans 0.03899844 1/16 5/9703
122 Polymyositis, Idiopathic 0.03899844 1/16 5/9703
181 MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 0.03899844 1/16 5/9703
19 Communicating Hydrocephalus 0.04185858 1/16 7/9703
42 liposarcoma 0.04185858 1/16 6/9703
89 Liposarcoma, Dedifferentiated 0.04185858 1/16 6/9703
105 Hydrocephalus Ex-Vacuo 0.04185858 1/16 7/9703
118 Post-Traumatic Hydrocephalus 0.04185858 1/16 7/9703
121 Obstructive Hydrocephalus 0.04185858 1/16 7/9703
135 Abnormal muscle tone 0.04185858 1/16 6/9703
150 Liposarcoma, well differentiated 0.04185858 1/16 6/9703
154 Cerebral ventriculomegaly 0.04185858 1/16 7/9703
162 Hyperphosphatasia with Mental Retardation 0.04185858 1/16 7/9703
178 Fetal Cerebral Ventriculomegaly 0.04185858 1/16 7/9703
192 Postaxial polydactyly type A 0.04185858 1/16 6/9703
197 Neurodevelopmental delay 0.04185858 1/16 7/9703
32 Congenital Hydrocephalus 0.04537085 1/16 8/9703
91 Embryonal Rhabdomyosarcoma 0.04537085 1/16 8/9703
179 Aqueductal Stenosis 0.04537085 1/16 8/9703
1 Adenocarcinoma 0.04712929 2/16 116/9703
31 Hydrocephalus 0.04712929 1/16 9/9703
80 Adenocarcinoma, Basal Cell 0.04712929 2/16 116/9703
81 Adenocarcinoma, Oxyphilic 0.04712929 2/16 116/9703
82 Carcinoma, Cribriform 0.04712929 2/16 116/9703
83 Carcinoma, Granular Cell 0.04712929 2/16 116/9703
84 Adenocarcinoma, Tubular 0.04712929 2/16 116/9703
78 Polydactyly 0.04718806 2/16 117/9703
Artery_Coronary
Number of cTWAS Genes in Tissue: 37CTD-3224K15.3 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDRP11-1277A3.3 gene(s) from the input list not found in DisGeNET CURATEDSLC2A12 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDRFTN1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
25 Hepatosplenomegaly 0.01510734 1/19 1/9703
32 Jaundice, Obstructive 0.01510734 1/19 1/9703
80 Lipoblastomatosis 0.01510734 1/19 1/9703
83 Epidermolysis bullosa simplex, Ogna type 0.01510734 1/19 1/9703
87 Delayed developmental milestones 0.01510734 1/19 1/9703
103 Cholecystolithiasis 0.01510734 1/19 1/9703
108 Lipoblastoma 0.01510734 1/19 1/9703
116 Pseudoxanthoma Elasticum-Like Disorder with Multiple Coagulation Factor Deficiency 0.01510734 1/19 1/9703
122 Combined Oxidative Phosphorylation Deficiency 3 0.01510734 1/19 1/9703
123 Cholestasis, progressive familial intrahepatic 3 0.01510734 1/19 1/9703
129 Low phospholipid-associated cholelithiasis 0.01510734 1/19 1/9703
131 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.01510734 1/19 1/9703
136 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.01510734 1/19 1/9703
138 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB 0.01510734 1/19 1/9703
141 CHOLESTASIS, INTRAHEPATIC, OF PREGNANCY 3 0.01510734 1/19 1/9703
146 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.01510734 1/19 1/9703
148 ROBINOW SYNDROME, AUTOSOMAL DOMINANT 3 0.01510734 1/19 1/9703
149 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.01510734 1/19 1/9703
153 MYOPIA 25, AUTOSOMAL DOMINANT 0.01510734 1/19 1/9703
154 PONTOCEREBELLAR HYPOPLASIA, TYPE 2F 0.01510734 1/19 1/9703
157 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01510734 1/19 1/9703
12 Cholelithiasis 0.01980999 1/19 2/9703
34 Lambert-Eaton Myasthenic Syndrome 0.01980999 1/19 2/9703
76 Cutis Laxa, Autosomal Recessive, Type I 0.01980999 1/19 2/9703
77 Vitamin D-dependent rickets, type 1 0.01980999 1/19 2/9703
84 Cutis laxa, recessive, type I 0.01980999 1/19 2/9703
89 Myasthenic Syndrome 0.01980999 1/19 2/9703
112 Salivary Gland Pleomorphic Adenoma 0.01980999 1/19 2/9703
118 VITAMIN K-DEPENDENT CLOTTING FACTORS, COMBINED DEFICIENCY OF, 1 0.01980999 1/19 2/9703
126 Alcohol Toxicity 0.01980999 1/19 2/9703
130 Epidermolysis Bullosa Simplex With Pyloric Atresia 0.01980999 1/19 2/9703
147 Abnormality of the respiratory system 0.01980999 1/19 2/9703
21 Fibrosis 0.02005752 2/19 50/9703
114 Cirrhosis 0.02005752 2/19 50/9703
74 Progressive intrahepatic cholestasis (disorder) 0.02209297 1/19 3/9703
85 Urolithiasis 0.02209297 1/19 3/9703
86 Radiolabeled somatostatin analog study 0.02209297 1/19 3/9703
100 Acute schizophrenia 0.02209297 1/19 3/9703
120 Epidermolysis bullosa with pyloric atresia 0.02209297 1/19 3/9703
124 POSTAXIAL POLYDACTYLY, TYPE B 0.02209297 1/19 3/9703
145 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.02209297 1/19 3/9703
152 Polydactyly, Postaxial, Type A1 0.02209297 1/19 3/9703
160 Cholestasis, benign recurrent intrahepatic 1 0.02209297 1/19 3/9703
48 Rickets 0.02300889 1/19 4/9703
65 POLYDACTYLY, POSTAXIAL 0.02300889 1/19 4/9703
73 Robinow Syndrome 0.02300889 1/19 4/9703
97 End Stage Liver Disease 0.02300889 1/19 4/9703
99 MENTAL RETARDATION, X-LINKED, SNYDER-ROBINSON TYPE 0.02300889 1/19 4/9703
117 Perisylvian syndrome 0.02300889 1/19 4/9703
121 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.02300889 1/19 4/9703
132 Pontocerebellar Hypoplasia Type 2 0.02300889 1/19 4/9703
133 Chronic Liver Failure 0.02300889 1/19 4/9703
140 Familial intrahepatic cholestasis of pregnancy 0.02300889 1/19 4/9703
156 Robinow Syndrome, Autosomal Dominant 0.02300889 1/19 4/9703
159 Cholestasis, progressive familial intrahepatic 1 0.02300889 1/19 4/9703
53 Polymyositis 0.02724818 1/19 5/9703
72 Polymyositis Ossificans 0.02724818 1/19 5/9703
91 Polymyositis, Idiopathic 0.02724818 1/19 5/9703
11 Cholangitis, Sclerosing 0.03007485 1/19 6/9703
17 Cutis Laxa 0.03007485 1/19 6/9703
35 Fibroid Tumor 0.03007485 1/19 6/9703
79 Aplasia Cutis Congenita 0.03007485 1/19 6/9703
144 Postaxial polydactyly type A 0.03007485 1/19 6/9703
2 Aortic Aneurysm 0.03025279 1/19 7/9703
9 Cholangitis 0.03025279 1/19 7/9703
16 Communicating Hydrocephalus 0.03025279 1/19 7/9703
52 Uterine Fibroids 0.03025279 1/19 7/9703
78 Hydrocephalus Ex-Vacuo 0.03025279 1/19 7/9703
88 Post-Traumatic Hydrocephalus 0.03025279 1/19 7/9703
90 Obstructive Hydrocephalus 0.03025279 1/19 7/9703
113 Cerebral ventriculomegaly 0.03025279 1/19 7/9703
119 Hyperphosphatasia with Mental Retardation 0.03025279 1/19 7/9703
134 Fetal Cerebral Ventriculomegaly 0.03025279 1/19 7/9703
28 Congenital Hydrocephalus 0.03274816 1/19 8/9703
75 Cholestasis of pregnancy 0.03274816 1/19 8/9703
93 Primary sclerosing cholangitis 0.03274816 1/19 8/9703
135 Aqueductal Stenosis 0.03274816 1/19 8/9703
15 Colorectal Neoplasms 0.03303843 3/19 277/9703
27 Hydrocephalus 0.03587573 1/19 9/9703
59 Russell-Silver syndrome 0.03932719 1/19 10/9703
66 Macrocephaly 0.04268629 1/19 11/9703
29 Hypercalcemia 0.04914076 1/19 13/9703
44 Milk-Alkali Syndrome 0.04914076 1/19 13/9703
Esophagus_Gastroesophageal_Junction
Number of cTWAS Genes in Tissue: 34WRAP73 gene(s) from the input list not found in DisGeNET CURATEDTMEM129 gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDZNF3 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZBTB38 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDPLCD3 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
14 Hepatosplenomegaly 0.01064594 1/20 1/9703
54 Middle Cerebral Artery Syndrome 0.01064594 2/20 34/9703
60 Glutaric aciduria, type 1 0.01064594 1/20 1/9703
68 Delayed developmental milestones 0.01064594 1/20 1/9703
73 Preterm premature rupture of membranes (disorder) 0.01064594 1/20 1/9703
74 Middle Cerebral Artery Thrombosis 0.01064594 2/20 34/9703
75 Middle Cerebral Artery Occlusion 0.01064594 2/20 34/9703
76 Infarction, Middle Cerebral Artery 0.01064594 2/20 34/9703
79 Middle Cerebral Artery Embolus 0.01064594 2/20 34/9703
80 Left Middle Cerebral Artery Infarction 0.01064594 2/20 34/9703
81 Embolic Infarction, Middle Cerebral Artery 0.01064594 2/20 34/9703
82 Thrombotic Infarction, Middle Cerebral Artery 0.01064594 2/20 34/9703
83 Right Middle Cerebral Artery Infarction 0.01064594 2/20 34/9703
84 Myasthenic Syndromes, Congenital 0.01064594 2/20 24/9703
86 Congenital Myasthenic Syndromes, Presynaptic 0.01064594 2/20 19/9703
101 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.01064594 1/20 1/9703
102 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.01064594 1/20 1/9703
104 Combined Oxidative Phosphorylation Deficiency 3 0.01064594 1/20 1/9703
105 ATAXIA, SPASTIC, 1, AUTOSOMAL DOMINANT 0.01064594 1/20 1/9703
110 OSTEOGENESIS IMPERFECTA, TYPE X 0.01064594 1/20 1/9703
113 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB 0.01064594 1/20 1/9703
118 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.01064594 1/20 1/9703
119 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.01064594 1/20 1/9703
123 MYOPIA 25, AUTOSOMAL DOMINANT 0.01064594 1/20 1/9703
124 CILIARY DYSKINESIA, PRIMARY, 35 0.01064594 1/20 1/9703
125 PONTOCEREBELLAR HYPOPLASIA, TYPE 2F 0.01064594 1/20 1/9703
128 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01064594 1/20 1/9703
58 Cutis Laxa, Autosomal Recessive, Type I 0.01647530 1/20 2/9703
61 Vitamin D-dependent rickets, type 1 0.01647530 1/20 2/9703
67 Cutis laxa, recessive, type I 0.01647530 1/20 2/9703
100 MALE PSEUDOHERMAPHRODITISM: DEFICIENCY OF TESTICULAR 17,20-DESMOLASE 0.01647530 1/20 2/9703
111 Obesity, Hyperphagia, and Developmental Delay 0.01647530 1/20 2/9703
116 Abnormality of the respiratory system 0.01647530 1/20 2/9703
26 Neoplasms, Hormone-Dependent 0.02327796 1/20 3/9703
89 Acute schizophrenia 0.02327796 1/20 3/9703
33 Rickets 0.02713104 1/20 4/9703
65 Partial atrioventricular canal 0.02713104 1/20 4/9703
99 Myasthenic Syndrome, Congenital, with Facial Dysmorphism, associated with Acetylcholine Receptor Deficiency 0.02713104 1/20 4/9703
109 Pontocerebellar Hypoplasia Type 2 0.02713104 1/20 4/9703
117 MYASTHENIC SYNDROME, CONGENITAL, 1A, SLOW-CHANNEL 0.02713104 1/20 4/9703
15 Hermaphroditism 0.02823385 1/20 5/9703
31 Pseudohermaphroditism 0.02823385 1/20 5/9703
42 Polymyositis 0.02823385 1/20 5/9703
56 Polymyositis Ossificans 0.02823385 1/20 5/9703
57 Ambiguous Genitalia 0.02823385 1/20 5/9703
70 Polymyositis, Idiopathic 0.02823385 1/20 5/9703
107 Intersex Conditions 0.02823385 1/20 5/9703
108 Sex Differentiation Disorders 0.02823385 1/20 5/9703
6 Cutis Laxa 0.03315671 1/20 6/9703
2 Aortic Aneurysm 0.03641547 1/20 7/9703
36 Disorders of Sex Development 0.03641547 1/20 7/9703
103 Hyperphosphatasia with Mental Retardation 0.03641547 1/20 7/9703
112 Heterotaxy Syndrome 0.04079252 1/20 8/9703
Breast_Mammary_Tissue
Number of cTWAS Genes in Tissue: 23NFATC4 gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDC17orf82 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
139 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.02370645 1/15 1/9703
140 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.02370645 1/15 1/9703
144 Combined Oxidative Phosphorylation Deficiency 3 0.02370645 1/15 1/9703
153 CARDIOMYOPATHY, DILATED, 1GG 0.02370645 1/15 1/9703
154 LOEYS-DIETZ SYNDROME 3 0.02370645 1/15 1/9703
157 PARAGANGLIOMAS 5 0.02370645 1/15 1/9703
163 DEAFNESS, AUTOSOMAL DOMINANT 27 0.02370645 1/15 1/9703
164 WILMS TUMOR, SUSCEPTIBILITY TO 0.02370645 1/15 1/9703
166 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.02370645 1/15 1/9703
169 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.02370645 1/15 1/9703
170 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.02370645 1/15 1/9703
180 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.02370645 1/15 1/9703
2 Aneurysm, Dissecting 0.03965480 1/15 5/9703
3 Aortic Aneurysm 0.03965480 1/15 7/9703
13 Communicating Hydrocephalus 0.03965480 1/15 7/9703
