Systolic blood pressure automated reading - all weights
wesleycrouse
2022-02-28
Last updated: 2022-04-12
Checks: 7 0
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 107bb6d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
working directory clean
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-a-360_allweights.Rmd) and HTML (docs/ukb-a-360_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 107bb6d | wesleycrouse | 2022-04-12 | gene set enrichment for supplied gene sets |
| html | 95e0f8e | wesleycrouse | 2022-04-07 | scroll bar |
| Rmd | a4575d7 | wesleycrouse | 2022-04-07 | formating |
| Rmd | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | f7e9822 | wesleycrouse | 2022-04-06 | testing subsections |
| Rmd | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | d0f6e53 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| Rmd | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| html | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| html | b999e70 | wesleycrouse | 2022-04-04 | fixing display of results |
| html | dd02af5 | wesleycrouse | 2022-04-04 | kegg |
| html | 34ca036 | wesleycrouse | 2022-04-04 | kegg for individual tissues |
| Rmd | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | f426350 | wesleycrouse | 2022-04-04 | kegg enrichment |
| html | 364b716 | wesleycrouse | 2022-04-02 | TWAS FP based on confidence sets |
| Rmd | c9809d4 | wesleycrouse | 2022-04-01 | additional TWAS FP analyses |
| html | a9dcb4d | wesleycrouse | 2022-04-01 | alternative TWAS FP figures based on confidence sets |
| Rmd | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| html | 33c0201 | wesleycrouse | 2022-03-30 | reporting numbers of genes |
| html | 1c45bb5 | wesleycrouse | 2022-03-30 | fixing formatting |
| html | ad4604a | wesleycrouse | 2022-03-30 | Adding numbers to results |
| html | 18f0b45 | wesleycrouse | 2022-03-24 | format |
| Rmd | 16289f6 | wesleycrouse | 2022-03-24 | improving layout |
| Rmd | 8397beb | wesleycrouse | 2022-03-24 | format |
| html | 8397beb | wesleycrouse | 2022-03-24 | format |
| html | 27e1022 | wesleycrouse | 2022-03-24 | layout |
| html | 85641ef | wesleycrouse | 2022-03-24 | layout |
| Rmd | 717e77e | wesleycrouse | 2022-03-23 | adjusting heatmaps |
| html | 717e77e | wesleycrouse | 2022-03-23 | adjusting heatmaps |
| html | ae26765 | wesleycrouse | 2022-03-23 | plots |
| html | aefd338 | wesleycrouse | 2022-03-23 | adjusting heatmap |
| html | 64ee362 | wesleycrouse | 2022-03-23 | adjusting gene-tissue heatmap |
| html | b5e392d | wesleycrouse | 2022-03-23 | tables |
| Rmd | 1b1fcaf | wesleycrouse | 2022-03-23 | chart for tissue specificity |
| Rmd | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | 10b99b6 | wesleycrouse | 2022-03-23 | gene by tissue heatmap |
| html | bbf031d | wesleycrouse | 2022-03-23 | adjusting sections |
| Rmd | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | e7a699d | wesleycrouse | 2022-03-22 | false positives and novel genes |
| html | 60f39e6 | wesleycrouse | 2022-03-22 | additional traits |
| Rmd | 073f2a3 | wesleycrouse | 2022-03-22 | enrichment analysis for all weights |
| Rmd | ba908fe | wesleycrouse | 2022-03-21 | more traits for all weight analysis |
options(width=1000)trait_id <- "ukb-a-360"
trait_name <- "Systolic blood pressure automated reading"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions and effects
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
# ctwas_res$region_cs_tag <- paste(ctwas_res$region_tag, ctwas_res$cs_index, sep="_")
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes_index <- ctwas_gene_res$susie_pip>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_genes_index]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#