25 Fibroid Tumor 0.03965480 1/15 6/9703
38 Degenerative polyarthritis 0.03965480 2/15 93/9703
42 Rickets 0.03965480 1/15 4/9703
43 Schizophrenia 0.03965480 5/15 883/9703
47 Uterine Fibroids 0.03965480 1/15 7/9703
49 Polymyositis 0.03965480 1/15 5/9703
50 Osteoarthrosis Deformans 0.03965480 2/15 93/9703
66 POLYDACTYLY, POSTAXIAL 0.03965480 1/15 4/9703
74 Polymyositis Ossificans 0.03965480 1/15 5/9703
76 Vitamin D-dependent rickets, type 1 0.03965480 1/15 2/9703
77 Hydrocephalus Ex-Vacuo 0.03965480 1/15 7/9703
84 Dissection of aorta 0.03965480 1/15 5/9703
85 Partial atrioventricular canal 0.03965480 1/15 4/9703
88 Hereditary gingival fibromatosis 0.03965480 1/15 3/9703
94 Post-Traumatic Hydrocephalus 0.03965480 1/15 7/9703
96 Obstructive Hydrocephalus 0.03965480 1/15 7/9703
97 Polymyositis, Idiopathic 0.03965480 1/15 5/9703
116 Acute schizophrenia 0.03965480 1/15 3/9703
127 Cerebral ventriculomegaly 0.03965480 1/15 7/9703
131 Loeys-Dietz Aortic Aneurysm Syndrome 0.03965480 1/15 5/9703
132 Myasthenic Syndrome, Congenital, with Facial Dysmorphism, associated with Acetylcholine Receptor Deficiency 0.03965480 1/15 4/9703
134 Perisylvian syndrome 0.03965480 1/15 4/9703
141 Mitochondrial Complex II Deficiency 0.03965480 1/15 4/9703
142 Hyperphosphatasia with Mental Retardation 0.03965480 1/15 7/9703
143 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.03965480 1/15 4/9703
145 POSTAXIAL POLYDACTYLY, TYPE B 0.03965480 1/15 3/9703
147 Alcohol Toxicity 0.03965480 1/15 2/9703
148 Loeys-Dietz Syndrome 0.03965480 1/15 7/9703
151 Fetal Cerebral Ventriculomegaly 0.03965480 1/15 7/9703
162 Postaxial polydactyly type A 0.03965480 1/15 6/9703
165 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.03965480 1/15 3/9703
168 MYASTHENIC SYNDROME, CONGENITAL, 1A, SLOW-CHANNEL 0.03965480 1/15 4/9703
171 Dissection, Blood Vessel 0.03965480 1/15 5/9703
175 Polydactyly, Postaxial, Type A1 0.03965480 1/15 3/9703
181 Loeys-Dietz Syndrome, Type 1a 0.03965480 1/15 5/9703
21 Congenital Hydrocephalus 0.04272368 1/15 8/9703
152 Aqueductal Stenosis 0.04272368 1/15 8/9703
155 Heterotaxy Syndrome 0.04272368 1/15 8/9703
20 Hydrocephalus 0.04465901 1/15 9/9703
48 Uterine Neoplasms 0.04465901 1/15 9/9703
106 Encephalopathy, Subacute Necrotizing, Infantile 0.04465901 1/15 9/9703
107 Encephalopathy, Subacute Necrotizing, Juvenile 0.04465901 1/15 9/9703
57 Uterine Cancer 0.04873044 1/15 10/9703Gene sets curated by Macarthur Lab
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_tissue=ctwas_genes_tissue)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_tissue]})
##########
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_tissue)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
} Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 41
gene_set nset ngenes percent padj
1 gwascatalog 3890 23 0.005912596 0.004355315
2 clinvar_path_likelypath 1811 12 0.006626173 0.029346845
3 mgi_essential 1426 6 0.004207574 0.549524616
4 fda_approved_drug_targets 211 1 0.004739336 0.631752203
5 core_essentials_hart 181 0 0.000000000 1.000000000
Thyroid
Number of cTWAS Genes in Tissue: 31
gene_set nset ngenes percent padj
1 gwascatalog 4081 18 0.004410684 0.009352283
2 clinvar_path_likelypath 1878 7 0.003727370 0.373602418
3 mgi_essential 1463 5 0.003417635 0.442077395
4 core_essentials_hart 189 0 0.000000000 1.000000000
5 fda_approved_drug_targets 213 0 0.000000000 1.000000000
Artery_Coronary
Number of cTWAS Genes in Tissue: 37
gene_set nset ngenes percent padj
1 gwascatalog 3581 23 0.006422787 0.0005888015
2 clinvar_path_likelypath 1684 10 0.005938242 0.0969191927
3 mgi_essential 1317 6 0.004555809 0.4227349690
4 fda_approved_drug_targets 190 1 0.005263158 0.5809707169
5 core_essentials_hart 176 0 0.000000000 1.0000000000
Esophagus_Gastroesophageal_Junction
Number of cTWAS Genes in Tissue: 34
gene_set nset ngenes percent padj
1 clinvar_path_likelypath 1743 12 0.006884682 0.01187034
2 gwascatalog 3797 17 0.004477219 0.05656378
3 mgi_essential 1369 7 0.005113221 0.13132242
4 fda_approved_drug_targets 192 2 0.010416667 0.13132242
5 core_essentials_hart 176 0 0.000000000 1.00000000
Breast_Mammary_Tissue
Number of cTWAS Genes in Tissue: 23
gene_set nset ngenes percent padj
1 gwascatalog 3793 15 0.0039546533 0.004193528
2 clinvar_path_likelypath 1762 8 0.0045402951 0.031610768
3 mgi_essential 1400 1 0.0007142857 1.000000000
4 core_essentials_hart 175 0 0.0000000000 1.000000000
5 fda_approved_drug_targets 204 0 0.0000000000 1.000000000Summary of results across tissues
weight_groups <- as.data.frame(matrix(c("Adipose_Subcutaneous", "Adipose",
"Adipose_Visceral_Omentum", "Adipose",
"Adrenal_Gland", "Endocrine",
"Artery_Aorta", "Cardiovascular",
"Artery_Coronary", "Cardiovascular",
"Artery_Tibial", "Cardiovascular",
"Brain_Amygdala", "CNS",
"Brain_Anterior_cingulate_cortex_BA24", "CNS",
"Brain_Caudate_basal_ganglia", "CNS",
"Brain_Cerebellar_Hemisphere", "CNS",
"Brain_Cerebellum", "CNS",
"Brain_Cortex", "CNS",
"Brain_Frontal_Cortex_BA9", "CNS",
"Brain_Hippocampus", "CNS",
"Brain_Hypothalamus", "CNS",
"Brain_Nucleus_accumbens_basal_ganglia", "CNS",
"Brain_Putamen_basal_ganglia", "CNS",
"Brain_Spinal_cord_cervical_c-1", "CNS",
"Brain_Substantia_nigra", "CNS",
"Breast_Mammary_Tissue", "None",
"Cells_Cultured_fibroblasts", "Skin",
"Cells_EBV-transformed_lymphocytes", "Blood or Immune",
"Colon_Sigmoid", "Digestive",
"Colon_Transverse", "Digestive",
"Esophagus_Gastroesophageal_Junction", "Digestive",
"Esophagus_Mucosa", "Digestive",
"Esophagus_Muscularis", "Digestive",
"Heart_Atrial_Appendage", "Cardiovascular",
"Heart_Left_Ventricle", "Cardiovascular",
"Kidney_Cortex", "None",
"Liver", "None",
"Lung", "None",
"Minor_Salivary_Gland", "None",
"Muscle_Skeletal", "None",
"Nerve_Tibial", "None",
"Ovary", "None",
"Pancreas", "None",
"Pituitary", "Endocrine",
"Prostate", "None",
"Skin_Not_Sun_Exposed_Suprapubic", "Skin",
"Skin_Sun_Exposed_Lower_leg", "Skin",
"Small_Intestine_Terminal_Ileum", "Digestive",
"Spleen", "Blood or Immune",
"Stomach", "Digestive",
"Testis", "Endocrine",
"Thyroid", "Endocrine",
"Uterus", "None",
"Vagina", "None",
"Whole_Blood", "Blood or Immune"),
nrow=49, ncol=2, byrow=T), stringsAsFactors=F)
colnames(weight_groups) <- c("weight", "group")
#display tissue groups
print(weight_groups) weight group
1 Adipose_Subcutaneous Adipose
2 Adipose_Visceral_Omentum Adipose
3 Adrenal_Gland Endocrine
4 Artery_Aorta Cardiovascular
5 Artery_Coronary Cardiovascular
6 Artery_Tibial Cardiovascular
7 Brain_Amygdala CNS
8 Brain_Anterior_cingulate_cortex_BA24 CNS
9 Brain_Caudate_basal_ganglia CNS
10 Brain_Cerebellar_Hemisphere CNS
11 Brain_Cerebellum CNS
12 Brain_Cortex CNS
13 Brain_Frontal_Cortex_BA9 CNS
14 Brain_Hippocampus CNS
15 Brain_Hypothalamus CNS
16 Brain_Nucleus_accumbens_basal_ganglia CNS
17 Brain_Putamen_basal_ganglia CNS
18 Brain_Spinal_cord_cervical_c-1 CNS
19 Brain_Substantia_nigra CNS
20 Breast_Mammary_Tissue None
21 Cells_Cultured_fibroblasts Skin
22 Cells_EBV-transformed_lymphocytes Blood or Immune
23 Colon_Sigmoid Digestive
24 Colon_Transverse Digestive
25 Esophagus_Gastroesophageal_Junction Digestive
26 Esophagus_Mucosa Digestive
27 Esophagus_Muscularis Digestive
28 Heart_Atrial_Appendage Cardiovascular
29 Heart_Left_Ventricle Cardiovascular
30 Kidney_Cortex None
31 Liver None
32 Lung None
33 Minor_Salivary_Gland None
34 Muscle_Skeletal None
35 Nerve_Tibial None
36 Ovary None
37 Pancreas None
38 Pituitary Endocrine
39 Prostate None
40 Skin_Not_Sun_Exposed_Suprapubic Skin
41 Skin_Sun_Exposed_Lower_leg Skin
42 Small_Intestine_Terminal_Ileum Digestive
43 Spleen Blood or Immune
44 Stomach Digestive
45 Testis Endocrine
46 Thyroid Endocrine
47 Uterus None
48 Vagina None
49 Whole_Blood Blood or Immunegroups <- unique(weight_groups$group)
df_group <- list()
for (i in 1:length(groups)){
group <- groups[i]
weights <- weight_groups$weight[weight_groups$group==group]
df_group[[group]] <- list(ctwas=unique(unlist(lapply(df[weights], function(x){x$ctwas}))),
background=unique(unlist(lapply(df[weights], function(x){x$gene_pips$genename}))))
}
output <- output[sapply(weight_groups$weight, match, output$weight),,drop=F]
output$group <- weight_groups$group
output$n_ctwas_group <- sapply(output$group, function(x){length(df_group[[x]][["ctwas"]])})
output$n_ctwas_group[output$group=="None"] <- 0
#barplot of number of cTWAS genes in each tissue
output <- output[order(-output$n_ctwas),,drop=F]
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(output$n_ctwas, names.arg=output$weight, las=2, ylab="Number of cTWAS Genes", cex.names=0.6, main="Number of cTWAS Genes by Tissue")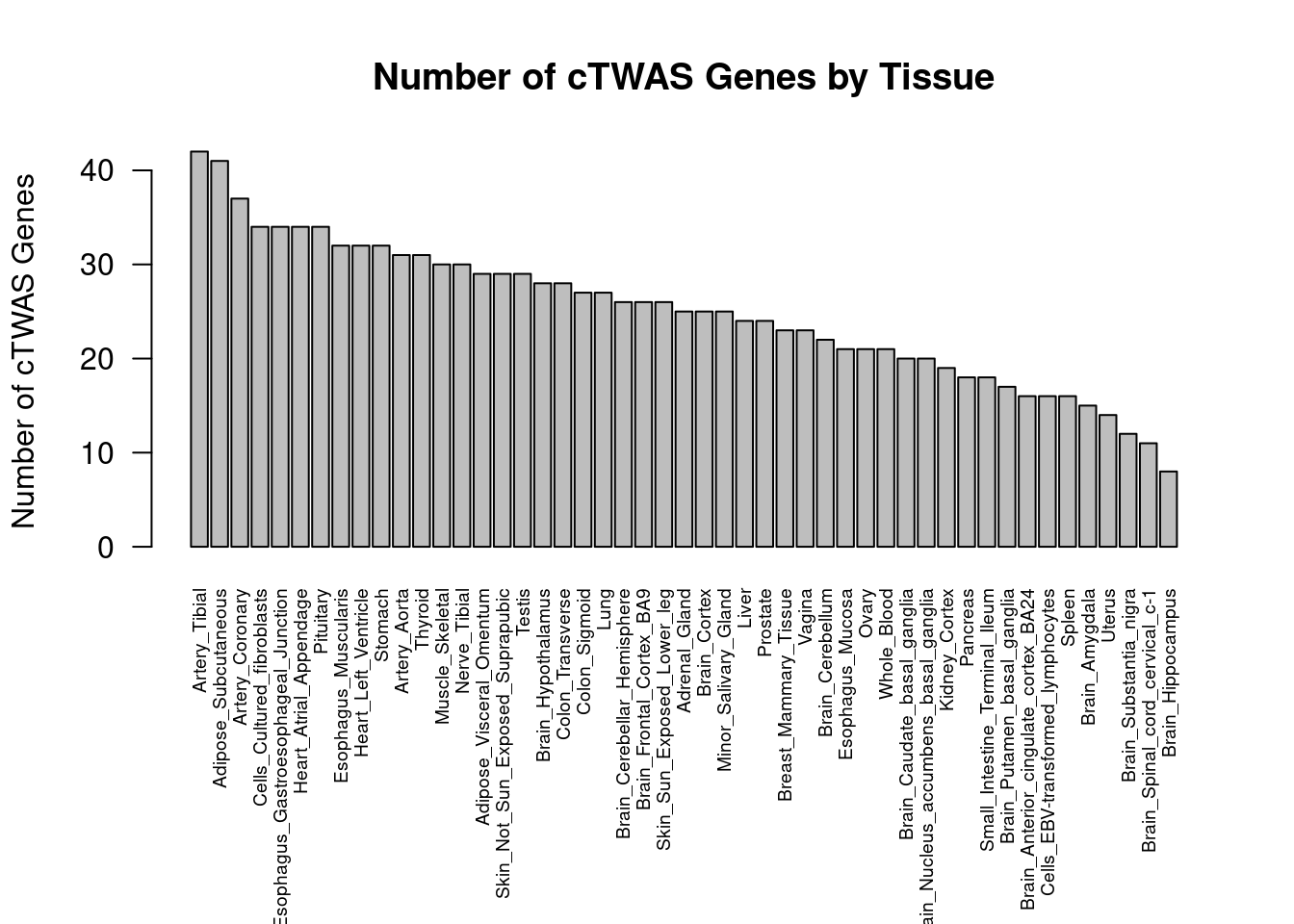
#barplot of number of cTWAS genes in each tissue
df_plot <- -sort(-sapply(groups[groups!="None"], function(x){length(df_group[[x]][["ctwas"]])}))
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(df_plot, las=2, ylab="Number of cTWAS Genes", main="Number of cTWAS Genes by Tissue Group")
Enrichment analysis for cTWAS genes in each tissue group
GO
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_group, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Adipose
Number of cTWAS Genes in Tissue Group: 61
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)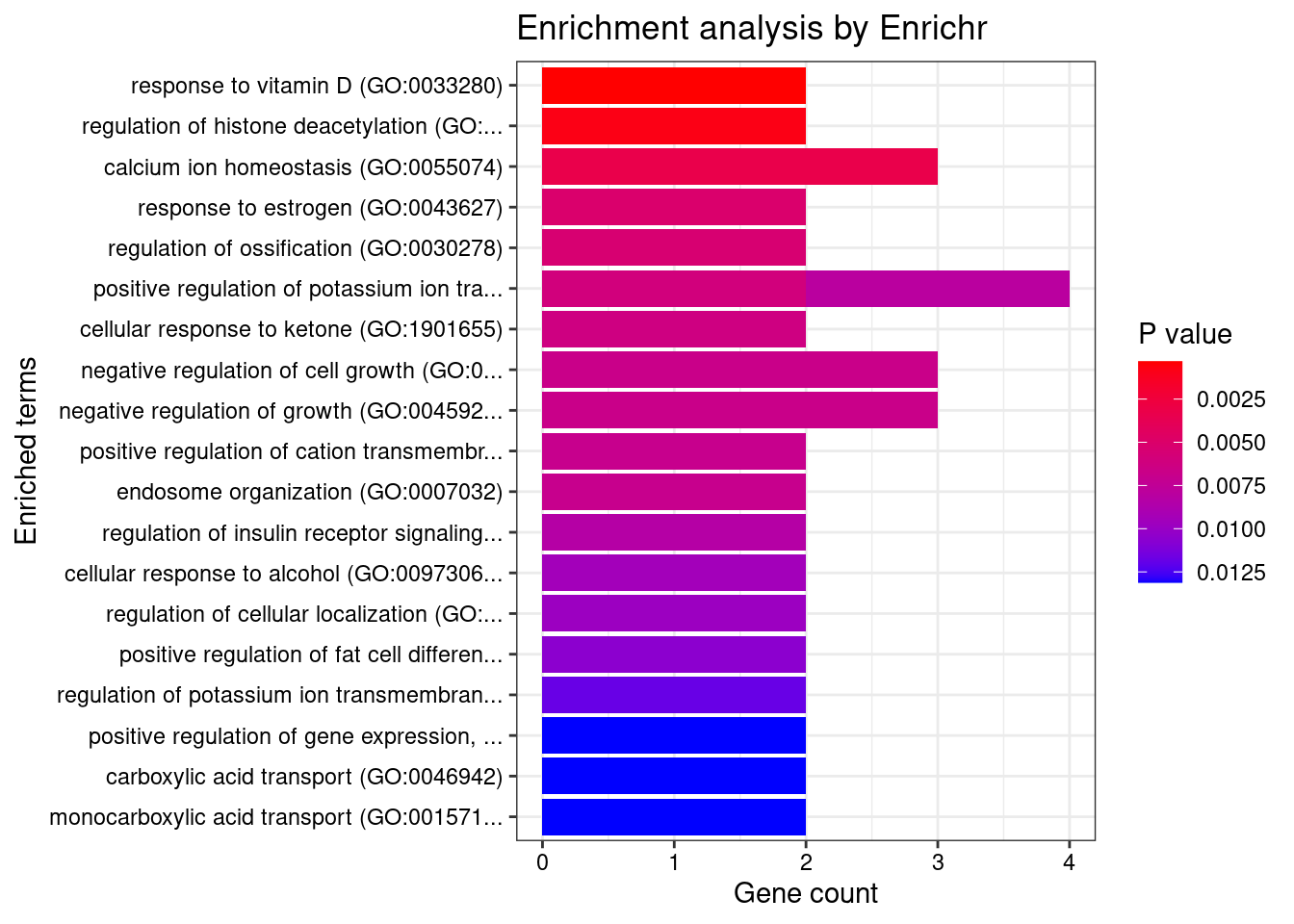
Endocrine
Number of cTWAS Genes in Tissue Group: 90
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)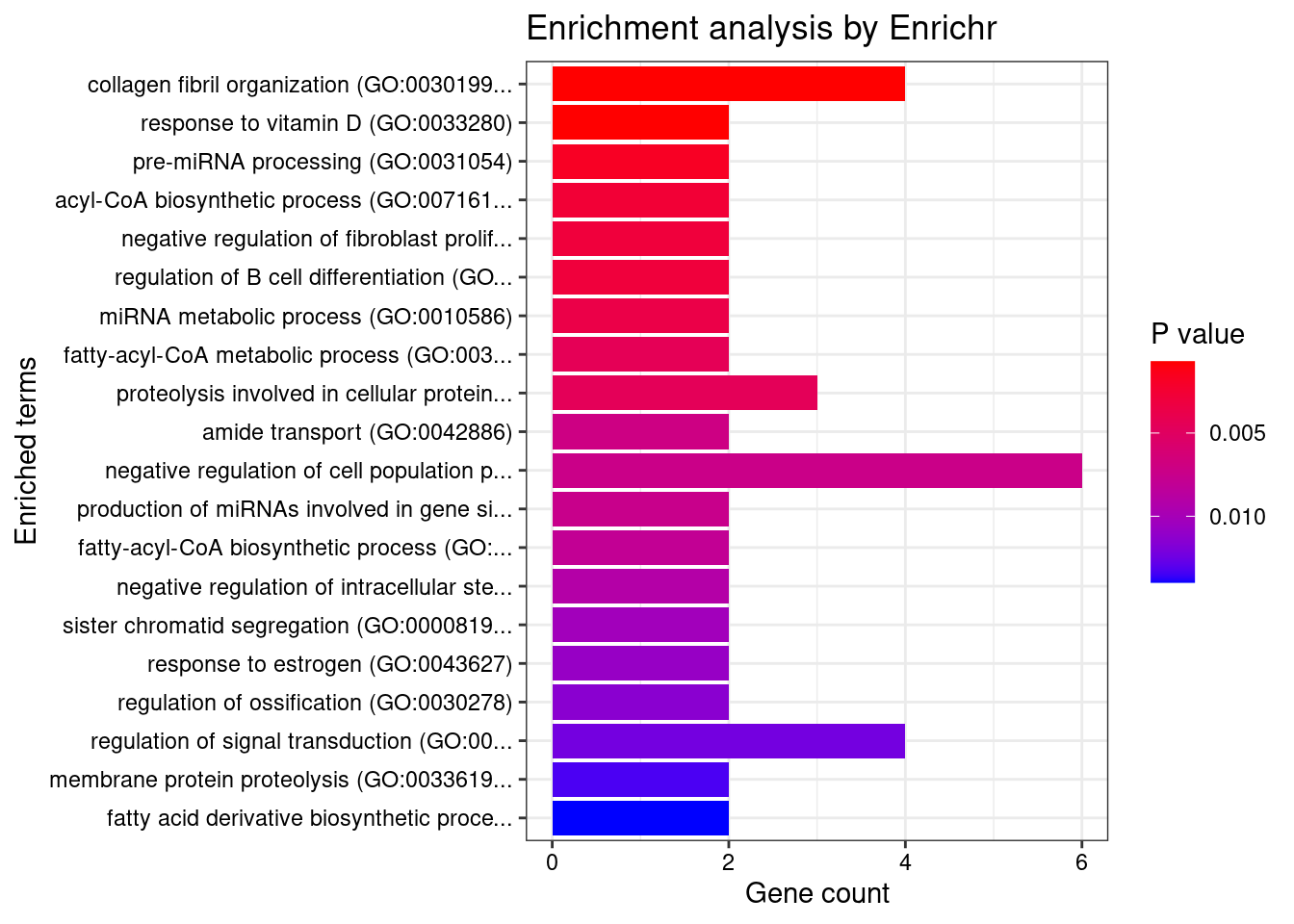
Cardiovascular
Number of cTWAS Genes in Tissue Group: 111
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)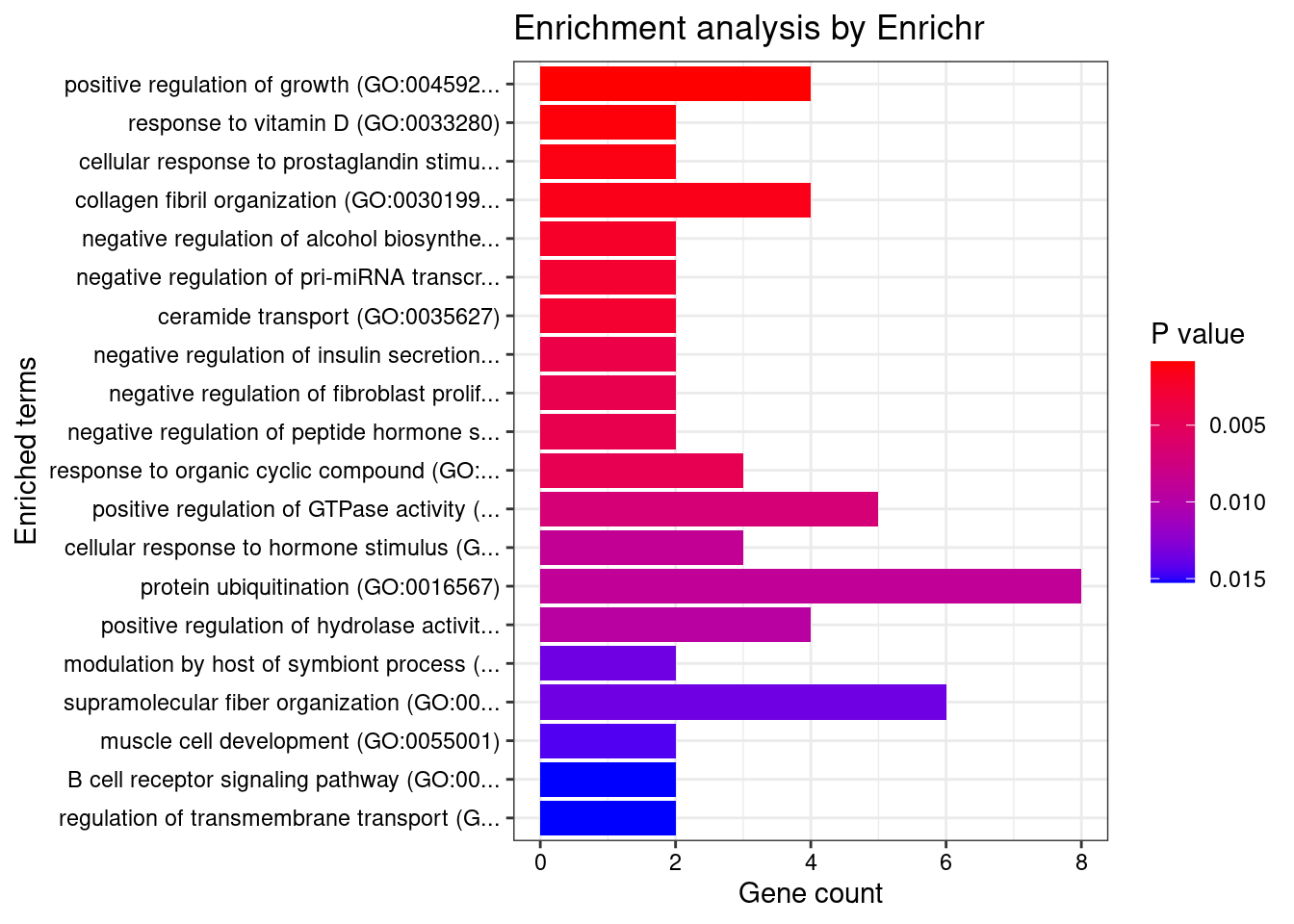
CNS
Number of cTWAS Genes in Tissue Group: 107
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)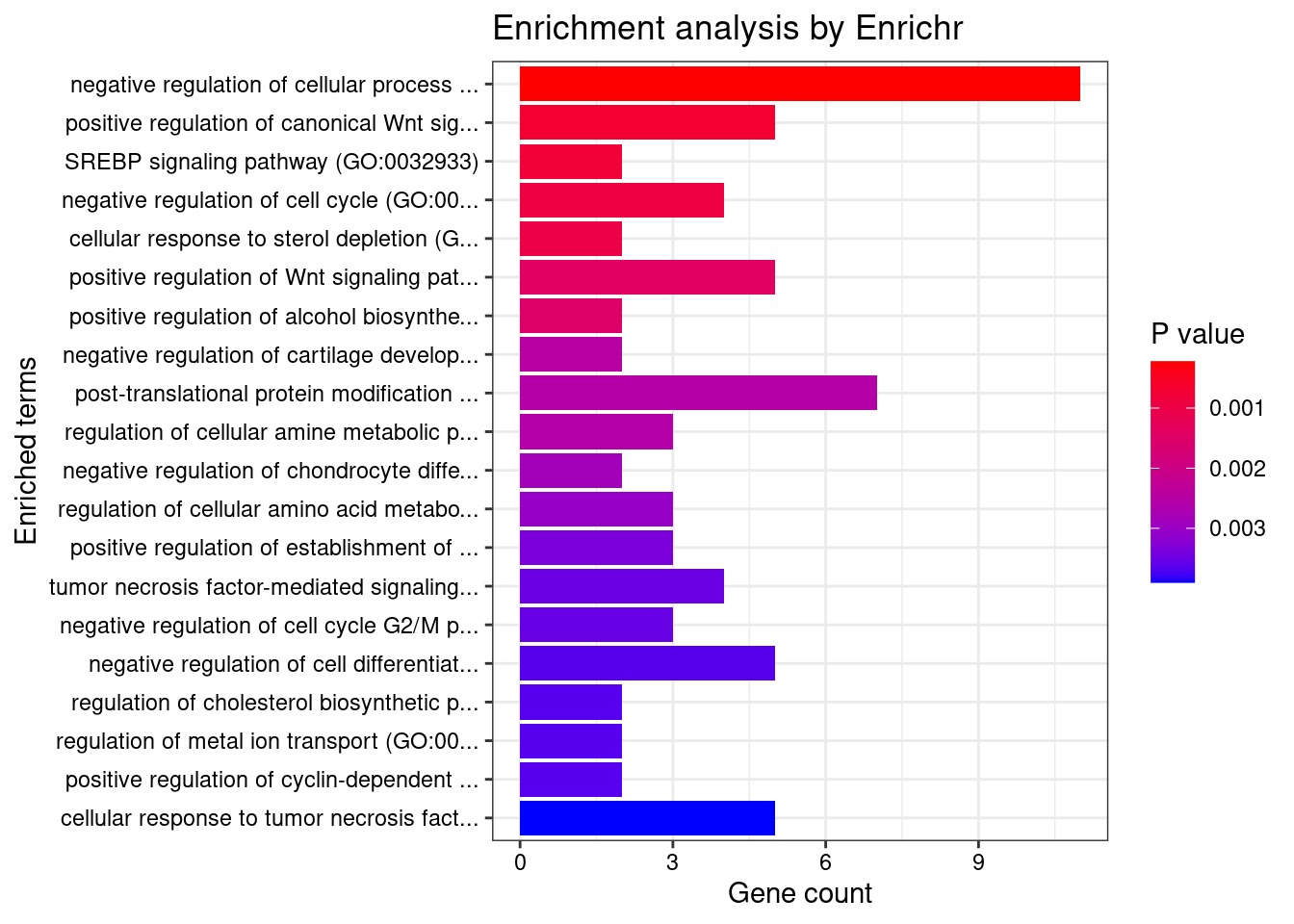
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
None
Number of cTWAS Genes in Tissue Group: 138
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)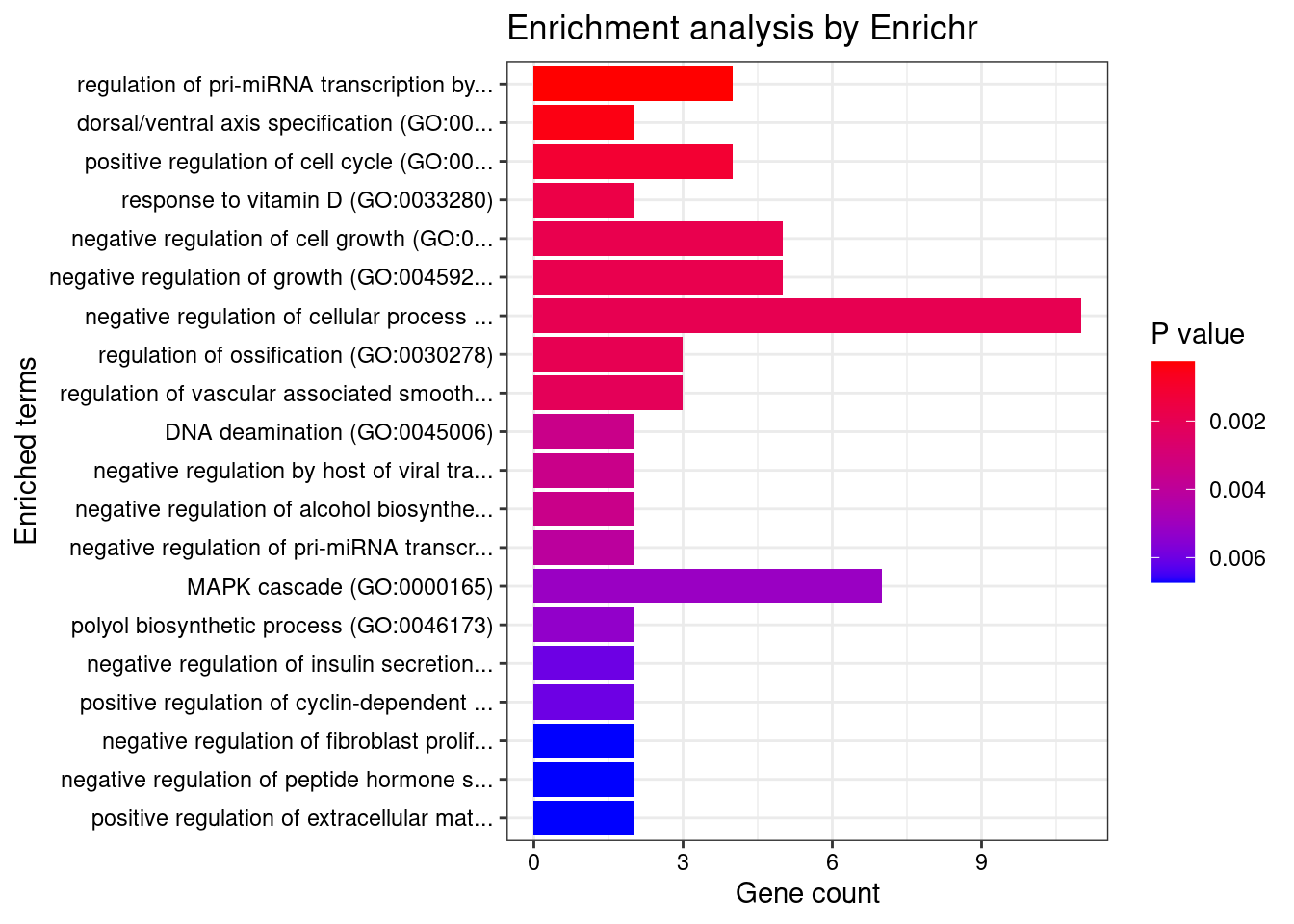
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
Skin
Number of cTWAS Genes in Tissue Group: 68
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 negative regulation of alcohol biosynthetic process (GO:1902931) 4/13 5.181704e-05 CYP27B1;REST;GFI1;SCAP
2 negative regulation of pri-miRNA transcription by RNA polymerase II (GO:1902894) 3/14 4.042809e-03 REST;PDGFB;NFATC4
3 negative regulation of vitamin D biosynthetic process (GO:0010957) 2/5 2.289426e-02 CYP27B1;GFI1
4 regulation of calcidiol 1-monooxygenase activity (GO:0060558) 2/7 3.184132e-02 CYP27B1;GFI1
5 negative regulation of monooxygenase activity (GO:0032769) 2/8 3.184132e-02 CYP27B1;GFI1
6 SREBP signaling pathway (GO:0032933) 2/8 3.184132e-02 SCAP;SREBF2
7 cellular response to sterol depletion (GO:0071501) 2/9 3.501341e-02 SCAP;SREBF2
8 regulation of pri-miRNA transcription by RNA polymerase II (GO:1902893) 3/45 3.653955e-02 REST;PDGFB;NFATC4
9 positive regulation of alcohol biosynthetic process (GO:1902932) 2/11 3.728071e-02 SCAP;PTH1R
10 protein kinase C signaling (GO:0070528) 2/11 3.728071e-02 PLVAP;PDGFB
11 regulation of alternative mRNA splicing, via spliceosome (GO:0000381) 3/54 4.544815e-02 REST;CELF6;SRSF12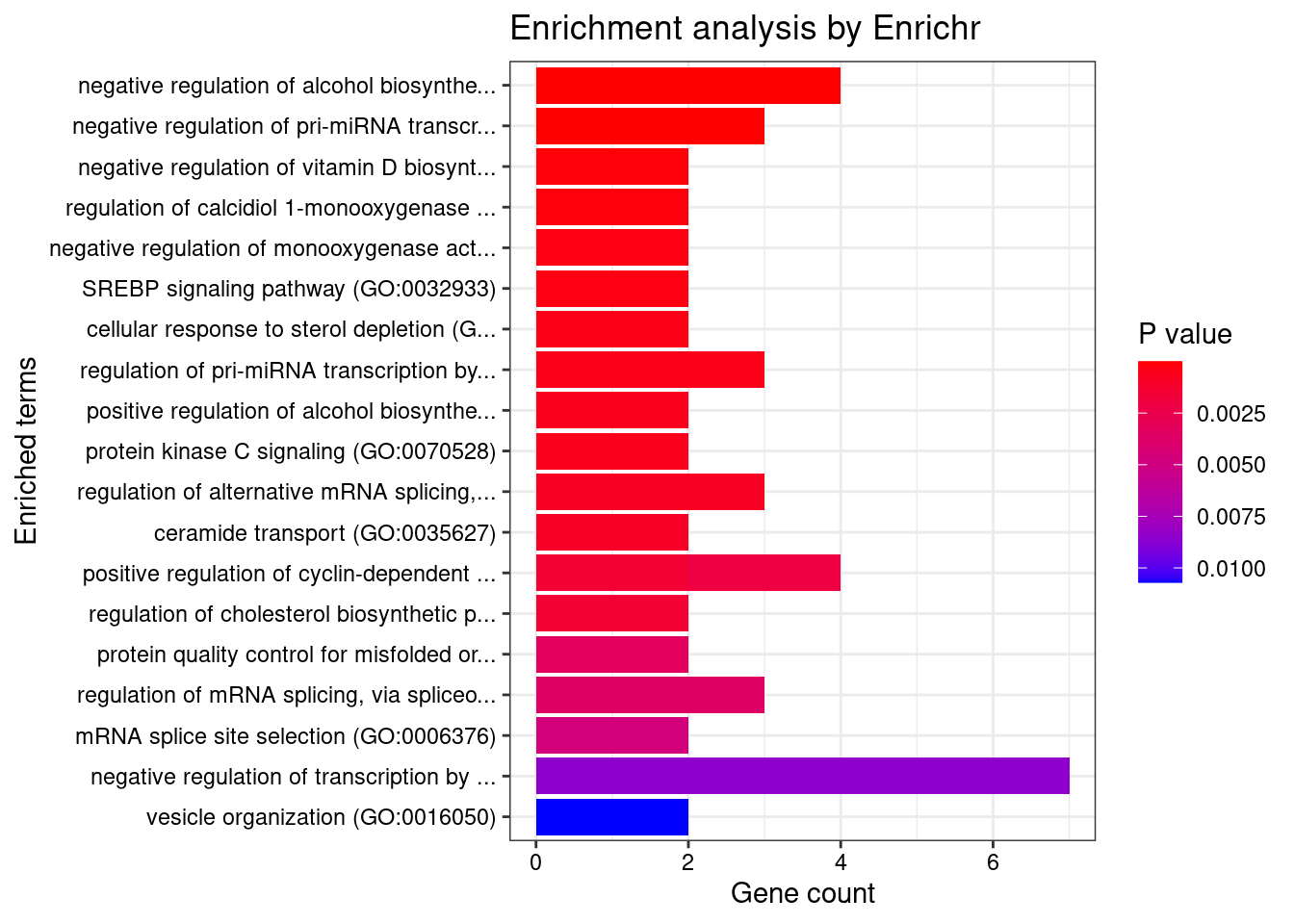
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
Blood or Immune
Number of cTWAS Genes in Tissue Group: 45