# twas_genes_index <- abs(ctwas_gene_res$z) > sig_thresh
# twas_genes <- ctwas_gene_res$genename[twas_genes_index]
#
# #gene PIPs and z scores
# gene_pips <- ctwas_gene_res[,c("genename", "region_tag", "susie_pip", "z", "region_cs_tag")]
#
# #total PIPs by region
# regions <- unique(ctwas_gene_res$region_tag)
# region_pips <- data.frame(region=regions, stringsAsFactors=F)
# region_pips$gene_pip <- sapply(regions, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_tag==x])})
# region_pips$snp_pip <- sapply(regions, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_tag==x])})
# region_pips$snp_maxz <- sapply(regions, function(x){max(abs(ctwas_snp_res$z[ctwas_snp_res$region_tag==x]))})
#
# #total PIPs by causal set
# regions_cs <- unique(ctwas_gene_res$region_cs_tag)
# region_cs_pips <- data.frame(region_cs=regions_cs, stringsAsFactors=F)
# region_cs_pips$gene_pip <- sapply(regions_cs, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_cs_tag==x])})
# region_cs_pips$snp_pip <- sapply(regions_cs, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_cs_tag==x])})
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes,
# gene_pips=gene_pips,
# region_pips=region_pips,
# sig_thresh=sig_thresh,
# region_cs_pips=region_cs_pips)
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL,
stringsAsFactors=F)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)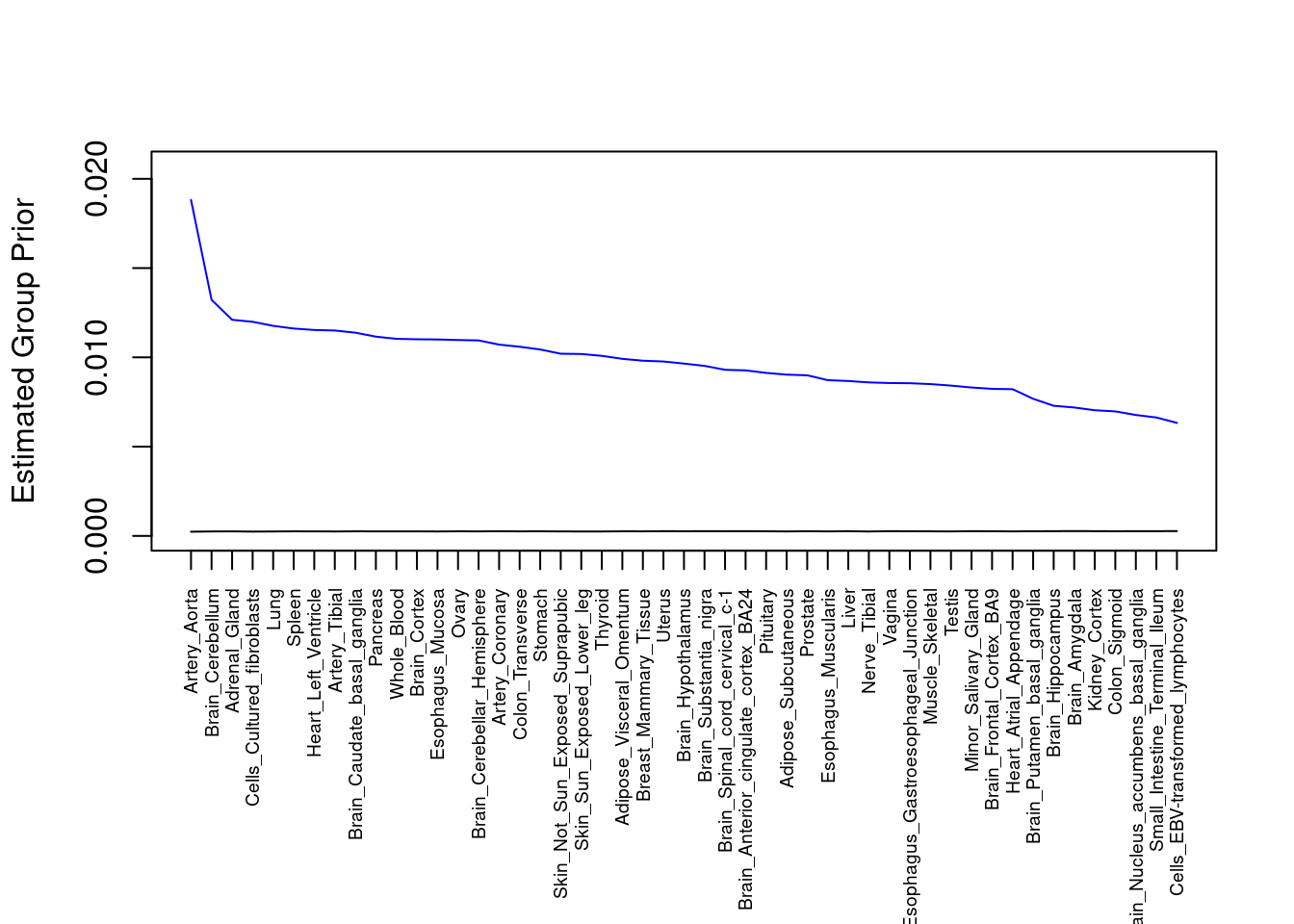
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)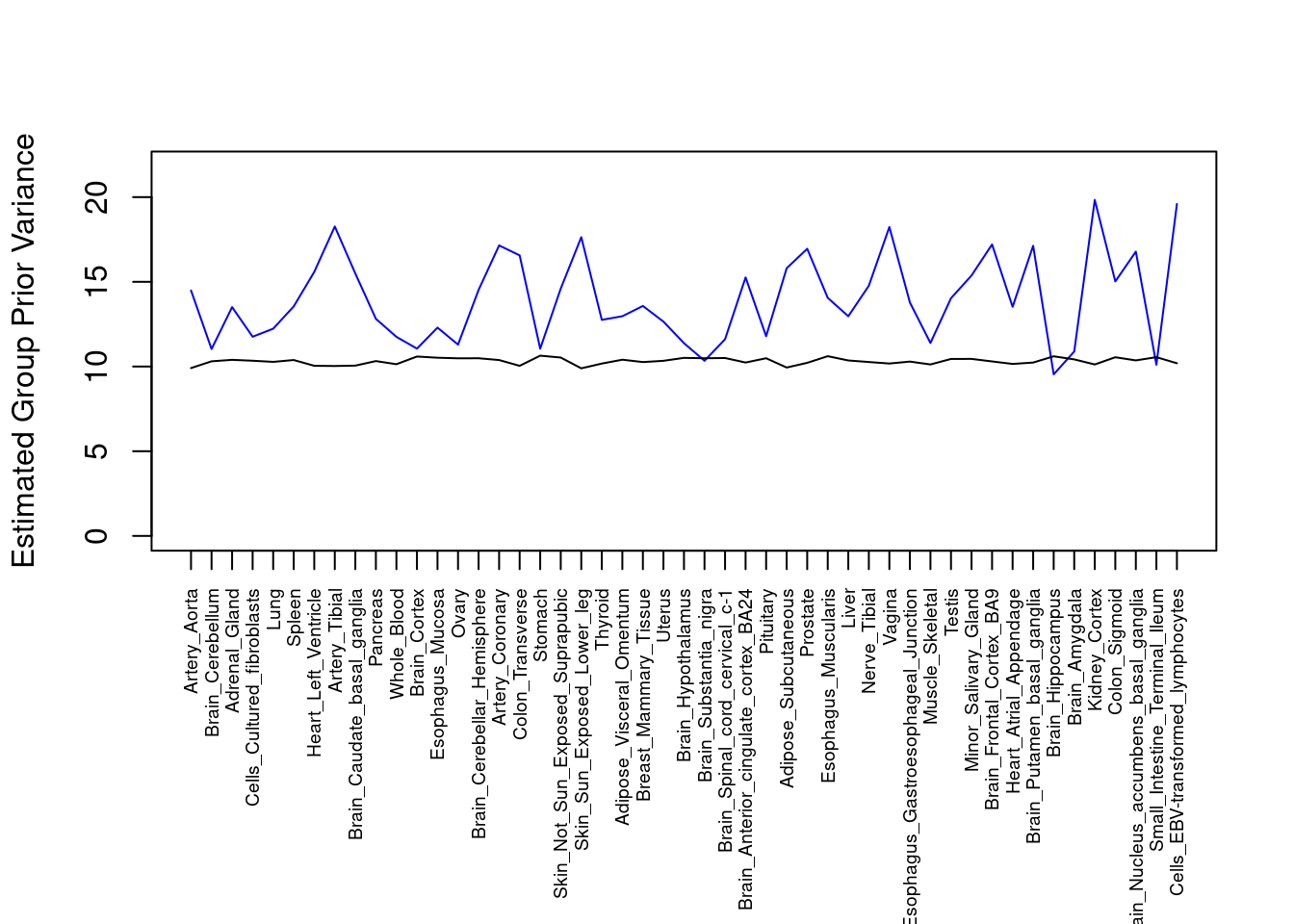
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)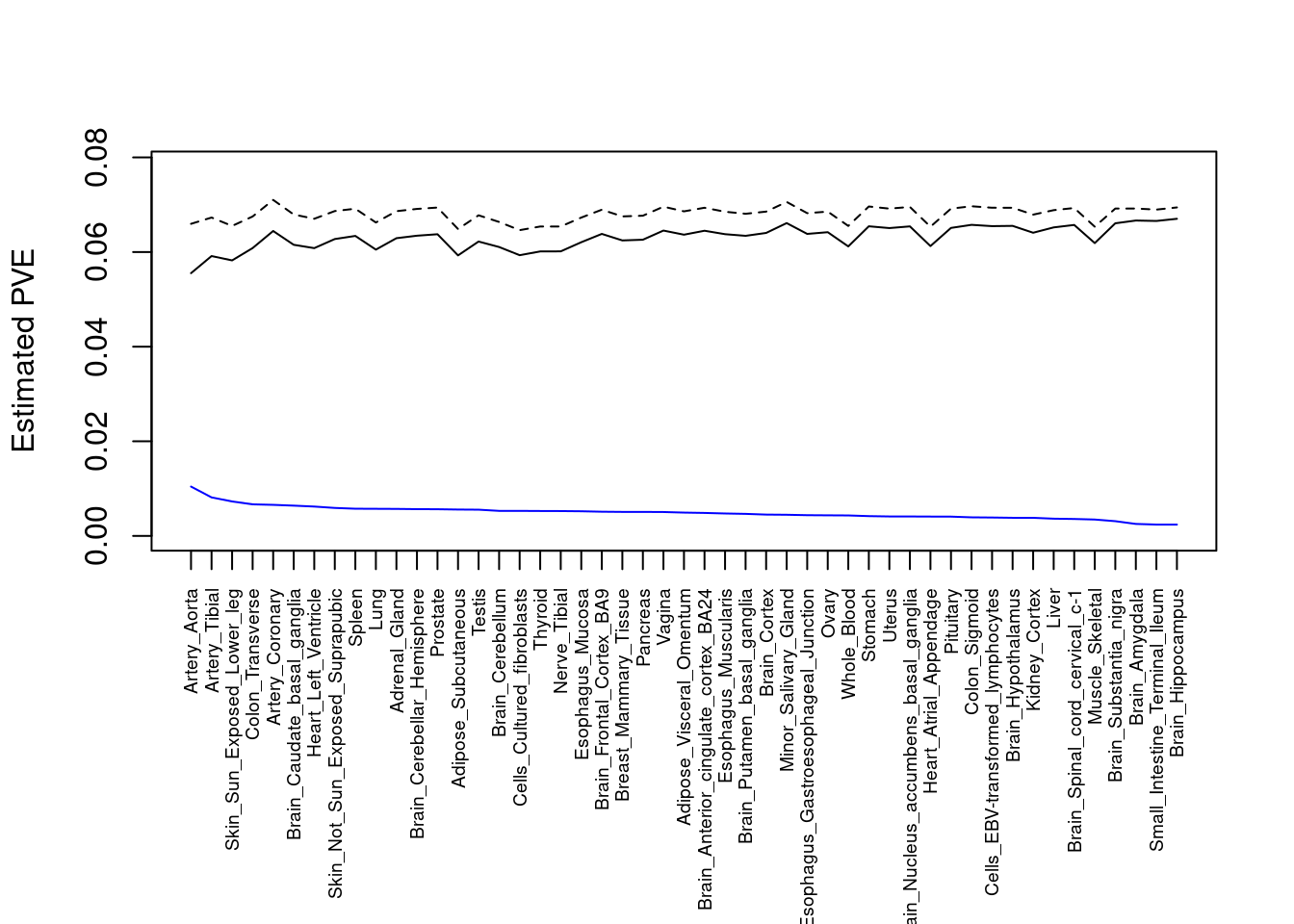
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")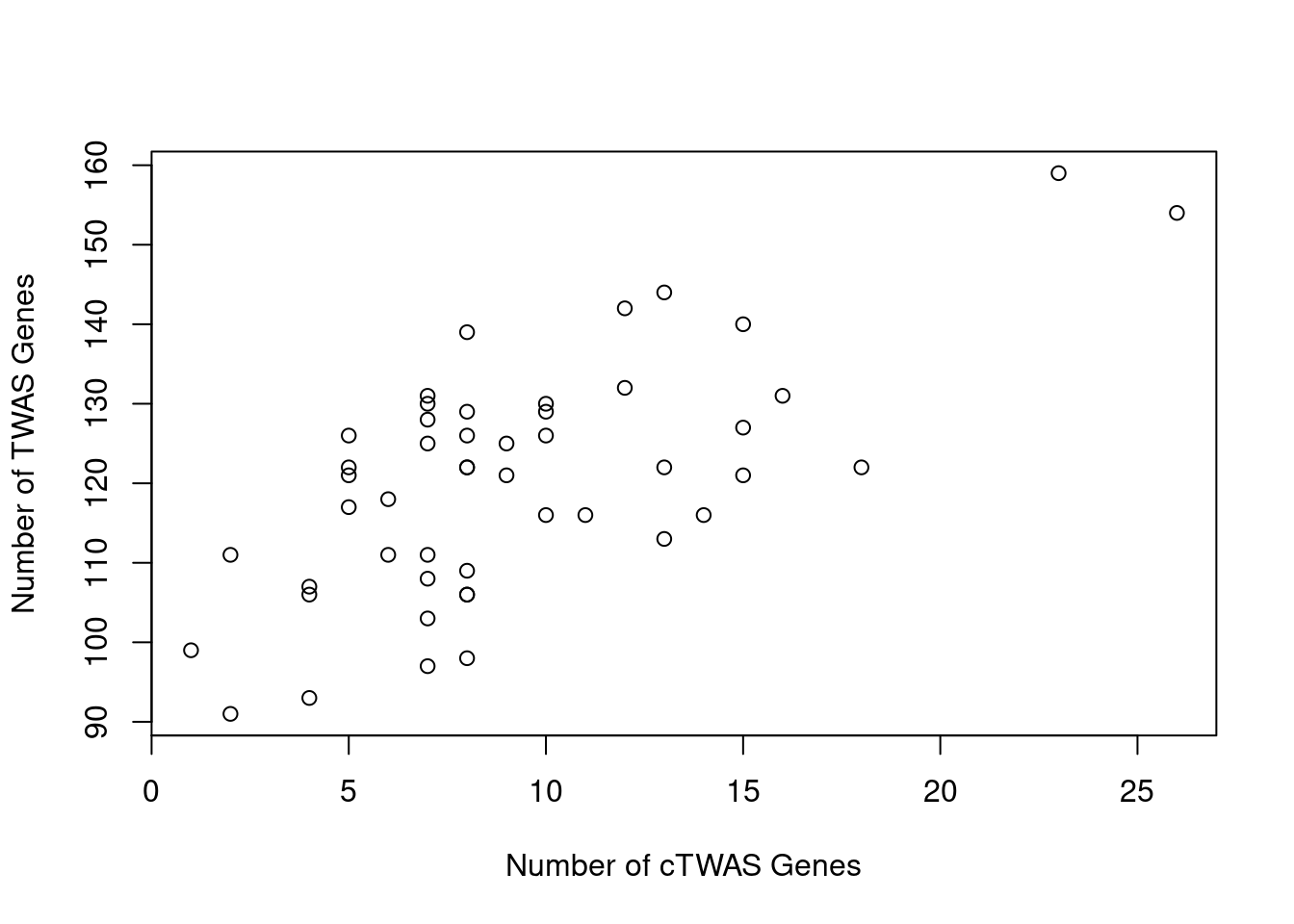
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 140#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 688Enrichment analysis for cTWAS genes
GO
#enrichment for cTWAS genes using enrichR
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)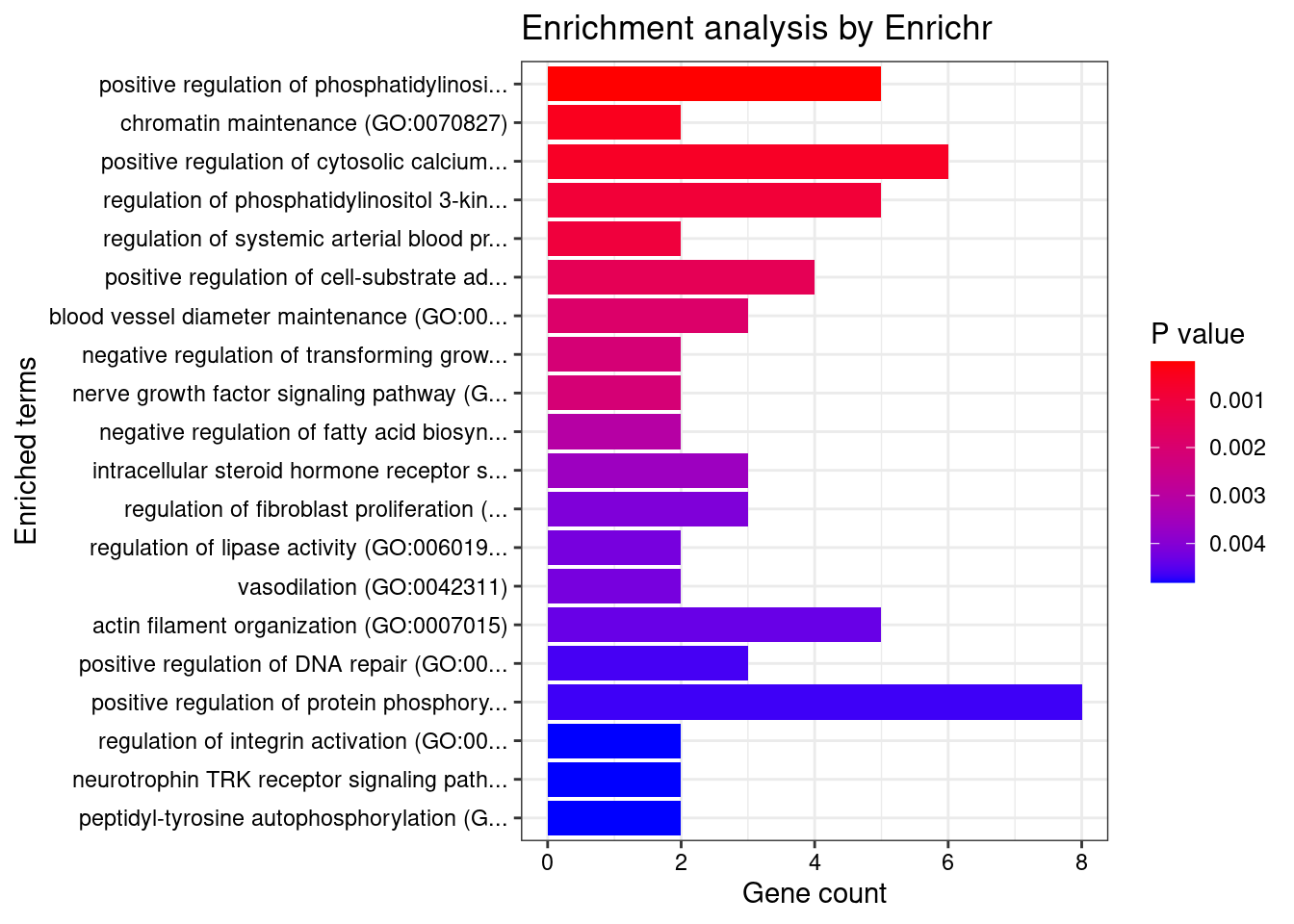
GO_Cellular_Component_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Molecular_Function_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
KEGG
#enrichment for cTWAS genes using KEGG
library(WebGestaltR)******************************************* ** Welcome to WebGestaltR ! ** *******************************************background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#listGeneSet()
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...enrichResult[,c("description", "size", "overlap", "FDR", "userId")] description size overlap FDR userId
1 Renin secretion 62 5 0.03047657 EDNRA;ADRB1;PPP3R1;AGT;PTGER4DisGeNET
#enrichment for cTWAS genes using DisGeNET
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <- disease_enrichment(entities=ctwas_genes, vocabulary = "HGNC", database = "CURATED")TMEM175 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDZNF692 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDC20orf187 gene(s) from the input list not found in DisGeNET CURATEDSLC2A4RG gene(s) from the input list not found in DisGeNET CURATEDKIAA1462 gene(s) from the input list not found in DisGeNET CURATEDRP11-286N22.10 gene(s) from the input list not found in DisGeNET CURATEDRP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDCPXM1 gene(s) from the input list not found in DisGeNET CURATEDRP5-965G21.3 gene(s) from the input list not found in DisGeNET CURATEDSOX13 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDLINC01169 gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDFAM212A gene(s) from the input list not found in DisGeNET CURATEDGDF7 gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDCAMK1D gene(s) from the input list not found in DisGeNET CURATEDSPIRE1 gene(s) from the input list not found in DisGeNET CURATEDNDUFAF8 gene(s) from the input list not found in DisGeNET CURATEDMEX3A gene(s) from the input list not found in DisGeNET CURATEDNPW gene(s) from the input list not found in DisGeNET CURATEDUSP36 gene(s) from the input list not found in DisGeNET CURATEDGIT2 gene(s) from the input list not found in DisGeNET CURATEDSHB gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDCDC16 gene(s) from the input list not found in DisGeNET CURATEDKIF13B gene(s) from the input list not found in DisGeNET CURATEDZNF598 gene(s) from the input list not found in DisGeNET CURATEDCTDNEP1 gene(s) from the input list not found in DisGeNET CURATEDASCC2 gene(s) from the input list not found in DisGeNET CURATEDDDI2 gene(s) from the input list not found in DisGeNET CURATEDTMEM176B gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDCTD-2349P21.5 gene(s) from the input list not found in DisGeNET CURATEDSTK38L gene(s) from the input list not found in DisGeNET CURATEDLRRC10B gene(s) from the input list not found in DisGeNET CURATEDEFR3B gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDRP11-1055B8.3 gene(s) from the input list not found in DisGeNET CURATEDC22orf31 gene(s) from the input list not found in DisGeNET CURATEDYEATS2 gene(s) from the input list not found in DisGeNET CURATEDARL4A gene(s) from the input list not found in DisGeNET CURATEDRP4-758J18.13 gene(s) from the input list not found in DisGeNET CURATEDSENP3 gene(s) from the input list not found in DisGeNET CURATEDSHISA8 gene(s) from the input list not found in DisGeNET CURATEDTTC33 gene(s) from the input list not found in DisGeNET CURATEDPGBD2 gene(s) from the input list not found in DisGeNET CURATEDTMEM179B gene(s) from the input list not found in DisGeNET CURATEDARHGEF25 gene(s) from the input list not found in DisGeNET CURATEDZNF827 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDRGS19 gene(s) from the input list not found in DisGeNET CURATEDRP11-757G1.6 gene(s) from the input list not found in DisGeNET CURATEDLINC00930 gene(s) from the input list not found in DisGeNET CURATEDSSPO gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDMAP6D1 gene(s) from the input list not found in DisGeNET CURATEDNPNT gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDCIB4 gene(s) from the input list not found in DisGeNET CURATEDZBTB46 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDC17orf82 gene(s) from the input list not found in DisGeNET CURATEDGLTP gene(s) from the input list not found in DisGeNET CURATEDCTDSPL gene(s) from the input list not found in DisGeNET CURATEDNUDT16L1 gene(s) from the input list not found in DisGeNET CURATEDSF3B3 gene(s) from the input list not found in DisGeNET CURATEDLINC01451 gene(s) from the input list not found in DisGeNET CURATEDRP11-405A12.2 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATEDif (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
} Description FDR Ratio BgRatio
13 Alopecia 9.720447e-05 5/65 19/9703
301 Left Ventricular Hypertrophy 8.122766e-03 4/65 25/9703
84 Diabetes Mellitus 1.486101e-02 4/65 32/9703
318 Androgenetic Alopecia 1.597778e-02 3/65 15/9703
391 Female pattern alopecia (disorder) 1.597778e-02 3/65 15/9703
741 Alopecia, Male Pattern 1.597778e-02 3/65 15/9703
131 HSAN Type IV 2.043190e-02 2/65 4/9703
205 Hereditary Sensory and Autonomic Neuropathies 2.043190e-02 2/65 4/9703
291 Hereditary Sensory Radicular Neuropathy 2.043190e-02 2/65 4/9703
297 Pseudopelade 2.043190e-02 3/65 19/9703
412 Lung Injury 2.269056e-02 3/65 21/9703
677 Chronic Lung Injury 2.269056e-02 3/65 21/9703
130 Hereditary Sensory Autonomic Neuropathy, Type 2 2.608142e-02 2/65 5/9703