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 response to vitamin D (GO:0033280) 2/9 0.02819488 CYP27B1;SFRP1
2 negative regulation of cell growth (GO:0030308) 4/126 0.02819488 CYP27B1;SFRP1;SMAD3;PSRC1
3 negative regulation of growth (GO:0045926) 4/126 0.02819488 CYP27B1;SFRP1;SMAD3;PSRC1
4 positive regulation of canonical Wnt signaling pathway (GO:0090263) 4/130 0.02819488 PPM1B;SFRP1;PSMC3;PSMB10
5 positive regulation of Wnt signaling pathway (GO:0030177) 4/153 0.04197457 PPM1B;SFRP1;PSMC3;PSMB10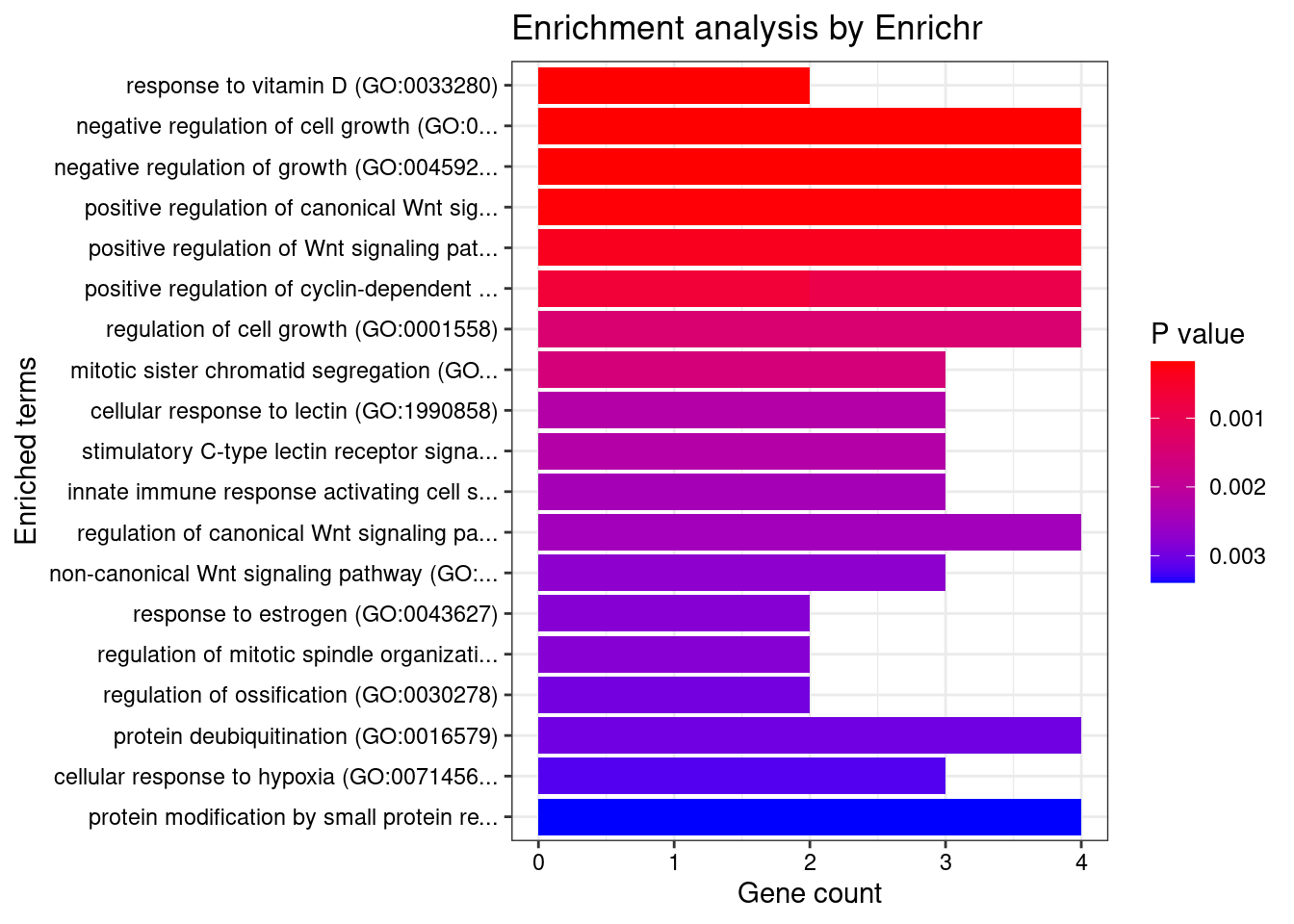
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
Digestive
Number of cTWAS Genes in Tissue Group: 110
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)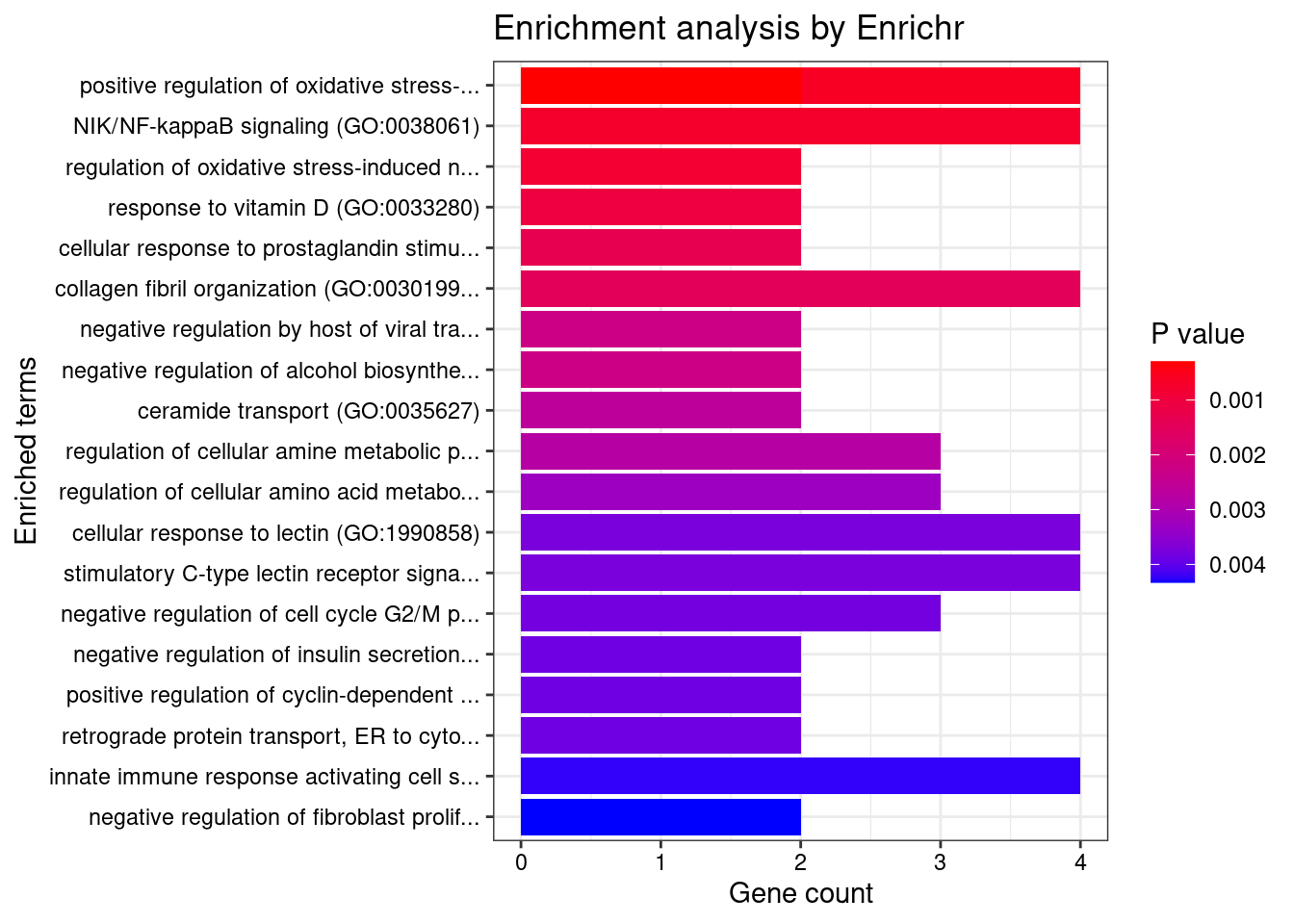
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
KEGG
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_group, referenceGene=background_group,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 61
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Endocrine
Number of cTWAS Genes in Tissue Group: 90
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Cardiovascular
Number of cTWAS Genes in Tissue Group: 111
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
CNS
Number of cTWAS Genes in Tissue Group: 107
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
None
Number of cTWAS Genes in Tissue Group: 138
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Skin
Number of cTWAS Genes in Tissue Group: 68
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Blood or Immune
Number of cTWAS Genes in Tissue Group: 45
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Digestive
Number of cTWAS Genes in Tissue Group: 110
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_group, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 61ARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDSLC16A9 gene(s) from the input list not found in DisGeNET CURATEDRP11-322E11.5 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDHOOK2 gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDTMCC1 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDAPOBEC3G gene(s) from the input list not found in DisGeNET CURATEDC2orf40 gene(s) from the input list not found in DisGeNET CURATEDSPCS2 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDCCL4L2 gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDRAD18 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
89 Rickets 0.008233706 2/30 4/9703
193 Myasthenic Syndromes, Congenital 0.008233706 3/30 24/9703
6 Sertoli-Leydig cell tumor of ovary 0.024904311 1/30 1/9703
38 Gynandroblastoma 0.024904311 1/30 1/9703
81 Noonan Syndrome 0.024904311 2/30 24/9703
92 Sertoli Cell Tumor 0.024904311 1/30 1/9703
126 Sertoli-Leydig Cell Tumor 0.024904311 1/30 1/9703
129 Hereditary orotic aciduria 0.024904311 1/30 1/9703
139 Jansen type metaphyseal chondrodysplasia 0.024904311 1/30 1/9703
141 Orotic aciduria 0.024904311 1/30 1/9703
142 Hereditary orotic aciduria, type 1 0.024904311 1/30 1/9703
165 Epidermolysis bullosa simplex, Ogna type 0.024904311 1/30 1/9703
195 Congenital Myasthenic Syndromes, Presynaptic 0.024904311 2/30 19/9703
212 Pleuropulmonary blastoma 0.024904311 1/30 1/9703
227 ABCD syndrome 0.024904311 1/30 1/9703
228 HIRSCHSPRUNG DISEASE, SUSCEPTIBILITY TO, 2 0.024904311 1/30 1/9703
229 Eiken Skeletal Dysplasia 0.024904311 1/30 1/9703
233 Failure of Tooth Eruption, Primary 0.024904311 1/30 1/9703
235 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.024904311 1/30 1/9703
236 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.024904311 1/30 1/9703
239 Chondrodysplasia, blomstrand type 0.024904311 1/30 1/9703
241 Combined Oxidative Phosphorylation Deficiency 3 0.024904311 1/30 1/9703
242 Rhabdomyosarcoma, Embryonal, 2 0.024904311 1/30 1/9703
248 ATAXIA, SPASTIC, 1, AUTOSOMAL DOMINANT 0.024904311 1/30 1/9703
256 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.024904311 1/30 1/9703
260 AGAMMAGLOBULINEMIA 6, AUTOSOMAL RECESSIVE 0.024904311 1/30 1/9703
261 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.024904311 1/30 1/9703
271 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.024904311 1/30 1/9703
276 DICER1 syndrome 0.024904311 1/30 1/9703
279 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.024904311 1/30 1/9703
280 AGAMMAGLOBULINEMIA 6 0.024904311 1/30 1/9703
284 ROBINOW SYNDROME, AUTOSOMAL DOMINANT 3 0.024904311 1/30 1/9703
285 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.024904311 1/30 1/9703
286 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.024904311 1/30 1/9703
287 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.024904311 1/30 1/9703
293 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.024904311 1/30 1/9703
298 GLOBAL DEVELOPMENTAL DELAY, LUNG CYSTS, OVERGROWTH, AND WILMS TUMOR 0.024904311 1/30 1/9703
255 Bilateral Wilms Tumor 0.027947875 2/30 29/9703
57 Lambert-Eaton Myasthenic Syndrome 0.037554375 1/30 2/9703
78 Nephroblastoma 0.037554375 2/30 36/9703
127 Sex Cord-Stromal Tumor 0.037554375 1/30 2/9703
143 Vitamin D-dependent rickets, type 1 0.037554375 1/30 2/9703
154 Euthyroid Goiter 0.037554375 1/30 2/9703
155 Malignant Granulosa Cell Tumor 0.037554375 1/30 2/9703
171 Myasthenic Syndrome 0.037554375 1/30 2/9703
206 Congenital hypoplastic anemia 0.037554375 1/30 2/9703
221 NUT midline carcinoma 0.037554375 1/30 2/9703
245 Alcohol Toxicity 0.037554375 1/30 2/9703
251 Epidermolysis Bullosa Simplex With Pyloric Atresia 0.037554375 1/30 2/9703
55 Pulmonary Hypertension 0.039955530 2/30 40/9703
15 Malignant Neoplasms 0.041051109 3/30 128/9703
173 Thyroid carcinoma 0.046239764 2/30 44/9703
53 Hyperparathyroidism 0.046713943 1/30 3/9703
198 Acute schizophrenia 0.046713943 1/30 3/9703
238 Epidermolysis bullosa with pyloric atresia 0.046713943 1/30 3/9703
243 POSTAXIAL POLYDACTYLY, TYPE B 0.046713943 1/30 3/9703
270 Ovarian clear cell carcinoma 0.046713943 1/30 3/9703
278 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.046713943 1/30 3/9703
291 Polydactyly, Postaxial, Type A1 0.046713943 1/30 3/9703
20 Colonic Neoplasms 0.048924562 3/30 152/9703
22 Colorectal Neoplasms 0.048924562 4/30 277/9703
30 Enchondromatosis 0.048924562 1/30 4/9703
35 Fibrosis 0.048924562 2/30 50/9703
60 Acute Erythroblastic Leukemia 0.048924562 1/30 4/9703
128 POLYDACTYLY, POSTAXIAL 0.048924562 1/30 4/9703
138 Robinow Syndrome 0.048924562 1/30 4/9703
158 Partial atrioventricular canal 0.048924562 1/30 4/9703
220 Cirrhosis 0.048924562 2/30 50/9703
226 Myasthenic Syndrome, Congenital, with Facial Dysmorphism, associated with Acetylcholine Receptor Deficiency 0.048924562 1/30 4/9703
231 Perisylvian syndrome 0.048924562 1/30 4/9703
232 WAARDENBURG SYNDROME, TYPE 4A 0.048924562 1/30 4/9703
240 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.048924562 1/30 4/9703
268 Male Germ Cell Tumor 0.048924562 1/30 4/9703
283 MYASTHENIC SYNDROME, CONGENITAL, 1A, SLOW-CHANNEL 0.048924562 1/30 4/9703
292 Robinow Syndrome, Autosomal Dominant 0.048924562 1/30 4/9703
16 Malignant tumor of colon 0.049746517 3/30 159/9703
Endocrine
Number of cTWAS Genes in Tissue Group: 90SLC16A9 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATEDC17orf74 gene(s) from the input list not found in DisGeNET CURATEDFAM208A gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDPPP2R3C gene(s) from the input list not found in DisGeNET CURATEDPLCD4 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDSH3D21 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDRP11-503L19.1 gene(s) from the input list not found in DisGeNET CURATEDPODN gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDSPAG8 gene(s) from the input list not found in DisGeNET CURATEDNFE2L1 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDTBC1D3H gene(s) from the input list not found in DisGeNET CURATEDUSP49 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDEZH1 gene(s) from the input list not found in DisGeNET CURATEDOVAAL gene(s) from the input list not found in DisGeNET CURATEDRP4-798A10.4 gene(s) from the input list not found in DisGeNET CURATEDRP11-140I16.3 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDSYNGR2 gene(s) from the input list not found in DisGeNET CURATEDMRPS9 gene(s) from the input list not found in DisGeNET CURATEDCNOT9 gene(s) from the input list not found in DisGeNET CURATEDDCST2 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDCCDC57 gene(s) from the input list not found in DisGeNET CURATEDSPRYD3 gene(s) from the input list not found in DisGeNET CURATEDAF067845.3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
7 Sertoli-Leydig cell tumor of ovary 0.04742547 1/49 1/9703
20 Malignant Neoplasms 0.04742547 5/49 128/9703
54 Gynandroblastoma 0.04742547 1/49 1/9703
60 Hepatosplenomegaly 0.04742547 1/49 1/9703
127 Sertoli Cell Tumor 0.04742547 1/49 1/9703
167 Sertoli-Leydig Cell Tumor 0.04742547 1/49 1/9703
190 Coffin-Siris syndrome 0.04742547 2/49 13/9703
194 Glutaric aciduria, type 1 0.04742547 1/49 1/9703
222 Epidermolysis bullosa simplex, Ogna type 0.04742547 1/49 1/9703
244 Preterm premature rupture of membranes (disorder) 0.04742547 1/49 1/9703
275 Pleuropulmonary blastoma 0.04742547 1/49 1/9703
277 DEVELOPMENTAL DYSPLASIA OF THE HIP 1 0.04742547 1/49 1/9703
291 Angel shaped phalangoepiphyseal dysplasia 0.04742547 1/49 1/9703
295 EPIPHYSEAL DYSPLASIA, MULTIPLE, 3 0.04742547 1/49 1/9703
297 Striatal Degeneration, Autosomal Dominant 0.04742547 1/49 1/9703
301 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.04742547 1/49 1/9703
302 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.04742547 1/49 1/9703
312 Combined Oxidative Phosphorylation Deficiency 3 0.04742547 1/49 1/9703
313 Rhabdomyosarcoma, Embryonal, 2 0.04742547 1/49 1/9703
315 Wegener-Like Granulomatosis 0.04742547 1/49 1/9703
329 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.04742547 1/49 1/9703
335 AGAMMAGLOBULINEMIA 6, AUTOSOMAL RECESSIVE 0.04742547 1/49 1/9703
336 CRANIOECTODERMAL DYSPLASIA 2 0.04742547 1/49 1/9703
337 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.04742547 1/49 1/9703
338 OSTEOGENESIS IMPERFECTA, TYPE X 0.04742547 1/49 1/9703
340 SHORT-RIB THORACIC DYSPLASIA 7 WITH OR WITHOUT POLYDACTYLY 0.04742547 1/49 1/9703
341 PIGMENTED NODULAR ADRENOCORTICAL DISEASE, PRIMARY, 3 0.04742547 1/49 1/9703
348 Epiphyseal Dysplasia, Multiple, with Myopathy 0.04742547 1/49 1/9703
350 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.04742547 1/49 1/9703
358 DICER1 syndrome 0.04742547 1/49 1/9703
361 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.04742547 1/49 1/9703
362 AGAMMAGLOBULINEMIA 6 0.04742547 1/49 1/9703
364 Abnormality of brain morphology 0.04742547 1/49 1/9703
365 Abnormality of the liver 0.04742547 1/49 1/9703
367 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.04742547 1/49 1/9703
374 MYOPIA 25, AUTOSOMAL DOMINANT 0.04742547 1/49 1/9703
377 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.04742547 1/49 1/9703
382 SHORT-RIB THORACIC DYSPLASIA 7/20 WITH POLYDACTYLY, DIGENIC 0.04742547 1/49 1/9703
383 COFFIN-SIRIS SYNDROME 7 0.04742547 1/49 1/9703
384 TRICHOHEPATONEURODEVELOPMENTAL SYNDROME 0.04742547 1/49 1/9703
385 GLOBAL DEVELOPMENTAL DELAY, LUNG CYSTS, OVERGROWTH, AND WILMS TUMOR 0.04742547 1/49 1/9703
Cardiovascular