69 Coronary heart disease 2.917585e-02 3/65 24/9703
26 Cardiac Arrhythmia 3.079646e-02 3/65 25/9703
505 Sensory Neuropathy, Hereditary 3.164928e-02 2/65 6/9703
17 Congenital Pain Insensitivity 3.233266e-02 2/65 7/9703
68 Coronary Arteriosclerosis 3.233266e-02 4/65 65/9703
132 Hereditary Sensory Autonomic Neuropathy, Type 5 3.233266e-02 2/65 7/9703
143 Hypokalemia 3.233266e-02 2/65 7/9703
155 Lead Poisoning 3.233266e-02 2/65 7/9703
668 Coronary Artery Disease 3.233266e-02 4/65 65/9703
129 Hereditary Sensory Autonomic Neuropathy, Type 1 4.074377e-02 2/65 8/9703
11 Alkalosis 4.374477e-02 1/65 1/9703
36 Birth Weight 4.374477e-02 2/65 14/9703
48 Cardiomyopathy, Alcoholic 4.374477e-02 1/65 1/9703
50 Cardiovascular Diseases 4.374477e-02 3/65 36/9703
51 Carotid Artery Diseases 4.374477e-02 2/65 11/9703
74 Hearing Loss, Sudden 4.374477e-02 1/65 1/9703
87 Diabetic Angiopathies 4.374477e-02 2/65 16/9703
89 Diabetic Retinopathy 4.374477e-02 2/65 11/9703
117 Congenital Heart Defects 4.374477e-02 3/65 44/9703
120 Heart failure 4.374477e-02 4/65 110/9703
121 Congestive heart failure 4.374477e-02 4/65 110/9703
135 Hyperemia 4.374477e-02 2/65 13/9703
138 Hypertensive disease 4.374477e-02 6/65 190/9703
142 Hypertrophy 4.374477e-02 2/65 18/9703
156 Left-Sided Heart Failure 4.374477e-02 4/65 110/9703
157 Chronic Lymphocytic Leukemia 4.374477e-02 3/65 55/9703
158 Childhood Acute Lymphoblastic Leukemia 4.374477e-02 3/65 52/9703
159 L2 Acute Lymphoblastic Leukemia 4.374477e-02 3/65 50/9703
180 Microangiopathy, Diabetic 4.374477e-02 2/65 16/9703
200 nervous system disorder 4.374477e-02 3/65 53/9703
225 Psychosis, Brief Reactive 4.374477e-02 2/65 14/9703
236 Schizophreniform Disorders 4.374477e-02 2/65 14/9703
244 Spasmophilia 4.374477e-02 1/65 1/9703
256 Tetany 4.374477e-02 1/65 1/9703
258 Thrombosis 4.374477e-02 3/65 49/9703
266 Venous Engorgement 4.374477e-02 2/65 13/9703
306 Myocardial Ischemia 4.374477e-02 5/65 176/9703
326 Reactive Hyperemia 4.374477e-02 2/65 13/9703
342 Microvascular Angina 4.374477e-02 1/65 1/9703
362 Heart Failure, Right-Sided 4.374477e-02 4/65 110/9703
373 Female Pseudohermaphroditism 4.374477e-02 1/65 1/9703
403 Tetany, Neonatal 4.374477e-02 1/65 1/9703
421 Active Hyperemia 4.374477e-02 2/65 13/9703
431 Vitamin D-Dependent Rickets, Type 2A 4.374477e-02 1/65 1/9703
440 Erythrocyte Mean Corpuscular Hemoglobin Test 4.374477e-02 2/65 13/9703
474 Sleep Apnea, Obstructive 4.374477e-02 1/65 1/9703
480 Maxillofacial Abnormalities 4.374477e-02 1/65 1/9703
482 Mood Disorders 4.374477e-02 5/65 168/9703
485 Endomyocardial Fibrosis 4.374477e-02 2/65 9/9703
489 Carotid Atherosclerosis 4.374477e-02 2/65 11/9703
494 External Carotid Artery Diseases 4.374477e-02 2/65 11/9703
519 Internal Carotid Artery Diseases 4.374477e-02 2/65 11/9703
520 Arterial Diseases, Common Carotid 4.374477e-02 2/65 11/9703
556 Upper Airway Resistance Sleep Apnea Syndrome 4.374477e-02 1/65 1/9703
568 Acute Cerebrovascular Accidents 4.374477e-02 3/65 54/9703
578 Blood Coagulation Disorders, Inherited 4.374477e-02 1/65 1/9703
580 Trichomegaly 4.374477e-02 1/65 1/9703
591 Tetanilla 4.374477e-02 1/65 1/9703
594 Microsatellite Instability 4.374477e-02 2/65 15/9703
603 Deafness, Sudden 4.374477e-02 1/65 1/9703
607 Glucose Metabolism Disorders 4.374477e-02 1/65 1/9703
608 Finding of Mean Corpuscular Hemoglobin 4.374477e-02 2/65 13/9703
636 Replication Error Phenotype 4.374477e-02 2/65 15/9703
638 NEPHROLITHIASIS, CALCIUM OXALATE 4.374477e-02 1/65 1/9703
640 Disproportionate tall stature 4.374477e-02 1/65 1/9703
642 Band Heterotopia of Brain 4.374477e-02 1/65 1/9703
643 Cerebral Autosomal Recessive Arteriopathy with Subcortical Infarcts and Leukoencephalopathy 4.374477e-02 1/65 1/9703