Number of cTWAS Genes in Tissue Group: 111ARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATEDSAV1 gene(s) from the input list not found in DisGeNET CURATEDTMEM129 gene(s) from the input list not found in DisGeNET CURATEDC1QTNF1 gene(s) from the input list not found in DisGeNET CURATEDSLC2A12 gene(s) from the input list not found in DisGeNET CURATEDUBE2Z gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDC2orf49 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDRP5-1085F17.3 gene(s) from the input list not found in DisGeNET CURATEDZBTB38 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATEDRAD18 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDSPAG8 gene(s) from the input list not found in DisGeNET CURATEDUBE2Q1 gene(s) from the input list not found in DisGeNET CURATEDZNF219 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDCTD-3224K15.3 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDRP11-1277A3.3 gene(s) from the input list not found in DisGeNET CURATEDTRIM34 gene(s) from the input list not found in DisGeNET CURATEDAPOLD1 gene(s) from the input list not found in DisGeNET CURATEDCCDC169 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDAPOBEC3G gene(s) from the input list not found in DisGeNET CURATEDMETTL21B gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDMIR193BHG gene(s) from the input list not found in DisGeNET CURATEDC1QTNF4 gene(s) from the input list not found in DisGeNET CURATEDFAM129A gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDARRDC2 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDSPRYD3 gene(s) from the input list not found in DisGeNET CURATEDRFTN1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
256 Sex Cord-Stromal Tumor 0.02294675 2/59 2/9703
CNS
Number of cTWAS Genes in Tissue Group: 107JMJD4 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDCCDC127 gene(s) from the input list not found in DisGeNET CURATEDPARPBP gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPRSS27 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDPSRC1 gene(s) from the input list not found in DisGeNET CURATEDBET1L gene(s) from the input list not found in DisGeNET CURATEDTULP4 gene(s) from the input list not found in DisGeNET CURATEDHEMK1 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDC14orf80 gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDSAV1 gene(s) from the input list not found in DisGeNET CURATEDRP5-1085F17.3 gene(s) from the input list not found in DisGeNET CURATEDTPCN2 gene(s) from the input list not found in DisGeNET CURATEDNGDN gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDNEU3 gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDCRIP1 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDAL132709.1 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDUBE2Q1 gene(s) from the input list not found in DisGeNET CURATEDRNF169 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDRP11-78O7.2 gene(s) from the input list not found in DisGeNET CURATEDLINC01124 gene(s) from the input list not found in DisGeNET CURATEDCNOT9 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDMIR193BHG gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDEIF4EBP3 gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDAPOBEC3G gene(s) from the input list not found in DisGeNET CURATEDGRIN3B gene(s) from the input list not found in DisGeNET CURATEDUPK1A gene(s) from the input list not found in DisGeNET CURATEDCCDC169 gene(s) from the input list not found in DisGeNET CURATEDNUPR2 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
241 Coffin-Siris syndrome 0.01949747 3/53 13/9703
130 Osteoarthritis of hip 0.03782120 3/53 20/9703
1 Adenocarcinoma 0.04927223 4/53 116/9703
6 Aortic Aneurysm 0.04927223 2/53 7/9703
36 Colorectal Neoplasms 0.04927223 7/53 277/9703
51 Dyschondroplasias 0.04927223 2/53 19/9703
59 Gastrinoma 0.04927223 1/53 1/9703
60 Glucagonoma 0.04927223 1/53 1/9703
70 Hepatosplenomegaly 0.04927223 1/53 1/9703
71 HIV Infections 0.04927223 4/53 103/9703
81 insulinoma 0.04927223 1/53 1/9703
86 Fibroid Tumor 0.04927223 2/53 6/9703
113 Multiple Epiphyseal Dysplasia 0.04927223 2/53 20/9703
160 Uterine Fibroids 0.04927223 2/53 7/9703
193 Adenocarcinoma, Basal Cell 0.04927223 4/53 116/9703
194 Adenocarcinoma, Oxyphilic 0.04927223 4/53 116/9703
195 Carcinoma, Cribriform 0.04927223 4/53 116/9703
196 Carcinoma, Granular Cell 0.04927223 4/53 116/9703
197 Adenocarcinoma, Tubular 0.04927223 4/53 116/9703
233 Islet Cell Tumor 0.04927223 1/53 1/9703
240 Jansen type metaphyseal chondrodysplasia 0.04927223 1/53 1/9703
249 AIDS with Kaposi's sarcoma 0.04927223 1/53 1/9703
254 Carcinoid tumor of lung 0.04927223 1/53 1/9703
266 Malignant mesothelioma 0.04927223 4/53 109/9703
278 Delayed developmental milestones 0.04927223 1/53 1/9703
342 DEVELOPMENTAL DYSPLASIA OF THE HIP 1 0.04927223 1/53 1/9703
358 Angel shaped phalangoepiphyseal dysplasia 0.04927223 1/53 1/9703
362 EPIPHYSEAL DYSPLASIA, MULTIPLE, 3 0.04927223 1/53 1/9703
364 Eiken Skeletal Dysplasia 0.04927223 1/53 1/9703
369 Failure of Tooth Eruption, Primary 0.04927223 1/53 1/9703
370 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.04927223 1/53 1/9703
371 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.04927223 1/53 1/9703
374 Chondrodysplasia, blomstrand type 0.04927223 1/53 1/9703
392 PARATHYROID ADENOMA, SOMATIC 0.04927223 1/53 1/9703
404 LOEYS-DIETZ SYNDROME 3 0.04927223 1/53 1/9703
407 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB 0.04927223 1/53 1/9703
413 Epiphyseal Dysplasia, Multiple, with Myopathy 0.04927223 1/53 1/9703
416 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.04927223 1/53 1/9703
417 Congenital pontocerebellar hypoplasia type 8 0.04927223 1/53 1/9703
429 MENTAL RETARDATION, AUTOSOMAL DOMINANT 27 0.04927223 1/53 1/9703
430 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.04927223 1/53 1/9703
431 LIPOMA, SOMATIC 0.04927223 1/53 1/9703
432 ANGIOFIBROMA, SOMATIC 0.04927223 1/53 1/9703
433 ADRENAL ADENOMA, SOMATIC 0.04927223 1/53 1/9703
441 MYOPIA 25, AUTOSOMAL DOMINANT 0.04927223 1/53 1/9703
442 PONTOCEREBELLAR HYPOPLASIA, TYPE 2F 0.04927223 1/53 1/9703
443 DEVELOPMENTAL DELAY WITH SHORT STATURE, DYSMORPHIC FEATURES, AND SPARSE HAIR 0.04927223 1/53 1/9703
444 HIV Coinfection 0.04927223 4/53 103/9703
447 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.04927223 1/53 1/9703
453 KLHL9-related early-onset distal myopathy 0.04927223 1/53 1/9703
459 COFFIN-SIRIS SYNDROME 7 0.04927223 1/53 1/9703
None
Number of cTWAS Genes in Tissue Group: 138SHE gene(s) from the input list not found in DisGeNET CURATEDJMJD4 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPRSS27 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDPAGR1 gene(s) from the input list not found in DisGeNET CURATEDRAD18 gene(s) from the input list not found in DisGeNET CURATEDDYRK4 gene(s) from the input list not found in DisGeNET CURATEDTRAPPC2B gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDC14orf80 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDFOXN2 gene(s) from the input list not found in DisGeNET CURATEDAPOLD1 gene(s) from the input list not found in DisGeNET CURATEDNGDN gene(s) from the input list not found in DisGeNET CURATEDPRRT3 gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDBCL2L2 gene(s) from the input list not found in DisGeNET CURATEDUSP37 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDSYTL2 gene(s) from the input list not found in DisGeNET CURATEDRFTN1 gene(s) from the input list not found in DisGeNET CURATEDKCNJ12 gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDCNOT9 gene(s) from the input list not found in DisGeNET CURATEDMIR193BHG gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDCTD-3224K15.3 gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDAPOBEC3G gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDTM2D1 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDCOPZ2 gene(s) from the input list not found in DisGeNET CURATEDPSRC1 gene(s) from the input list not found in DisGeNET CURATEDEFCAB8 gene(s) from the input list not found in DisGeNET CURATEDC1QTNF1 gene(s) from the input list not found in DisGeNET CURATEDHEMK1 gene(s) from the input list not found in DisGeNET CURATEDRP11-509I21.2 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDSPAG8 gene(s) from the input list not found in DisGeNET CURATEDPTK6 gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDCCDC187 gene(s) from the input list not found in DisGeNET CURATEDVAMP5 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDCRIP1 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDCCDC57 gene(s) from the input list not found in DisGeNET CURATEDLPCAT1 gene(s) from the input list not found in DisGeNET CURATEDFAM180B gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDSLC45A4 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDC17orf82 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDUSP39 gene(s) from the input list not found in DisGeNET CURATEDGRIN3B gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
5 Aortic Aneurysm 0.04348183 2/69 7/9703
6 Sertoli-Leydig cell tumor of ovary 0.04348183 1/69 1/9703
33 Delusional disorder 0.04348183 1/69 1/9703
52 Gastrinoma 0.04348183 1/69 1/9703
57 Glucagonoma 0.04348183 1/69 1/9703
58 Gynandroblastoma 0.04348183 1/69 1/9703
65 Hepatosplenomegaly 0.04348183 1/69 1/9703
74 insulinoma 0.04348183 1/69 1/9703
81 Fibroid Tumor 0.04348183 2/69 6/9703
104 Nephroblastoma 0.04348183 3/69 36/9703
116 Acrocephaly 0.04348183 2/69 16/9703
119 Periapical Periodontitis 0.04348183 1/69 1/9703
120 Pituitary Adenoma 0.04348183 2/69 9/9703
130 Rickets 0.04348183 2/69 4/9703
135 Sertoli Cell Tumor 0.04348183 1/69 1/9703
146 Uterine Fibroids 0.04348183 2/69 7/9703
173 Congenital absent nipple 0.04348183 1/69 1/9703
183 Carcinoma, Neuroendocrine 0.04348183 2/69 12/9703
185 Sertoli-Leydig Cell Tumor 0.04348183 1/69 1/9703
191 Brachycephaly 0.04348183 2/69 17/9703
201 Islet Cell Tumor 0.04348183 1/69 1/9703
207 Jansen type metaphyseal chondrodysplasia 0.04348183 1/69 1/9703
208 Coffin-Siris syndrome 0.04348183 2/69 13/9703
209 Scaphycephaly 0.04348183 2/69 16/9703
210 Trigonocephaly 0.04348183 2/69 16/9703
227 Carcinoid tumor of lung 0.04348183 1/69 1/9703
232 Lipoblastomatosis 0.04348183 1/69 1/9703
261 Congenital absence of breast with absent nipple 0.04348183 1/69 1/9703
265 Delayed developmental milestones 0.04348183 1/69 1/9703
271 Periodontitis, Acute Nonsuppurative 0.04348183 1/69 1/9703
286 Preterm premature rupture of membranes (disorder) 0.04348183 1/69 1/9703
339 Lipoblastoma 0.04348183 1/69 1/9703
344 Pleuropulmonary blastoma 0.04348183 1/69 1/9703
360 Synostotic Posterior Plagiocephaly 0.04348183 2/69 16/9703
361 CHARCOT-MARIE-TOOTH DISEASE, TYPE 4H 0.04348183 1/69 1/9703
366 Eiken Skeletal Dysplasia 0.04348183 1/69 1/9703
374 Failure of Tooth Eruption, Primary 0.04348183 1/69 1/9703
376 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.04348183 1/69 1/9703
377 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.04348183 1/69 1/9703
380 Chondrodysplasia, blomstrand type 0.04348183 1/69 1/9703
381 Metopic synostosis 0.04348183 2/69 16/9703
383 Combined Oxidative Phosphorylation Deficiency 3 0.04348183 1/69 1/9703
384 Spondyloepimetaphyseal Dysplasia With Abnormal Dentition 0.04348183 1/69 1/9703
385 Rhabdomyosarcoma, Embryonal, 2 0.04348183 1/69 1/9703
390 ATAXIA, SPASTIC, 1, AUTOSOMAL DOMINANT 0.04348183 1/69 1/9703
393 PARATHYROID ADENOMA, SOMATIC 0.04348183 1/69 1/9703
394 Spondylocheirodysplasia, Ehlers-Danlos Syndrome-Like 0.04348183 1/69 1/9703
397 Bilateral Wilms Tumor 0.04348183 3/69 29/9703
399 Synostotic Anterior Plagiocephaly 0.04348183 2/69 16/9703
405 CRANIOECTODERMAL DYSPLASIA 2 0.04348183 1/69 1/9703
406 CARDIOMYOPATHY, DILATED, 1GG 0.04348183 1/69 1/9703
407 LOEYS-DIETZ SYNDROME 3 0.04348183 1/69 1/9703
408 OSTEOGENESIS IMPERFECTA, TYPE X 0.04348183 1/69 1/9703
411 SHORT-RIB THORACIC DYSPLASIA 7 WITH OR WITHOUT POLYDACTYLY 0.04348183 1/69 1/9703
412 PARAGANGLIOMAS 5 0.04348183 1/69 1/9703
413 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB 0.04348183 1/69 1/9703
423 AORTIC VALVE DISEASE 2 0.04348183 1/69 1/9703
425 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.04348183 1/69 1/9703
431 DICER1 syndrome 0.04348183 1/69 1/9703
436 DEAFNESS, AUTOSOMAL DOMINANT 27 0.04348183 1/69 1/9703
437 WILMS TUMOR, SUSCEPTIBILITY TO 0.04348183 1/69 1/9703
440 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.04348183 1/69 1/9703
441 BREASTS AND/OR NIPPLES, APLASIA OR HYPOPLASIA OF, 2 0.04348183 1/69 1/9703
442 SPINOCEREBELLAR ATAXIA, AUTOSOMAL RECESSIVE 17 0.04348183 1/69 1/9703
443 LIPOMA, SOMATIC 0.04348183 1/69 1/9703
444 ANGIOFIBROMA, SOMATIC 0.04348183 1/69 1/9703
445 ADRENAL ADENOMA, SOMATIC 0.04348183 1/69 1/9703
449 ROBINOW SYNDROME, AUTOSOMAL DOMINANT 3 0.04348183 1/69 1/9703
450 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.04348183 1/69 1/9703
451 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.04348183 1/69 1/9703
458 PONTOCEREBELLAR HYPOPLASIA, TYPE 2F 0.04348183 1/69 1/9703
462 CRANIOSYNOSTOSIS 7 0.04348183 1/69 1/9703
466 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.04348183 1/69 1/9703
467 Craniosynostosis, Type 1 0.04348183 2/69 16/9703
474 Familial thoracic aortic aneurysm and aortic dissection 0.04348183 3/69 53/9703
479 SHORT-RIB THORACIC DYSPLASIA 7/20 WITH POLYDACTYLY, DIGENIC 0.04348183 1/69 1/9703
480 COFFIN-SIRIS SYNDROME 7 0.04348183 1/69 1/9703
481 NEPHROTIC SYNDROME, TYPE 19 0.04348183 1/69 1/9703
483 GLOBAL DEVELOPMENTAL DELAY, LUNG CYSTS, OVERGROWTH, AND WILMS TUMOR 0.04348183 1/69 1/9703
313 Congenital Myasthenic Syndromes, Presynaptic 0.04760818 2/69 19/9703
166 Myocardial Ischemia 0.04887126 5/69 176/9703
Skin
Number of cTWAS Genes in Tissue Group: 68UPK1A gene(s) from the input list not found in DisGeNET CURATEDCCDC127 gene(s) from the input list not found in DisGeNET CURATEDMUCL1 gene(s) from the input list not found in DisGeNET CURATEDSLC45A4 gene(s) from the input list not found in DisGeNET CURATEDKIAA1644 gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDHOOK2 gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDNFATC4 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDRNF167 gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDZYG11B gene(s) from the input list not found in DisGeNET CURATEDJMJD4 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDTATDN2 gene(s) from the input list not found in DisGeNET CURATEDTC2N gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDZCCHC24 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDZNF827 gene(s) from the input list not found in DisGeNET CURATEDRFX3-AS1 gene(s) from the input list not found in DisGeNET CURATEDNATD1 gene(s) from the input list not found in DisGeNET CURATEDAPOLD1 gene(s) from the input list not found in DisGeNET CURATEDCRIP1 gene(s) from the input list not found in DisGeNET CURATEDUGGT2 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCELF6 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDSMIM24 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
292 Liver carcinoma 0.006976647 9/31 507/9703
100 Rickets 0.010542685 2/31 4/9703
296 Bilateral Wilms Tumor 0.012149405 3/31 29/9703
87 Nephroblastoma 0.017537488 3/31 36/9703
4 Sertoli-Leydig cell tumor of ovary 0.026527736 1/31 1/9703
48 Gynandroblastoma 0.026527736 1/31 1/9703
53 Hepatosplenomegaly 0.026527736 1/31 1/9703
63 Jaundice, Obstructive 0.026527736 1/31 1/9703
104 Sertoli Cell Tumor 0.026527736 1/31 1/9703
135 Dermatofibrosarcoma 0.026527736 1/31 1/9703
138 Sertoli-Leydig Cell Tumor 0.026527736 1/31 1/9703
159 Jansen type metaphyseal chondrodysplasia 0.026527736 1/31 1/9703
182 Dermatofibrosarcoma Protuberans, Myxoid 0.026527736 1/31 1/9703
183 Pigmented Dermatofibrosarcoma Protuberans (Bednar Tumor) 0.026527736 1/31 1/9703
252 Cholecystolithiasis 0.026527736 1/31 1/9703
260 Pleuropulmonary blastoma 0.026527736 1/31 1/9703
275 Eiken Skeletal Dysplasia 0.026527736 1/31 1/9703
276 Neutropenia, Nonimmune Chronic Idiopathic, Adult 0.026527736 1/31 1/9703
278 Failure of Tooth Eruption, Primary 0.026527736 1/31 1/9703
279 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.026527736 1/31 1/9703
280 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.026527736 1/31 1/9703
282 Chondrodysplasia, blomstrand type 0.026527736 1/31 1/9703
283 BARDET-BIEDL SYNDROME 7 0.026527736 1/31 1/9703
286 Cholestasis, progressive familial intrahepatic 3 0.026527736 1/31 1/9703
287 Rhabdomyosarcoma, Embryonal, 2 0.026527736 1/31 1/9703
293 Low phospholipid-associated cholelithiasis 0.026527736 1/31 1/9703
294 Pontocerebellar Hypoplasia Type 2C 0.026527736 1/31 1/9703
295 Neutropenia, Severe Congenital, Autosomal Dominant 2 0.026527736 1/31 1/9703
306 Osteogenesis Imperfecta, Type VI 0.026527736 1/31 1/9703
315 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.026527736 1/31 1/9703
316 CHOLESTASIS, INTRAHEPATIC, OF PREGNANCY 3 0.026527736 1/31 1/9703
319 Metastatic Dermatofibrosarcoma Protuberans 0.026527736 1/31 1/9703
320 Giant Cell Fibroblastoma 0.026527736 1/31 1/9703
323 BASAL GANGLIA CALCIFICATION, IDIOPATHIC, 5 0.026527736 1/31 1/9703
325 DICER1 syndrome 0.026527736 1/31 1/9703
328 DEAFNESS, AUTOSOMAL DOMINANT 27 0.026527736 1/31 1/9703
329 WILMS TUMOR, SUSCEPTIBILITY TO 0.026527736 1/31 1/9703
332 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.026527736 1/31 1/9703
336 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.026527736 1/31 1/9703
337 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.026527736 1/31 1/9703
346 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.026527736 1/31 1/9703
353 KLHL9-related early-onset distal myopathy 0.026527736 1/31 1/9703
357 GLOBAL DEVELOPMENTAL DELAY, LUNG CYSTS, OVERGROWTH, AND WILMS TUMOR 0.026527736 1/31 1/9703
24 Cholelithiasis 0.041415595 1/31 2/9703
31 Corneal Ulcer 0.041415595 1/31 2/9703
47 Gingival Hyperplasia 0.041415595 1/31 2/9703
139 Sex Cord-Stromal Tumor 0.041415595 1/31 2/9703
169 Vitamin D-dependent rickets, type 1 0.041415595 1/31 2/9703
179 Euthyroid Goiter 0.041415595 1/31 2/9703
181 Malignant Granulosa Cell Tumor 0.041415595 1/31 2/9703
193 Dermatofibrosarcoma Protuberans 0.041415595 1/31 2/9703
290 Alcohol Toxicity 0.041415595 1/31 2/9703
331 DIARRHEA 7, PROTEIN-LOSING ENTEROPATHY TYPE 0.041415595 1/31 2/9703
333 Abnormality of the respiratory system 0.041415595 1/31 2/9703
356 DIARRHEA 10, PROTEIN-LOSING ENTEROPATHY TYPE 0.041415595 1/31 2/9703
16 Malignant Neoplasms 0.049114651 3/31 128/9703
58 Hyperparathyroidism 0.049442113 1/31 3/9703
96 Raynaud Disease 0.049442113 1/31 3/9703
97 Raynaud Phenomenon 0.049442113 1/31 3/9703
165 Progressive intrahepatic cholestasis (disorder) 0.049442113 1/31 3/9703
195 Hereditary gingival fibromatosis 0.049442113 1/31 3/9703
243 Acute schizophrenia 0.049442113 1/31 3/9703
288 POSTAXIAL POLYDACTYLY, TYPE B 0.049442113 1/31 3/9703
313 Ovarian clear cell carcinoma 0.049442113 1/31 3/9703
314 MENINGIOMA, FAMILIAL, SUSCEPTIBILITY TO 0.049442113 1/31 3/9703
330 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.049442113 1/31 3/9703
341 Polydactyly, Postaxial, Type A1 0.049442113 1/31 3/9703
348 Idiopathic basal ganglia calcification 1 0.049442113 1/31 3/9703
350 Cholestasis, benign recurrent intrahepatic 1 0.049442113 1/31 3/9703
29 Colorectal Neoplasms 0.049908213 4/31 277/9703
33 Deglutition Disorders 0.049908213 1/31 4/9703
40 Enchondromatosis 0.049908213 1/31 4/9703
46 Fibrosis 0.049908213 2/31 50/9703
72 Liver neoplasms 0.049908213 3/31 142/9703
141 POLYDACTYLY, POSTAXIAL 0.049908213 1/31 4/9703
163 Oropharyngeal Dysphagia 0.049908213 1/31 4/9703
164 Esophageal Dysphagia 0.049908213 1/31 4/9703
186 Partial atrioventricular canal 0.049908213 1/31 4/9703
187 Malignant neoplasm of liver 0.049908213 3/31 142/9703
218 End Stage Liver Disease 0.049908213 1/31 4/9703
271 Cirrhosis 0.049908213 2/31 50/9703
274 Myasthenic Syndrome, Congenital, with Facial Dysmorphism, associated with Acetylcholine Receptor Deficiency 0.049908213 1/31 4/9703
277 Perisylvian syndrome 0.049908213 1/31 4/9703
284 Neutropenia, Severe Congenital, Autosomal Dominant 1 0.049908213 1/31 4/9703
285 Megalanecephaly Polymicrogyria-Polydactyly Hydrocephalus Syndrome 0.049908213 1/31 4/9703
299 Pontocerebellar Hypoplasia Type 2 0.049908213 1/31 4/9703
300 Chronic Liver Failure 0.049908213 1/31 4/9703
310 Familial intrahepatic cholestasis of pregnancy 0.049908213 1/31 4/9703
312 Male Germ Cell Tumor 0.049908213 1/31 4/9703
335 MYASTHENIC SYNDROME, CONGENITAL, 1A, SLOW-CHANNEL 0.049908213 1/31 4/9703
349 Cholestasis, progressive familial intrahepatic 1 0.049908213 1/31 4/9703
Blood or Immune
Number of cTWAS Genes in Tissue Group: 45ATP5G1 gene(s) from the input list not found in DisGeNET CURATEDB4GALNT3 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDPAPD4 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDPSRC1 gene(s) from the input list not found in DisGeNET CURATEDUSP49 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDRNF181 gene(s) from the input list not found in DisGeNET CURATEDUBE2Q1 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDMETTL8 gene(s) from the input list not found in DisGeNET CURATEDARID3A gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDSYCE2 gene(s) from the input list not found in DisGeNET CURATEDMUCL1 gene(s) from the input list not found in DisGeNET CURATEDTC2N gene(s) from the input list not found in DisGeNET CURATEDRP5-1085F17.3 gene(s) from the input list not found in DisGeNET CURATEDHAGHL gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
46 Fibroid Tumor 0.01244424 2/23 6/9703
78 Uterine Fibroids 0.01244424 2/23 7/9703
39 HIV Infections 0.03095784 3/23 103/9703
123 Epidermolysis bullosa simplex, Ogna type 0.03095784 1/23 1/9703
151 Myasthenic Syndromes, Congenital 0.03095784 2/23 24/9703
178 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.03095784 1/23 1/9703
179 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.03095784 1/23 1/9703
183 Combined Oxidative Phosphorylation Deficiency 3 0.03095784 1/23 1/9703
187 ATAXIA, SPASTIC, 1, AUTOSOMAL DOMINANT 0.03095784 1/23 1/9703
194 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.03095784 1/23 1/9703
197 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.03095784 1/23 1/9703
198 LOEYS-DIETZ SYNDROME 3 0.03095784 1/23 1/9703
203 Congenital pontocerebellar hypoplasia type 8 0.03095784 1/23 1/9703
208 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.03095784 1/23 1/9703
209 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.03095784 1/23 1/9703
216 HIV Coinfection 0.03095784 3/23 103/9703
217 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.03095784 1/23 1/9703