644 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 4.374477e-02 1/65 1/9703
645 Glucocorticoid Receptor Deficiency 4.374477e-02 1/65 1/9703
646 Pseudohermaphroditism, Female, With Hypokalemia, Due To Glucocorticoid Resistance 4.374477e-02 1/65 1/9703
647 BODY COMPOSITION, BENEFICIAL 4.374477e-02 1/65 1/9703
651 Deafness, Autosomal Recessive 28 4.374477e-02 1/65 1/9703
653 HOMOCYSTINURIA DUE TO DEFICIENCY OF N(5,10)-METHYLENETETRAHYDROFOLATE REDUCTASE ACTIVITY 4.374477e-02 1/65 1/9703
654 Mthfr Deficiency, Thermolabile Type 4.374477e-02 1/65 1/9703
655 Methylenetetrahydrofolate reductase deficiency 4.374477e-02 1/65 1/9703
656 Macular Degeneration, Age-Related, 7 4.374477e-02 1/65 1/9703
657 Charcot-Marie-Tooth disease, Type 4B2 4.374477e-02 1/65 1/9703
658 SPINAL MUSCULAR ATROPHY WITH RESPIRATORY DISTRESS 1 4.374477e-02 1/65 1/9703
661 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 4.374477e-02 1/65 1/9703
669 Myocardial Failure 4.374477e-02 4/65 110/9703
672 Heart Decompensation 4.374477e-02 4/65 110/9703
680 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 4.374477e-02 1/65 1/9703
682 CILIARY DYSKINESIA, PRIMARY, 7 (disorder) 4.374477e-02 1/65 1/9703
686 Myopathy, Mitochondrial Progressive, With Congenital Cataract, Hearing Loss, And Developmental Delay 4.374477e-02 1/65 1/9703
694 46, XX Disorders of Sex Development 4.374477e-02 1/65 1/9703
698 MACULAR DEGENERATION, AGE-RELATED, 8 4.374477e-02 1/65 1/9703
699 HYPOMAGNESEMIA 6, RENAL 4.374477e-02 1/65 1/9703
712 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 4.374477e-02 1/65 1/9703
722 MACROCEPHALY AND EPILEPTIC ENCEPHALOPATHY 4.374477e-02 1/65 1/9703
723 PULMONARY HYPERTENSION, PRIMARY, 4 4.374477e-02 1/65 1/9703
724 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 4.374477e-02 1/65 1/9703
727 Phakomatosis cesiomarmorata 4.374477e-02 1/65 1/9703
732 Very long chain acyl-CoA dehydrogenase deficiency 4.374477e-02 1/65 1/9703
734 Glomerulopathy with fibronectin deposits 4.374477e-02 1/65 1/9703
735 SPASTIC PARAPLEGIA 45, AUTOSOMAL RECESSIVE 4.374477e-02 1/65 1/9703
736 CHARCOT-MARIE-TOOTH DISEASE, AXONAL, TYPE 2S 4.374477e-02 1/65 1/9703
737 HOMOCYSTINURIA DUE TO MTHFR DEFICIENCY 4.374477e-02 1/65 1/9703
743 CEREBRAL ARTERIOPATHY, AUTOSOMAL DOMINANT, WITH SUBCORTICAL INFARCTS AND LEUKOENCEPHALOPATHY, TYPE 2 4.374477e-02 1/65 1/9703
744 HYPOMAGNESEMIA, SEIZURES, AND MENTAL RETARDATION 4.374477e-02 1/65 1/9703
745 MANDIBULOFACIAL DYSOSTOSIS WITH ALOPECIA 4.374477e-02 1/65 1/9703
750 BAND HETEROTOPIA 4.374477e-02 1/65 1/9703
758 MENTAL RETARDATION, AUTOSOMAL DOMINANT 46 4.374477e-02 1/65 1/9703
759 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 4.374477e-02 1/65 1/9703
762 Ciliary Dyskinesia, Primary, 7, With Or Without Situs Inversus 4.374477e-02 1/65 1/9703
769 Familial isolated trichomegaly 4.374477e-02 1/65 1/9703
777 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 70 4.374477e-02 1/65 1/9703
79 Mental Depression 4.401299e-02 6/65 254/9703
92 Down Syndrome 4.401299e-02 2/65 19/9703
464 Down Syndrome, Partial Trisomy 21 4.401299e-02 2/65 19/9703
465 Trisomy 21, Meiotic Nondisjunction 4.401299e-02 2/65 19/9703
526 Trisomy 21, Mitotic Nondisjunction 4.401299e-02 2/65 19/9703
671 Precursor Cell Lymphoblastic Leukemia Lymphoma 4.872170e-02 3/65 61/9703Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes=ctwas_genes)
#background is union of genes analyzed in all tissue
background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background]})
####################
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
hyp_score_padj gene_set nset ngenes percent padj
1 gwascatalog 5959 66 0.011075684 2.721921e-06