44 Keloid 0.03762637 2/23 35/9703
24 Colorectal Neoplasms 0.04380721 4/23 277/9703
45 Lambert-Eaton Myasthenic Syndrome 0.04380721 1/23 2/9703
109 Vitamin D-dependent rickets, type 1 0.04380721 1/23 2/9703
131 Myasthenic Syndrome 0.04380721 1/23 2/9703
186 Alcohol Toxicity 0.04380721 1/23 2/9703
190 Epidermolysis Bullosa Simplex With Pyloric Atresia 0.04380721 1/23 2/9703
34 Fibrosis 0.04773551 2/23 50/9703
69 Raynaud Disease 0.04773551 1/23 3/9703
70 Raynaud Phenomenon 0.04773551 1/23 3/9703
173 Cirrhosis 0.04773551 2/23 50/9703
175 Uniparental disomy, paternal, chromosome 14 0.04773551 1/23 3/9703
181 Epidermolysis bullosa with pyloric atresia 0.04773551 1/23 3/9703
184 POSTAXIAL POLYDACTYLY, TYPE B 0.04773551 1/23 3/9703
207 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 1 0.04773551 1/23 3/9703
214 Polydactyly, Postaxial, Type A1 0.04773551 1/23 3/9703
Digestive
Number of cTWAS Genes in Tissue Group: 110ARHGEF26 gene(s) from the input list not found in DisGeNET CURATEDWRAP73 gene(s) from the input list not found in DisGeNET CURATEDEIF4EBP3 gene(s) from the input list not found in DisGeNET CURATEDDDX42 gene(s) from the input list not found in DisGeNET CURATEDSAV1 gene(s) from the input list not found in DisGeNET CURATEDTMEM129 gene(s) from the input list not found in DisGeNET CURATEDCFAP54 gene(s) from the input list not found in DisGeNET CURATEDMUCL1 gene(s) from the input list not found in DisGeNET CURATEDRHBDD1 gene(s) from the input list not found in DisGeNET CURATEDZNF3 gene(s) from the input list not found in DisGeNET CURATEDUBE2Z gene(s) from the input list not found in DisGeNET CURATEDPINLYP gene(s) from the input list not found in DisGeNET CURATEDDISP2 gene(s) from the input list not found in DisGeNET CURATEDZNF740 gene(s) from the input list not found in DisGeNET CURATEDHEMK1 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDTMCC1 gene(s) from the input list not found in DisGeNET CURATEDZBTB38 gene(s) from the input list not found in DisGeNET CURATEDCTSW gene(s) from the input list not found in DisGeNET CURATEDAMIGO1 gene(s) from the input list not found in DisGeNET CURATEDC2orf40 gene(s) from the input list not found in DisGeNET CURATEDNARFL gene(s) from the input list not found in DisGeNET CURATEDTHUMPD1 gene(s) from the input list not found in DisGeNET CURATEDSRSF12 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDCRIPAK gene(s) from the input list not found in DisGeNET CURATEDRAD18 gene(s) from the input list not found in DisGeNET CURATEDH1FX gene(s) from the input list not found in DisGeNET CURATEDPLCD3 gene(s) from the input list not found in DisGeNET CURATEDJMJD4 gene(s) from the input list not found in DisGeNET CURATEDDCST1 gene(s) from the input list not found in DisGeNET CURATEDPLEKHA3 gene(s) from the input list not found in DisGeNET CURATEDRGP1 gene(s) from the input list not found in DisGeNET CURATEDHIST1H2BD gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDPSMA1 gene(s) from the input list not found in DisGeNET CURATEDIL17RE gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDKRT80 gene(s) from the input list not found in DisGeNET CURATEDLINC01001 gene(s) from the input list not found in DisGeNET CURATEDPSRC1 gene(s) from the input list not found in DisGeNET CURATEDRP11-394I13.2 gene(s) from the input list not found in DisGeNET CURATEDMETTL21B gene(s) from the input list not found in DisGeNET CURATEDMPHOSPH6 gene(s) from the input list not found in DisGeNET CURATEDUNC50 gene(s) from the input list not found in DisGeNET CURATEDTMEM150A gene(s) from the input list not found in DisGeNET CURATEDZNF213 gene(s) from the input list not found in DisGeNET CURATEDQSOX2 gene(s) from the input list not found in DisGeNET CURATEDMIR193BHG gene(s) from the input list not found in DisGeNET CURATEDDCST2 gene(s) from the input list not found in DisGeNET CURATEDARHGEF40 gene(s) from the input list not found in DisGeNET CURATEDHOMEZ gene(s) from the input list not found in DisGeNET CURATEDC3orf18 gene(s) from the input list not found in DisGeNET CURATEDCNIH4 gene(s) from the input list not found in DisGeNET CURATEDEFCAB8 gene(s) from the input list not found in DisGeNET CURATEDPRSS27 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
56 Hepatosplenomegaly 0.04895429 1/54 1/9703
66 Jaundice, Obstructive 0.04895429 1/54 1/9703
165 Dyggve-Melchior-Clausen syndrome 0.04895429 1/54 1/9703
166 Coffin-Siris syndrome 0.04895429 2/54 13/9703
172 Glutaric aciduria, type 1 0.04895429 1/54 1/9703
204 Epidermolysis bullosa simplex, Ogna type 0.04895429 1/54 1/9703
207 Delayed developmental milestones 0.04895429 1/54 1/9703
222 Preterm premature rupture of membranes (disorder) 0.04895429 1/54 1/9703
243 Myasthenic Syndromes, Congenital 0.04895429 3/54 24/9703
245 Congenital Myasthenic Syndromes, Presynaptic 0.04895429 2/54 19/9703
255 Cholecystolithiasis 0.04895429 1/54 1/9703
283 EPIPHYSEAL DYSPLASIA, MULTIPLE, 3 0.04895429 1/54 1/9703
287 X-linked Dyggve-Melchior-Clausen syndrome 0.04895429 1/54 1/9703
295 ATRIOVENTRICULAR SEPTAL DEFECT, SUSCEPTIBILITY TO, 2 0.04895429 1/54 1/9703
296 Atrioventricular Septal Defect, Partial, with Heterotaxy Syndrome 0.04895429 1/54 1/9703
301 Radially deviated wrists 0.04895429 1/54 1/9703
303 Combined Oxidative Phosphorylation Deficiency 3 0.04895429 1/54 1/9703
304 Cholestasis, progressive familial intrahepatic 3 0.04895429 1/54 1/9703
309 ATAXIA, SPASTIC, 1, AUTOSOMAL DOMINANT 0.04895429 1/54 1/9703
313 Low phospholipid-associated cholelithiasis 0.04895429 1/54 1/9703
318 Epidermolysa bullosa simplex and limb girdle muscular dystrophy 0.04895429 1/54 1/9703
325 AGAMMAGLOBULINEMIA 6, AUTOSOMAL RECESSIVE 0.04895429 1/54 1/9703
326 CARDIOMYOPATHY, DILATED, 1GG 0.04895429 1/54 1/9703
327 MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 2Q 0.04895429 1/54 1/9703
328 OSTEOGENESIS IMPERFECTA, TYPE X 0.04895429 1/54 1/9703
333 PARAGANGLIOMAS 5 0.04895429 1/54 1/9703
335 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB 0.04895429 1/54 1/9703
342 Epiphyseal Dysplasia, Multiple, with Myopathy 0.04895429 1/54 1/9703
344 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.04895429 1/54 1/9703
345 CHOLESTASIS, INTRAHEPATIC, OF PREGNANCY 3 0.04895429 1/54 1/9703
350 DEAFNESS, AUTOSOMAL DOMINANT 27 0.04895429 1/54 1/9703
351 SMITH-MCCORT DYSPLASIA 1 0.04895429 1/54 1/9703
352 WILMS TUMOR, SUSCEPTIBILITY TO 0.04895429 1/54 1/9703
353 WHITE SPONGE NEVUS 1 0.04895429 1/54 1/9703
355 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3 0.04895429 1/54 1/9703
356 AGAMMAGLOBULINEMIA 6 0.04895429 1/54 1/9703
360 EPIDERMOLYSIS BULLOSA SIMPLEX WITH NAIL DYSTROPHY 0.04895429 1/54 1/9703
361 MYASTHENIC SYNDROME, CONGENITAL, 2C, ASSOCIATED WITH ACETYLCHOLINE RECEPTOR DEFICIENCY 0.04895429 1/54 1/9703
362 MYASTHENIC SYNDROME, CONGENITAL, 2A, SLOW-CHANNEL 0.04895429 1/54 1/9703
367 MYOPIA 25, AUTOSOMAL DOMINANT 0.04895429 1/54 1/9703
368 CILIARY DYSKINESIA, PRIMARY, 35 0.04895429 1/54 1/9703
369 PONTOCEREBELLAR HYPOPLASIA, TYPE 2F 0.04895429 1/54 1/9703
377 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.04895429 1/54 1/9703
387 COFFIN-SIRIS SYNDROME 7 0.04895429 1/54 1/9703Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_group=ctwas_genes_group)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_group]})
#hypergeometric test
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_group)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
#multiple testing correction
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 61
gene_set nset ngenes percent padj
1 gwascatalog 4611 34 0.007373672 0.000366677
2 clinvar_path_likelypath 2144 14 0.006529851 0.136376977
3 mgi_essential 1729 9 0.005205321 0.483179566
4 fda_approved_drug_targets 260 1 0.003846154 0.831680987
5 core_essentials_hart 212 0 0.000000000 1.000000000
Endocrine
Number of cTWAS Genes in Tissue Group: 90
gene_set nset ngenes percent padj
1 gwascatalog 5415 49 0.009048938 1.142949e-06
2 mgi_essential 2030 19 0.009359606 7.078294e-03
3 clinvar_path_likelypath 2493 19 0.007621340 3.964408e-02
4 fda_approved_drug_targets 302 2 0.006622517 5.271175e-01
5 core_essentials_hart 239 0 0.000000000 1.000000e+00
Cardiovascular
Number of cTWAS Genes in Tissue Group: 111
gene_set nset ngenes percent padj
1 gwascatalog 5199 60 0.011540681 4.225292e-06
2 fda_approved_drug_targets 286 7 0.024475524 8.280533e-03
3 clinvar_path_likelypath 2407 26 0.010801828 1.559600e-02
4 mgi_essential 1969 19 0.009649568 8.727542e-02
5 core_essentials_hart 243 1 0.004115226 8.103985e-01
CNS
Number of cTWAS Genes in Tissue Group: 107
gene_set nset ngenes percent padj
1 gwascatalog 5430 50 0.009208103 0.001387099
2 mgi_essential 2086 18 0.008628955 0.177775690
3 fda_approved_drug_targets 316 4 0.012658228 0.203615764
4 clinvar_path_likelypath 2528 19 0.007515823 0.218813045
5 core_essentials_hart 245 2 0.008163265 0.433377289
None
Number of cTWAS Genes in Tissue Group: 138
gene_set nset ngenes percent padj
1 gwascatalog 5644 69 0.012225372 4.953859e-06
2 mgi_essential 2139 22 0.010285180 1.173228e-01
3 clinvar_path_likelypath 2608 26 0.009969325 1.173228e-01
4 core_essentials_hart 256 1 0.003906250 8.533301e-01
5 fda_approved_drug_targets 320 2 0.006250000 8.533301e-01
Skin
Number of cTWAS Genes in Tissue Group: 68
gene_set nset ngenes percent padj
1 gwascatalog 5122 33 0.006442796 0.01168942
2 clinvar_path_likelypath 2345 15 0.006396588 0.14239718
3 mgi_essential 1930 10 0.005181347 0.39840492
4 fda_approved_drug_targets 275 2 0.007272727 0.39840492
5 core_essentials_hart 232 1 0.004310345 0.62345177
Blood or Immune
Number of cTWAS Genes in Tissue Group: 45
gene_set nset ngenes percent padj
1 gwascatalog 4798 22 0.004585244 0.06178366
2 clinvar_path_likelypath 2208 10 0.004528986 0.28090094
3 mgi_essential 1797 6 0.003338898 0.67011314
4 fda_approved_drug_targets 255 1 0.003921569 0.67011314
5 core_essentials_hart 222 0 0.000000000 1.00000000
Digestive
Number of cTWAS Genes in Tissue Group: 110
gene_set nset ngenes percent padj
1 gwascatalog 5418 56 0.010335917 5.458363e-05
2 clinvar_path_likelypath 2501 24 0.009596162 5.437542e-02
3 mgi_essential 2055 16 0.007785888 3.682654e-01
4 core_essentials_hart 246 2 0.008130081 5.765450e-01
5 fda_approved_drug_targets 310 2 0.006451613 5.850464e-01Analysis of TWAS False Positives by Region
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_tag, function(x){unlist(region_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from region total to get combined pip for other genes in region
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip < 0.5)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip < 0.5)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
Analysis of TWAS False Positives by Credible Set
cTWAS is using susie settings that mask credible sets consisting of variables with minimum pairwise correlations below a specified threshold. The default threshold is 0.5. I think this is intended to mask credible sets with “diffuse” support. As a consequence, many of the genes considered here (TWAS false positives; significant z score but low PIP) are not assigned to a credible set (have cs_index=0). For this reason, the first figure is not really appropriate for answering the question “are TWAS false positives due to SNPs or genes”.
The second figure includes only TWAS genes that are assigned to a reported causal set (i.e. they are in a “pure” causal set with high pairwise correlations). I think that this figure is closer to the intended analysis. However, it may be biased in some way because we have excluded many TWAS false positive genes that are in “impure” credible sets.
Some alternatives to these figures include the region-based analysis in the previous section; or re-analysis with lower/no minimum pairwise correlation threshold (“min_abs_corr” option in susie_get_cs) for reporting credible sets.
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
####################
#using only genes assigned to a credible set
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
#exclude genes that are not assigned to a credible set, cs_index==0
gene_pips <- gene_pips[as.numeric(sapply(gene_pips$region_cs_tag, function(x){rev(unlist(strsplit(x, "_")))[1]}))!=0,]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
cTWAS genes without genome-wide significant SNP nearby
novel_genes <- data.frame(genename=as.character(), weight=as.character(), susie_pip=as.numeric(), snp_maxz=as.numeric())
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$ctwas,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, sapply(gene_pips$region_tag, function(x){region_pips[x,"snp_maxz"]}))
names(gene_pips)[ncol(gene_pips)] <- "snp_maxz"
if (nrow(gene_pips)>0){
gene_pips$weight <- names(df)[i]
gene_pips <- gene_pips[gene_pips$snp_maxz < qnorm(1-(5E-8/2), lower=T),c("genename", "weight", "susie_pip", "snp_maxz")]
novel_genes <- rbind(novel_genes, gene_pips)
}
}
novel_genes_summary <- data.frame(genename=unique(novel_genes$genename))
novel_genes_summary$nweights <- sapply(novel_genes_summary$genename, function(x){length(novel_genes$weight[novel_genes$genename==x])})
novel_genes_summary$weights <- sapply(novel_genes_summary$genename, function(x){paste(novel_genes$weight[novel_genes$genename==x],collapse=", ")})
novel_genes_summary <- novel_genes_summary[order(-novel_genes_summary$nweights),]
novel_genes_summary[,c("genename","nweights")] genename nweights
8 AMIGO1 30
12 PPP1R21 26
9 DAP 22
3 MLF2 19
11 KRT80 10
13 PITRM1 9
2 ARHGEF26 6
16 HELQ 6
19 ABCB4 6
10 VAMP1 5
5 CNDP2 4
14 CTSH 4
25 GSTM1 4
30 PSRC1 4
21 UBE2Q1 3
26 SAV1 3
28 OSGIN1 3
33 MUCL1 3
6 LINC00908 2
15 FAM129A 2
17 RFTN1 2
22 GALNT5 2
23 CCDC169 2
27 DDX51 2
29 BCL2 2
36 C5orf63 2
38 MEF2D 2
48 FOXN2 2
1 UMPS 1
4 CCL4L2 1
7 BRD4 1
18 SLC2A12 1
20 MXI1 1
24 COL28A1 1
31 RNF167 1
32 KIAA1644 1
34 UNC50 1
35 CDH6 1
37 ASF1A 1
39 ELMO1 1
40 TRIM34 1
41 SCN8A 1
42 ZNF606 1
43 EPS8L3 1
44 SHE 1
45 PIK3CB 1
46 DYRK4 1
47 TM2D1 1
49 LPCAT1 1
50 FGD4 1
51 TRAPPC2B 1
52 UGGT2 1
53 TSEN34 1
54 PTPRB 1
55 RP4-798A10.4 1
56 RP11-503L19.1 1
57 AF067845.3 1
58 TBC1D3H 1
59 GRK3 1
60 HOXD12 1Tissue-specificity for cTWAS genes
gene_pips_by_weight <- data.frame(genename=as.character(ctwas_genes))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips
gene_pips <- gene_pips[match(ctwas_genes, gene_pips$genename),,drop=F]
#gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- -1
gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- 0 #missing values coded as PIP=0
gene_pips_by_weight <- cbind(gene_pips_by_weight, gene_pips$susie_pip)
names(gene_pips_by_weight)[ncol(gene_pips_by_weight)] <- names(df)[i]
}
gene_pips_by_weight <- as.matrix(gene_pips_by_weight[,-1])
rownames(gene_pips_by_weight) <- ctwas_genes
#number of tissues with PIP>0.5 for cTWAS genes
ctwas_frequency <- rowSums(gene_pips_by_weight>0.5)
hist(ctwas_frequency, col="grey", breaks=0:max(ctwas_frequency), xlim=c(0,ncol(gene_pips_by_weight)),
xlab="Number of Tissues with PIP>0.5",
ylab="Number of cTWAS Genes",
main="Tissue Specificity for cTWAS Genes")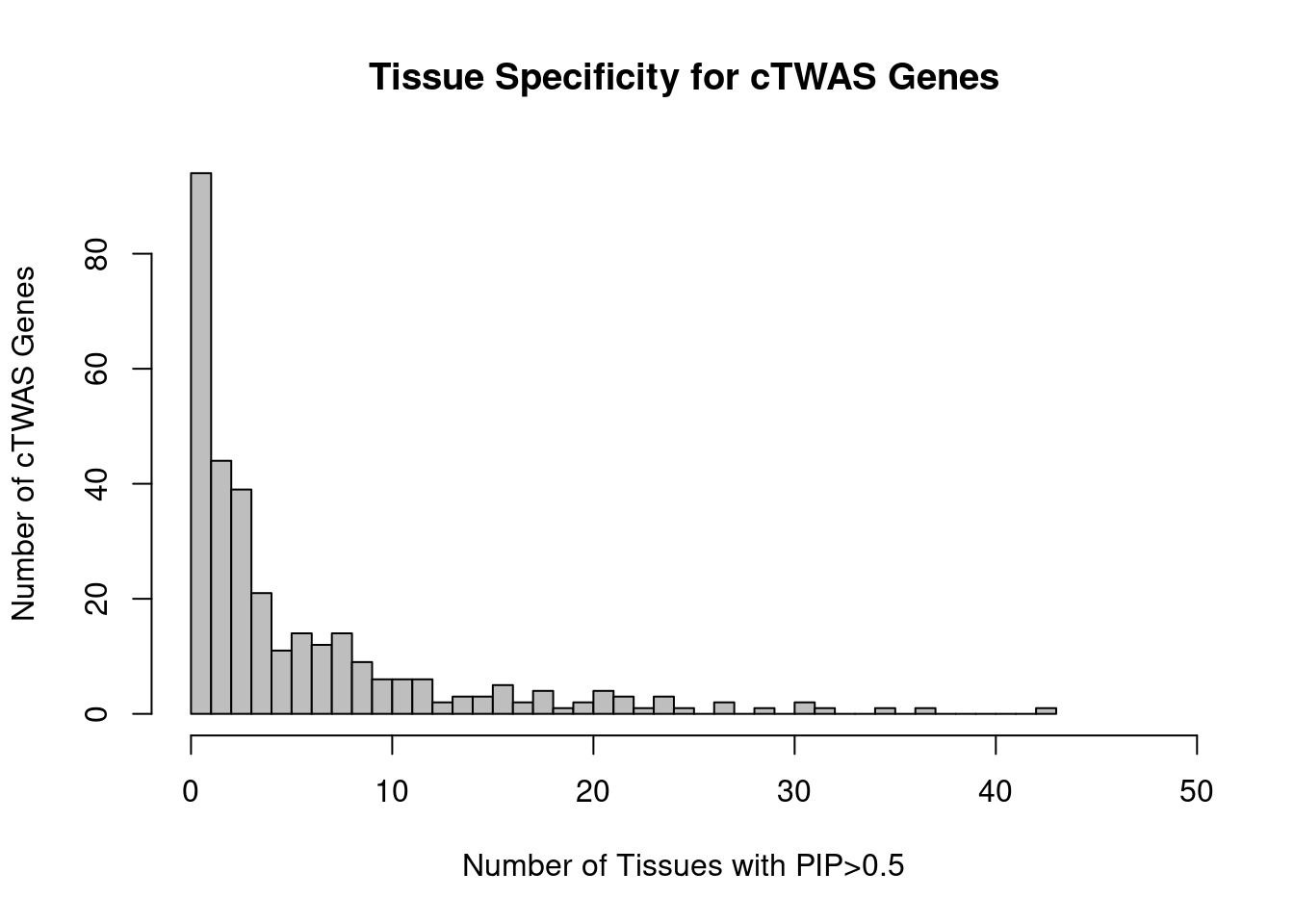
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
#heatmap of gene PIPs
cluster_ctwas_genes <- hclust(dist(gene_pips_by_weight))
cluster_ctwas_weights <- hclust(dist(t(gene_pips_by_weight)))
plot(cluster_ctwas_weights, cex=0.6)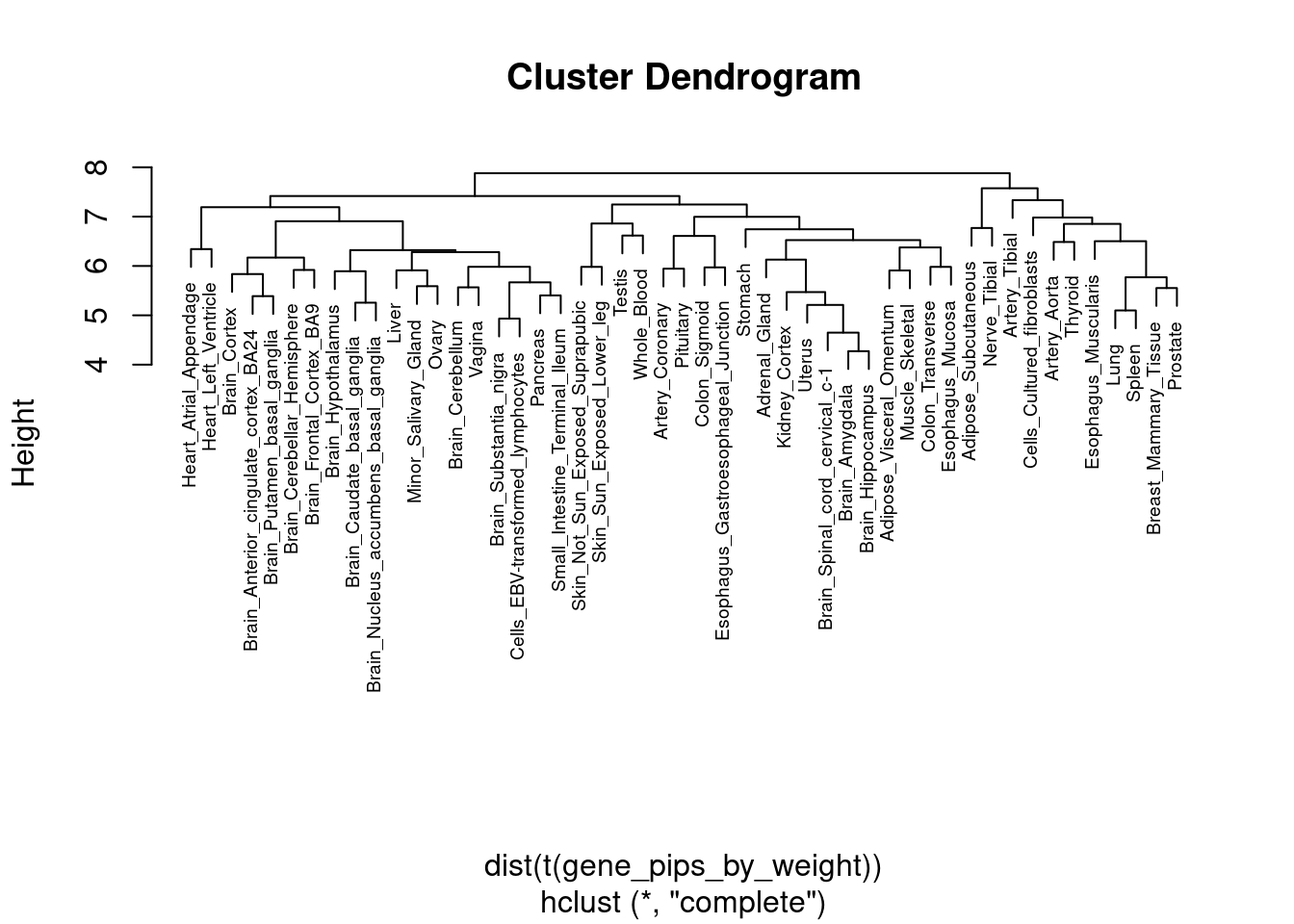
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
plot(cluster_ctwas_genes, cex=0.6, labels=F)
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
par(mar=c(14.1, 4.1, 4.1, 2.1))
image(t(gene_pips_by_weight[rev(cluster_ctwas_genes$order),rev(cluster_ctwas_weights$order)]),
axes=F)
mtext(text=colnames(gene_pips_by_weight)[cluster_ctwas_weights$order], side=1, line=0.3, at=seq(0,1,1/(ncol(gene_pips_by_weight)-1)), las=2, cex=0.8)
mtext(text=rownames(gene_pips_by_weight)[cluster_ctwas_genes$order], side=2, line=0.3, at=seq(0,1,1/(nrow(gene_pips_by_weight)-1)), las=1, cex=0.4)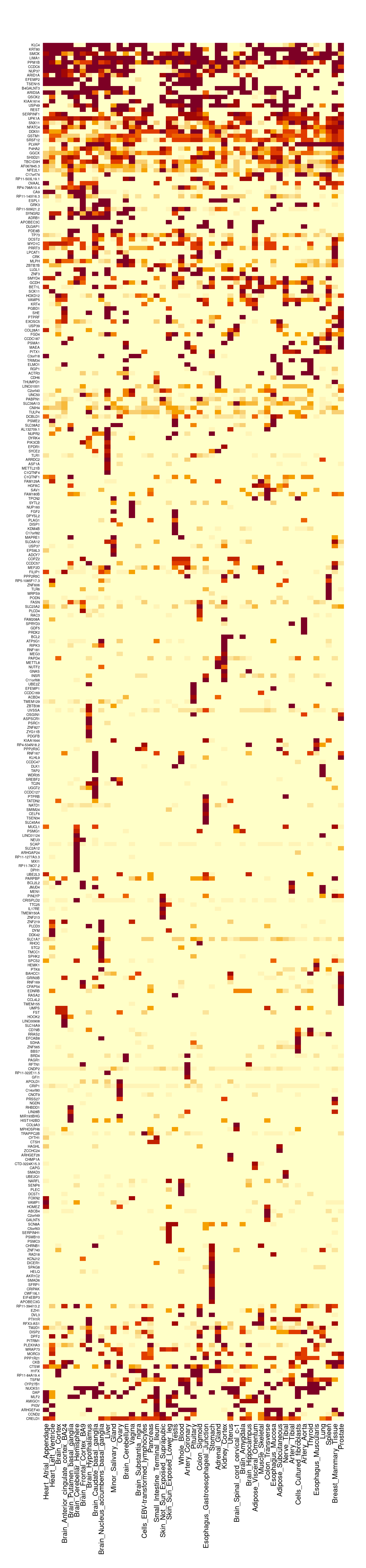
| Version | Author | Date |
|---|---|---|
| b67c150 | wesleycrouse | 2022-04-11 |
cTWAS genes with highest proportion of total PIP on a single tissue
#genes with highest porportion of PIP on a single tissue
gene_pips_proportion <- gene_pips_by_weight/rowSums(gene_pips_by_weight)
proportion_table <- data.frame(genename=as.character(rownames(gene_pips_proportion)))
proportion_table$max_pip_prop <- apply(gene_pips_proportion,1,max)
proportion_table$max_weight <- colnames(gene_pips_proportion)[apply(gene_pips_proportion,1,which.max)]
proportion_table[order(-proportion_table$max_pip_prop),] genename max_pip_prop max_weight
27 EDNRB 1.00000000 Adipose_Subcutaneous
162 TULP4 1.00000000 Brain_Hypothalamus
210 C3orf18 1.00000000 Esophagus_Muscularis
215 ASF1A 1.00000000 Heart_Atrial_Appendage
283 PGBD1 1.00000000 Small_Intestine_Terminal_Ileum
289 RP4-798A10.4 1.00000000 Testis
290 OVAAL 1.00000000 Testis
291 RP11-503L19.1 1.00000000 Testis
292 AF067845.3 1.00000000 Testis
296 C17orf74 1.00000000 Testis
297 TBC1D3H 1.00000000 Testis
313 HOXD12 1.00000000 Vagina
238 PIK3CB 1.00000000 Lung
246 SLC39A13 0.99999899 Muscle_Skeletal
187 PDGFB 0.99996036 Cells_Cultured_fibroblasts
155 LINC01124 0.99988355 Brain_Frontal_Cortex_BA9
276 CELF6 0.99941524 Skin_Not_Sun_Exposed_Suprapubic
124 EFEMP1 0.99939511 Brain_Anterior_cingulate_cortex_BA24
178 KDM4B 0.99920516 Breast_Mammary_Tissue
180 ZYG11B 0.99834287 Cells_Cultured_fibroblasts
317 RIPK3 0.99787631 Whole_Blood
249 FGF2 0.99529590 Nerve_Tibial
118 GNAS 0.99022021 Artery_Tibial
284 KRT4 0.98888521 Small_Intestine_Terminal_Ileum
208 IL17RE 0.98545829 Esophagus_Mucosa
314 RNF181 0.97939734 Whole_Blood
319 ATP5G1 0.97825612 Whole_Blood
228 EPS8L3 0.97626227 Kidney_Cortex
193 LINC01001 0.97402161 Colon_Sigmoid
299 NFE2L1 0.96224300 Testis
229 USP37 0.95364732 Kidney_Cortex
76 APOBEC3C 0.93693518 Adrenal_Gland
303 PODN 0.92810475 Thyroid
260 CCDC187 0.92766621 Pancreas
171 TPCN2 0.92125929 Brain_Putamen_basal_ganglia
304 MRPS9 0.92076899 Thyroid
167 SOX11 0.92019438 Brain_Nucleus_accumbens_basal_ganglia
158 PSMG1 0.86994691 Brain_Frontal_Cortex_BA9
236 SHE 0.86720955 Liver
11 RASA2 0.85426238 Adipose_Subcutaneous
278 SMIM24 0.84022526 Skin_Not_Sun_Exposed_Suprapubic
288 DYM 0.83374956 Stomach
308 FASN 0.79906068 Thyroid
255 PRRT3 0.78453487 Ovary
305 PLCD4 0.77680782 Thyroid
219 ARRDC2 0.76258112 Heart_Atrial_Appendage
60 SPHK2 0.73636060 Adipose_Visceral_Omentum
240 DYRK4 0.72489564 Lung
309 SLC23A2 0.71408420 Thyroid
151 DPH1 0.69830268 Brain_Cortex
202 ZNF3 0.69462456 Esophagus_Gastroesophageal_Junction
73 DLGAP1 0.68531468 Adrenal_Gland
307 RAC3 0.67744058 Thyroid
310 USP39 0.65731559 Uterus
182 ZNF827 0.65488323 Cells_Cultured_fibroblasts
204 LLGL1 0.64849166 Esophagus_Gastroesophageal_Junction
318 MEG3 0.64486210 Whole_Blood
258 MYO1C 0.64247274 Ovary
232 ADCY7 0.64139611 Kidney_Cortex
207 TMEM150A 0.63914138 Esophagus_Mucosa
286 SLC1A7 0.63611746 Stomach
223 TRIM34 0.62443627 Heart_Left_Ventricle
66 PDE8B 0.60526006 Adrenal_Gland
294 CA9 0.60244045 Testis
295 ESPL1 0.57967161 Testis
239 EPDR1 0.57274181 Lung
264 TAP2 0.57199230 Pituitary
312 VAMP5 0.57116493 Vagina
293 RP11-140I16.3 0.56996099 Testis
252 NUP160 0.53666494 Nerve_Tibial
243 RP11-509I21.2 0.52126557 Minor_Salivary_Gland
188 KIAA1644 0.50548007 Cells_Cultured_fibroblasts
194 ACTR3 0.50335463 Colon_Transverse
164 PSME2 0.50021635 Brain_Hypothalamus
113 CCDC169 0.50000000 Artery_Tibial
163 SLC38A2 0.50000000 Brain_Hypothalamus
173 CCDC127 0.50000000 Brain_Substantia_nigra
179 RP4-534N18.2 0.50000000 Cells_Cultured_fibroblasts
127 GDF5 0.49972864 Brain_Anterior_cingulate_cortex_BA24
242 WDR35 0.49946383 Pituitary
272 TATDN2 0.49944511 Skin_Sun_Exposed_Lower_leg
6 C2orf40 0.49585539 Colon_Sigmoid
311 EXOSC5 0.49565723 Uterus
44 ZBTB7B 0.49380918 Prostate
34 CCL4L2 0.48453305 Adipose_Subcutaneous
233 COPZ2 0.47987592 Kidney_Cortex
82 SPRYD3 0.47717908 Artery_Aorta
217 METTL21B 0.47064298 Heart_Atrial_Appendage
198 RGP1 0.46897299 Colon_Transverse
62 TP73 0.46468103 Adrenal_Gland
235 PTPRF 0.46350397 Liver
153 UBE2L3 0.45329250 Brain_Cortex
15 TMEM155 0.45156527 Adipose_Subcutaneous
301 GRK3 0.45011323 Testis
98 PLAG1 0.44989280 Artery_Coronary
150 PARPBP 0.44585336 Brain_Cortex
117 INSR 0.44535079 Artery_Tibial
168 BET1L 0.43967544 Brain_Nucleus_accumbens_basal_ganglia
275 TC2N 0.43095927 Whole_Blood
267 DLK1 0.42301694 Pituitary
250 DPYSL2 0.42123519 Nerve_Tibial
125 FILIP1 0.42073586 Brain_Caudate_basal_ganglia
109 TMEM129 0.41286456 Artery_Tibial
96 SLC2A12 0.40787288 Artery_Coronary
222 ELMO1 0.39381316 Heart_Left_Ventricle
287 DDX42 0.39203721 Stomach
195 MAEA 0.38274073 Colon_Transverse
39 HOOK2 0.36965833 Adipose_Subcutaneous
130 NEU3 0.36568560 Brain_Frontal_Cortex_BA9