2 fda_approved_drug_targets 350 7 0.020000000 1.952755e-02
3 mgi_essential 2299 24 0.010439321 2.052125e-02
4 clinvar_path_likelypath 2766 25 0.009038322 6.384479e-02
5 core_essentials_hart 265 3 0.011320755 2.454994e-01Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Cellular_Component_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Molecular_Function_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Enrichment analysis for cTWAS genes in top tissues separately
GO
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Artery_Aorta
Number of cTWAS Genes in Tissue: 26
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Artery_Tibial
Number of cTWAS Genes in Tissue: 23
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)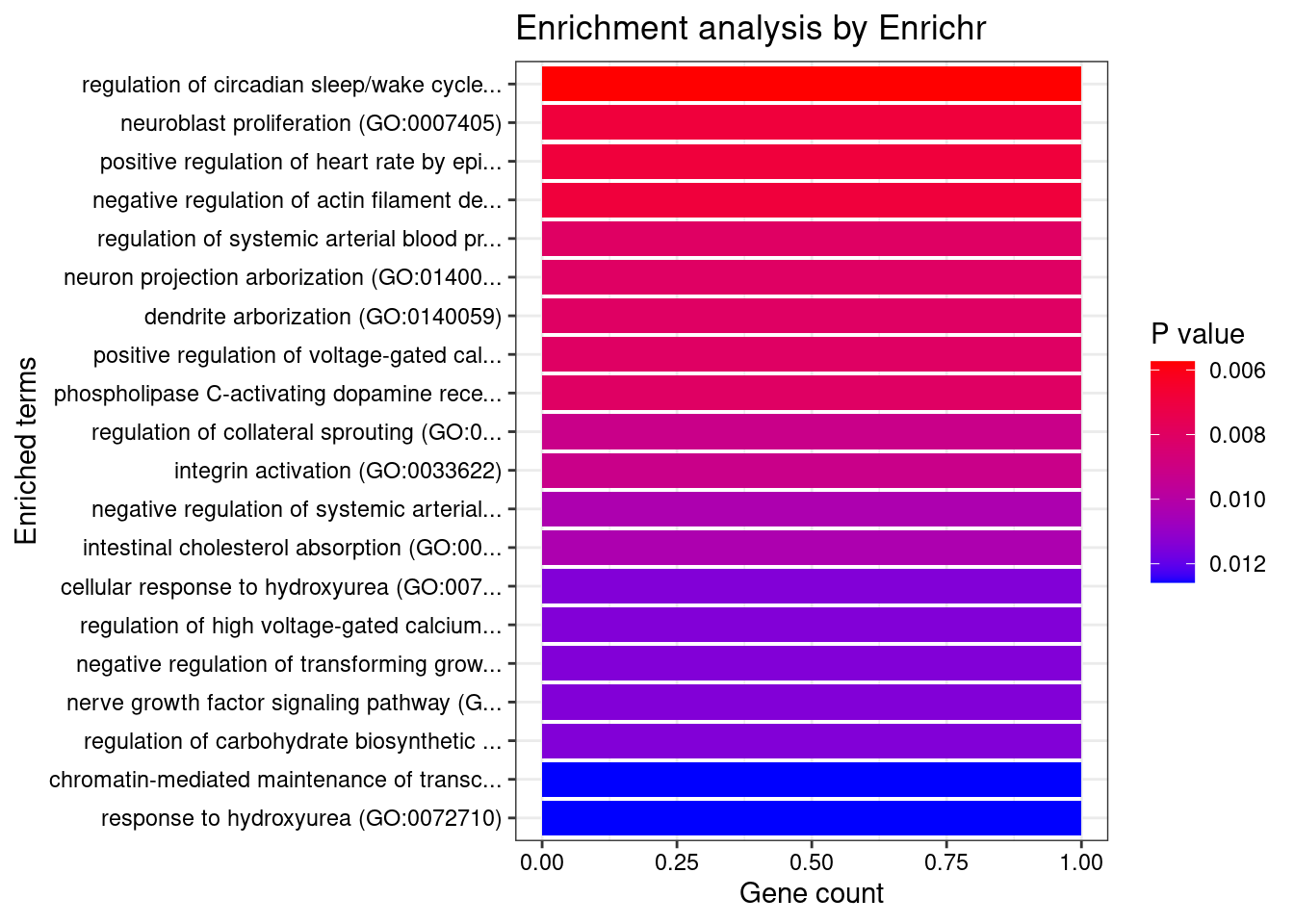
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Skin_Sun_Exposed_Lower_leg
Number of cTWAS Genes in Tissue: 15
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Colon_Transverse
Number of cTWAS Genes in Tissue: 9
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
| Version | Author | Date |
|---|---|---|
| 10b99b6 | wesleycrouse | 2022-03-23 |
Artery_Coronary
Number of cTWAS Genes in Tissue: 10
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 peptidyl-tyrosine autophosphorylation (GO:0038083) 2/15 0.004184002 NTRK1;AATK
2 autonomic nervous system development (GO:0048483) 2/21 0.004184002 NTRK1;FN1
3 peptidyl-tyrosine phosphorylation (GO:0018108) 2/69 0.028716927 NTRK1;AATK
4 positive regulation of phosphatidylinositol 3-kinase signaling (GO:0014068) 2/77 0.028716927 NTRK1;FN1
5 regulation of phosphatidylinositol 3-kinase signaling (GO:0014066) 2/106 0.042294647 NTRK1;FN1
6 protein autophosphorylation (GO:0046777) 2/159 0.042294647 NTRK1;AATK
7 regulation of MAPK cascade (GO:0043408) 2/166 0.042294647 NTRK1;FN1
8 cellular response to iron ion (GO:0071281) 1/6 0.042294647 HFE
9 negative regulation of CD8-positive, alpha-beta T cell activation (GO:2001186) 1/6 0.042294647 HFE
10 positive regulation of receptor binding (GO:1900122) 1/6 0.042294647 HFE
11 dendrite arborization (GO:0140059) 1/7 0.042294647 PHACTR1
12 neuron projection arborization (GO:0140058) 1/7 0.042294647 PHACTR1
13 positive regulation of peptide secretion (GO:0002793) 1/8 0.042294647 HFE
14 integrin activation (GO:0033622) 1/8 0.042294647 FN1
15 regulation of CD8-positive, alpha-beta T cell activation (GO:2001185) 1/9 0.042294647 HFE
16 negative regulation of T cell cytokine production (GO:0002725) 1/9 0.042294647 HFE
17 negative regulation of alpha-beta T cell activation (GO:0046636) 1/9 0.042294647 HFE
18 regulation of receptor binding (GO:1900120) 1/10 0.042294647 HFE
19 negative regulation of transforming growth factor beta production (GO:0071635) 1/10 0.042294647 FN1
20 nerve growth factor signaling pathway (GO:0038180) 1/10 0.042294647 NTRK1
21 negative regulation of receptor binding (GO:1900121) 1/10 0.042294647 HFE
22 regulation of coagulation (GO:0050818) 1/11 0.042928594 PROCR
23 regulation of ERK1 and ERK2 cascade (GO:0070372) 2/238 0.042928594 NTRK1;FN1
24 response to iron ion (GO:0010039) 1/13 0.042928594 HFE
25 negative regulation of T cell mediated immunity (GO:0002710) 1/14 0.042928594 HFE
26 negative regulation of coagulation (GO:0050819) 1/14 0.042928594 PROCR
27 regulation of protein phosphorylation (GO:0001932) 2/266 0.042928594 NTRK1;FN1
28 protein-containing complex assembly (GO:0065003) 2/267 0.042928594 HFE;FN1
29 stress fiber assembly (GO:0043149) 1/15 0.042928594 PHACTR1
30 neurotrophin TRK receptor signaling pathway (GO:0048011) 1/15 0.042928594 NTRK1
31 contractile actin filament bundle assembly (GO:0030038) 1/15 0.042928594 PHACTR1
32 sympathetic nervous system development (GO:0048485) 1/16 0.044339623 NTRK1