282 PTPRB 0.36477206 Skin_Sun_Exposed_Lower_leg
38 LINC00908 0.36172483 Artery_Aorta
211 PITX1 0.35479809 Esophagus_Muscularis
136 NUPR2 0.35237084 Brain_Cerebellar_Hemisphere
156 SCAP 0.34536305 Skin_Not_Sun_Exposed_Suprapubic
177 C17orf82 0.34339741 Breast_Mammary_Tissue
199 THUMPD1 0.33776913 Colon_Transverse
84 PABPN1 0.33719721 Artery_Aorta
77 FAM129A 0.33625461 Heart_Atrial_Appendage
224 APOLD1 0.33609825 Heart_Left_Ventricle
257 LPCAT1 0.33507790 Ovary
253 FAM180B 0.33442024 Nerve_Tibial
175 SDHA 0.33333333 Breast_Mammary_Tissue
189 MUCL1 0.33331502 Cells_EBV-transformed_lymphocytes
51 STC2 0.33036094 Stomach
203 SMYD4 0.32629837 Esophagus_Gastroesophageal_Junction
281 FST 0.32526850 Skin_Sun_Exposed_Lower_leg
279 TSEN34 0.32456144 Skin_Not_Sun_Exposed_Suprapubic
315 METTL8 0.32406708 Whole_Blood
48 TMCC1 0.32288773 Adipose_Visceral_Omentum
269 MLPH 0.32142471 Prostate
152 RP11-78O7.2 0.31980526 Brain_Cortex
154 SREBF2 0.31711735 Brain_Cortex
114 NUTF2 0.31249908 Artery_Tibial
53 SPCS2 0.30820361 Adipose_Visceral_Omentum
79 ARHGAP24 0.30622288 Artery_Aorta
316 PAPD4 0.30502118 Whole_Blood
18 SLC16A9 0.30313126 Adipose_Subcutaneous
116 UBE2Z 0.30234855 Artery_Tibial
115 ACBD4 0.30156234 Brain_Anterior_cingulate_cortex_BA24
266 PPP2R3C 0.29764060 Pituitary
88 RP5-1085F17.3 0.29729710 Brain_Caudate_basal_ganglia
107 ZBTB38 0.28716018 Esophagus_Gastroesophageal_Junction
268 CCDC47 0.28538071 Pituitary
186 MAPRE1 0.28333501 Cells_Cultured_fibroblasts
110 ADRB1 0.28029601 Artery_Tibial
74 GCDH 0.27799191 Esophagus_Gastroesophageal_Junction
121 ASPSCR1 0.27784135 Brain_Substantia_nigra
205 PLCD3 0.27510219 Esophagus_Mucosa
206 RHOC 0.26711316 Esophagus_Mucosa
111 C11orf68 0.26300603 Cells_Cultured_fibroblasts
271 CRK 0.26114402 Prostate
212 PSMA1 0.26086356 Esophagus_Muscularis
161 DCBLD1 0.25909860 Brain_Hypothalamus
306 FAM208A 0.25777860 Thyroid
300 SYNGR2 0.25692703 Testis
244 BAHCC1 0.25282345 Ovary
141 PRSS27 0.25185391 Liver
160 HEMK1 0.25000000 Brain_Hypothalamus
190 PRDX2 0.24981792 Thyroid
159 OSGIN1 0.24585487 Liver
54 SLC6A12 0.24326402 Nerve_Tibial
209 ZNF213 0.24284743 Esophagus_Mucosa
138 AL132709.1 0.24231450 Brain_Cortex
129 KLHL9 0.23634093 Brain_Caudate_basal_ganglia
176 BCL2L2 0.23493444 Breast_Mammary_Tissue
85 PPP2R5C 0.23259383 Brain_Anterior_cingulate_cortex_BA24
148 JMJD4 0.22945436 Brain_Cortex
251 SLC45A4 0.22132264 Nerve_Tibial
93 P4HA2 0.22026832 Esophagus_Gastroesophageal_Junction
285 SYCE2 0.21950750 Spleen
63 DCST2 0.21886148 Stomach
261 FGD4 0.21875227 Pancreas
145 RNF169 0.21667482 Brain_Cerebellum
191 CNIH4 0.21627376 Colon_Sigmoid
91 GGCX 0.21525521 Artery_Coronary
19 ZCCHC24 0.21252801 Adipose_Subcutaneous
214 HGFAC 0.20880812 Heart_Atrial_Appendage
133 CNOT9 0.20783220 Brain_Cerebellar_Hemisphere
147 GRIN3B 0.19990501 Vagina
131 NGDN 0.19971336 Brain_Caudate_basal_ganglia
226 ZNF219 0.19762645 Heart_Left_Ventricle
234 CCDC57 0.19743619 Thyroid
256 TLR1 0.19558557 Ovary
120 COL28A1 0.19287845 Brain_Amygdala
142 SNX11 0.19154706 Brain_Cerebellar_Hemisphere
216 C1QTNF4 0.19148537 Heart_Atrial_Appendage
137 SAV1 0.19032927 Brain_Cerebellar_Hemisphere
170 PSRC1 0.18980346 Brain_Putamen_basal_ganglia
169 BCL2 0.18766037 Heart_Left_Ventricle
35 CD79B 0.18639517 Adipose_Subcutaneous
196 TLR6 0.18537461 Colon_Transverse
10 UMPS 0.17923099 Adipose_Subcutaneous
274 UGGT2 0.17796864 Skin_Not_Sun_Exposed_Suprapubic
87 C1QTNF1 0.17714249 Artery_Aorta
108 UVSSA 0.17689908 Artery_Tibial
218 CYTH1 0.17216911 Pancreas
254 SYTL2 0.16942445 Nerve_Tibial
220 MEF2D 0.16902578 Heart_Left_Ventricle
95 RP11-1277A3.3 0.16881037 Artery_Coronary
71 LIMA1 0.16669385 Brain_Cerebellar_Hemisphere
20 RRAS2 0.16639635 Esophagus_Muscularis
69 PSMC3 0.16594050 Whole_Blood
263 TRAPPC2B 0.16584951 Pancreas
104 PSMB10 0.16191578 Artery_Coronary
197 CDH6 0.16173841 Colon_Transverse
132 CRIP1 0.15942707 Skin_Sun_Exposed_Lower_leg
86 TTC25 0.15795150 Artery_Aorta
30 HAGHL 0.15773110 Adipose_Subcutaneous
139 C14orf80 0.15688105 Liver
277 NATD1 0.15572129 Skin_Not_Sun_Exposed_Suprapubic
259 PTK6 0.15379920 Ovary
200 EFCAB8 0.15026759 Esophagus_Mucosa
12 ARHGEF26 0.14837181 Adipose_Subcutaneous
75 COL9A3 0.14776990 Brain_Putamen_basal_ganglia
280 GFI1 0.14753218 Skin_Sun_Exposed_Lower_leg
102 NFATC4 0.14742626 Cells_Cultured_fibroblasts
227 ZNF606 0.14408678 Heart_Left_Ventricle
165 CHMP1A 0.14303794 Brain_Hypothalamus
213 CFAP54 0.14297722 Esophagus_Muscularis
183 CRISPLD2 0.14243705 Cells_Cultured_fibroblasts
126 MIR193BHG 0.14184491 Heart_Atrial_Appendage
46 RHBDD1 0.13980161 Minor_Salivary_Gland
97 ABCB4 0.13845852 Heart_Left_Ventricle
128 HIST1H2BD 0.13730014 Brain_Caudate_basal_ganglia
185 RNF167 0.13646616 Cells_Cultured_fibroblasts
149 LIN28B 0.13598859 Brain_Putamen_basal_ganglia
192 UNC50 0.13526063 Colon_Sigmoid
37 CNDP2 0.13491533 Minor_Salivary_Gland
64 PPM1B 0.13287113 Prostate
103 HOMEZ 0.13118065 Artery_Tibial
122 PLVAP 0.12925231 Cells_Cultured_fibroblasts
101 B4GALNT3 0.12729034 Pituitary
29 DICER1 0.12548242 Skin_Sun_Exposed_Lower_leg
78 HELQ 0.12500782 Artery_Aorta
16 PLEC 0.12500000 Adipose_Subcutaneous
140 SMAD3 0.12468943 Brain_Cerebellar_Hemisphere
99 MXI1 0.12330430 Artery_Coronary
33 KCNJ12 0.12122439 Adipose_Subcutaneous
112 SERPINH1 0.11527330 Thyroid
144 MEN1 0.11498861 Lung
265 USP49 0.11417751 Whole_Blood
247 DISP1 0.11293918 Nerve_Tibial
92 RFTN1 0.11160918 Kidney_Cortex
135 SRSF12 0.11151742 Vagina
32 CHRNB1 0.11111111 Adipose_Subcutaneous
262 PAGR1 0.10674376 Pancreas
41 PINLYP 0.10486912 Brain_Cerebellum
184 SERPINF1 0.10361771 Cells_Cultured_fibroblasts
302 SH3D21 0.10346945 Thyroid
80 SPAG8 0.10303414 Minor_Salivary_Gland
181 BBS7 0.10042886 Cells_Cultured_fibroblasts
172 UPK1A 0.09908558 Brain_Spinal_cord_cervical_c-1
40 BRD4 0.09887228 Adipose_Subcutaneous
59 ARID3A 0.09884175 Artery_Coronary
36 RP11-322E11.5 0.09683359 Adipose_Subcutaneous
270 SMAD6 0.09660045 Prostate
4 KIAA1614 0.09556743 Heart_Left_Ventricle
24 ZNF740 0.09495440 Colon_Transverse
52 SFRP1 0.09256790 Adipose_Visceral_Omentum
72 CTSH 0.09163989 Muscle_Skeletal
237 ZNF565 0.09092697 Minor_Salivary_Gland
248 CAPG 0.08874514 Spleen
3 DCST1 0.08742374 Artery_Coronary
157 DDX51 0.08652058 Brain_Frontal_Cortex_BA9
81 AKR1C2 0.08496941 Artery_Tibial
100 EFEMP2 0.08431058 Artery_Coronary
166 SMOX 0.08307107 Esophagus_Gastroesophageal_Junction
31 MPHOSPH6 0.08306502 Small_Intestine_Terminal_Ileum
123 GSTM1 0.08229056 Whole_Blood
174 REST 0.08222969 Heart_Atrial_Appendage
225 SCN8A 0.08055760 Heart_Left_Ventricle
221 C2orf49 0.07746808 Heart_Left_Ventricle
94 CTD-3224K15.3 0.07610888 Vagina
245 FOXN2 0.07537144 Muscle_Skeletal
14 CRIPAK 0.07443102 Adipose_Subcutaneous
89 MORC3 0.07416462 Brain_Nucleus_accumbens_basal_ganglia
56 KRT80 0.07309426 Kidney_Cortex
201 C5orf63 0.07057393 Esophagus_Muscularis
47 RAD18 0.07032305 Adipose_Visceral_Omentum
105 UBE2Q1 0.06997934 Whole_Blood
55 VAMP1 0.06959050 Colon_Transverse
90 TSEN15 0.06798144 Esophagus_Gastroesophageal_Junction
146 NARFL 0.06758797 Thyroid
68 CCDC6 0.06520561 Thyroid
57 CKB 0.06421574 Ovary
143 SENP6 0.06260064 Testis
42 ARID1A 0.06250000 Adipose_Visceral_Omentum
25 CYP27B1 0.06127762 Adipose_Subcutaneous
134 EIF4EBP3 0.06126759 Brain_Cerebellar_Hemisphere
106 GALNT5 0.06095588 Artery_Tibial
83 NUP37 0.06011668 Artery_Aorta
231 CWF19L1 0.05862625 Kidney_Cortex
26 TSFM 0.05840201 Adipose_Subcutaneous
21 CTSW 0.05722425 Cells_Cultured_fibroblasts
9 PTH1R 0.05678032 Heart_Left_Ventricle
17 QSOX2 0.05136339 Colon_Transverse
5 NUCKS1 0.05091656 Adipose_Subcutaneous
45 RP11-394I13.2 0.04942327 Vagina
13 DVL3 0.04934170 Artery_Aorta
119 WRAP73 0.04797676 Pancreas
2 RP11-84A19.4 0.04761905 Adipose_Subcutaneous
23 MLF2 0.04670277 Lung
67 PITRM1 0.04667566 Brain_Caudate_basal_ganglia
61 APOBEC3G 0.04584605 Liver
49 H1FX 0.04543276 Brain_Substantia_nigra
70 DPF2 0.04502922 Heart_Left_Ventricle
230 KLC4 0.04467320 Kidney_Cortex
58 DISP2 0.04411442 Esophagus_Gastroesophageal_Junction
298 EZH1 0.04154933 Testis
8 CRELD1 0.04083409 Artery_Tibial
50 DAP 0.04001555 Heart_Left_Ventricle
241 TM2D1 0.03663434 Minor_Salivary_Gland
65 PPP1R21 0.03620137 Artery_Coronary
273 RFX3-AS1 0.03376957 Skin_Not_Sun_Exposed_Suprapubic
28 ARHGEF40 0.03301820 Thyroid
22 CCND2 0.03285179 Brain_Cortex
7 PLEKHA3 0.03205668 Heart_Left_Ventricle
1 PIGV 0.02702703 Adipose_Subcutaneous
43 AMIGO1 0.02498947 Esophagus_Muscularis
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.5 disgenet2r_0.99.2 WebGestaltR_0.4.4 enrichR_3.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.8 reshape2_1.4.3 purrr_0.3.4 lattice_0.20-38 colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 utf8_1.2.1 rlang_0.4.11 later_0.8.0 pillar_1.6.1 withr_2.4.1 glue_1.4.2 DBI_1.1.1 gdtools_0.1.9 rngtools_1.5 doRNG_1.8.2 plyr_1.8.4 foreach_1.5.1 lifecycle_1.0.0 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0 workflowr_1.6.2 codetools_0.2-16 evaluate_0.14 labeling_0.3 knitr_1.23 doParallel_1.0.16 httpuv_1.5.1 curl_3.3 parallel_3.6.1 fansi_0.5.0 Rcpp_1.0.6 readr_1.4.0 promises_1.0.1 scales_1.1.0 jsonlite_1.6 apcluster_1.4.8 farver_2.1.0 fs_1.3.1 hms_1.1.0 rjson_0.2.20 digest_0.6.20 stringi_1.4.3 dplyr_1.0.7 grid_3.6.1 rprojroot_2.0.2 tools_3.6.1 magrittr_2.0.1 tibble_3.1.2 crayon_1.4.1
[56] whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2 Matrix_1.2-18 svglite_1.2.2 rmarkdown_1.13 httr_1.4.1 iterators_1.0.13 R6_2.5.0 igraph_1.2.4.1 git2r_0.26.1 compiler_3.6.1