33 preassembly of GPI anchor in ER membrane (GO:0016254) 1/17 0.044339623 PIGV
34 negative regulation of cytokine production involved in immune response (GO:0002719) 1/17 0.044339623 HFE
35 regulation of transforming growth factor beta production (GO:0071634) 1/18 0.045596226 FN1
36 regulation of T cell cytokine production (GO:0002724) 1/19 0.046781913 HFE
37 positive regulation of hormone secretion (GO:0046887) 1/20 0.047902435 HFE
38 neurotrophin signaling pathway (GO:0038179) 1/21 0.048962936 NTRK1
39 cellular response to nerve growth factor stimulus (GO:1990090) 1/22 0.049968033 NTRK1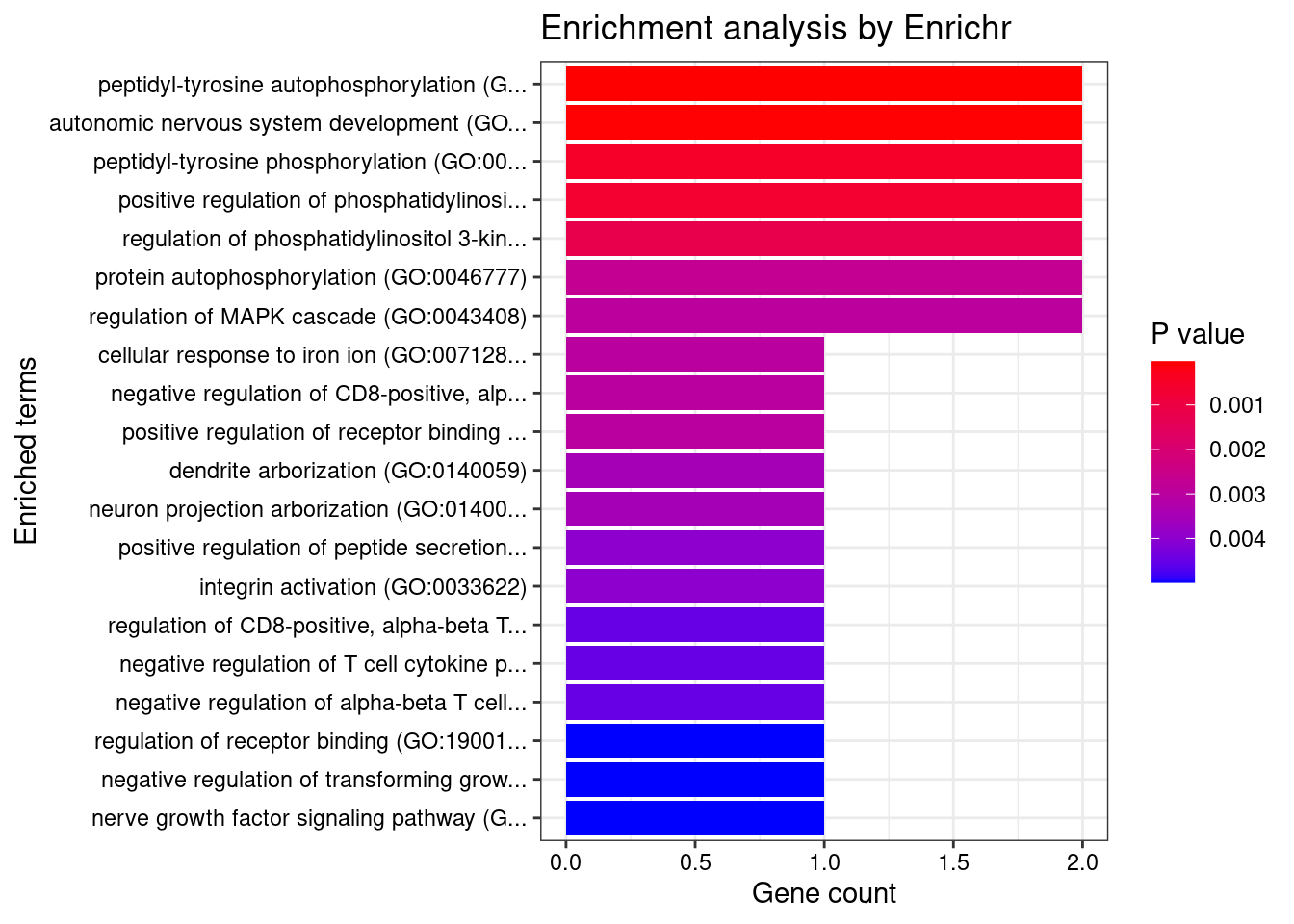
KEGG
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- NULL
try(enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_tissue, referenceGene=background_tissue,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F))
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
} Artery_Aorta
Number of cTWAS Genes in Tissue: 26
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Artery_Tibial
Number of cTWAS Genes in Tissue: 23
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Skin_Sun_Exposed_Lower_leg
Number of cTWAS Genes in Tissue: 15
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Colon_Transverse
Number of cTWAS Genes in Tissue: 9
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Artery_Coronary
Number of cTWAS Genes in Tissue: 10
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_tissue, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
} Artery_Aorta
Number of cTWAS Genes in Tissue: 26PAQR5 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDSHB gene(s) from the input list not found in DisGeNET CURATEDGDF7 gene(s) from the input list not found in DisGeNET CURATEDRP4-758J18.13 gene(s) from the input list not found in DisGeNET CURATEDLRRC10B gene(s) from the input list not found in DisGeNET CURATEDKIF13B gene(s) from the input list not found in DisGeNET CURATEDKIAA1462 gene(s) from the input list not found in DisGeNET CURATEDRP11-405A12.2 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDRP11-1055B8.3 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
36 Diabetes Mellitus 0.02481962 2/14 32/9703
100 Alcohol abuse 0.02481962 3/14 67/9703
162 Sleep Apnea, Obstructive 0.02481962 1/14 1/9703
179 Upper Airway Resistance Sleep Apnea Syndrome 0.02481962 1/14 1/9703
209 Band Heterotopia of Brain 0.02481962 1/14 1/9703
210 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 0.02481962 1/14 1/9703
212 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.02481962 1/14 1/9703
221 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.02481962 1/14 1/9703
234 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.02481962 1/14 1/9703
239 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 0.02481962 1/14 1/9703
242 Phakomatosis cesiomarmorata 0.02481962 1/14 1/9703
244 Glomerulopathy with fibronectin deposits 0.02481962 1/14 1/9703
246 MANDIBULOFACIAL DYSOSTOSIS WITH ALOPECIA 0.02481962 1/14 1/9703
247 BAND HETEROTOPIA 0.02481962 1/14 1/9703
258 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 70 0.02481962 1/14 1/9703
50 Congenital Heart Defects 0.02849908 2/14 44/9703
110 Variegate Porphyria 0.02976359 1/14 2/9703
127 Congenital hemangioma 0.02976359 1/14 2/9703
157 Spondylometaphyseal dysplasia, 'corner fracture' type 0.02976359 1/14 2/9703
196 Port-wine stain with oculocutaneous melanosis 0.02976359 1/14 2/9703
227 Porphyria, South African type 0.02976359 1/14 2/9703
237 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1 0.02976359 1/14 2/9703
241 Phakomatosis cesioflammea 0.02976359 1/14 2/9703
245 Autosomal dominant hypocalcemia 0.02976359 1/14 2/9703
250 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1, WITH BARTTER SYNDROME 0.02976359 1/14 2/9703
232 HEMOCHROMATOSIS, TYPE 1 0.03846162 1/14 3/9703
233 Ovarian clear cell carcinoma 0.03846162 1/14 3/9703
243 MIGRAINE WITH OR WITHOUT AURA, SUSCEPTIBILITY TO, 1 0.03846162 1/14 3/9703
249 Idiopathic basal ganglia calcification 1 0.03846162 1/14 3/9703
97 Wallerian Degeneration 0.04219293 1/14 4/9703
111 Porphyria Cutanea Tarda 0.04219293 1/14 4/9703
131 Mandibulofacial Dysostosis 0.04219293 1/14 4/9703
147 Malignant melanoma of iris 0.04219293 1/14 5/9703
148 Malignant melanoma of choroid 0.04219293 1/14 5/9703
154 Fahr's syndrome (disorder) 0.04219293 1/14 5/9703
155 Anterior Cerebral Circulation Infarction 0.04219293 1/14 5/9703
171 Breast Carcinoma 0.04219293 4/14 538/9703
180 Anterior Circulation Brain Infarction 0.04219293 1/14 5/9703
181 Brain Infarction, Posterior Circulation 0.04219293 1/14 5/9703
182 Venous Infarction, Brain 0.04219293 1/14 5/9703
183 Brain Infarction 0.04219293 1/14 5/9703
188 Genomic Instability 0.04219293 1/14 5/9703
211 Subcortical Band Heterotopia 0.04219293 1/14 4/9703
229 MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 0.04219293 1/14 5/9703
108 Streptococcal pneumonia 0.04947322 1/14 6/9703
Artery_Tibial
Number of cTWAS Genes in Tissue: 23CCNT2 gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDZNF692 gene(s) from the input list not found in DisGeNET CURATEDGDF7 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDZNF827 gene(s) from the input list not found in DisGeNET CURATEDDDI2 gene(s) from the input list not found in DisGeNET CURATEDZBTB46 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDRP11-405A12.2 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
31 Diabetes Mellitus 0.03137123 2/12 32/9703
230 Band Heterotopia of Brain 0.03137123 1/12 1/9703
231 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 0.03137123 1/12 1/9703
233 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.03137123 1/12 1/9703
250 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.03137123 1/12 1/9703
255 PULMONARY HYPERTENSION, PRIMARY, 4 0.03137123 1/12 1/9703
256 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 0.03137123 1/12 1/9703
259 Phakomatosis cesiomarmorata 0.03137123 1/12 1/9703
262 Glomerulopathy with fibronectin deposits 0.03137123 1/12 1/9703
265 BAND HETEROTOPIA 0.03137123 1/12 1/9703
279 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 70 0.03137123 1/12 1/9703
23 Corneal Ulcer 0.03630398 1/12 2/9703
118 Congenital hemangioma 0.03630398 1/12 2/9703
159 Spondylometaphyseal dysplasia, 'corner fracture' type 0.03630398 1/12 2/9703
221 Port-wine stain with oculocutaneous melanosis 0.03630398 1/12 2/9703
253 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1 0.03630398 1/12 2/9703
258 Phakomatosis cesioflammea 0.03630398 1/12 2/9703
264 Autosomal dominant hypocalcemia 0.03630398 1/12 2/9703
273 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1, WITH BARTTER SYNDROME 0.03630398 1/12 2/9703
28 Cystitis 0.04751682 1/12 4/9703
48 HSAN Type IV 0.04751682 1/12 4/9703
71 Hereditary Sensory and Autonomic Neuropathies 0.04751682 1/12 4/9703
79 Skin Ulcer 0.04751682 1/12 4/9703
92 Hereditary Sensory Radicular Neuropathy 0.04751682 1/12 4/9703
130 Overactive Detrusor 0.04751682 1/12 4/9703
214 Overactive Bladder 0.04751682 1/12 4/9703
232 Subcortical Band Heterotopia 0.04751682 1/12 4/9703
249 Ovarian clear cell carcinoma 0.04751682 1/12 3/9703
271 Idiopathic basal ganglia calcification 1 0.04751682 1/12 3/9703
Skin_Sun_Exposed_Lower_leg
Number of cTWAS Genes in Tissue: 15RP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDPGBD2 gene(s) from the input list not found in DisGeNET CURATEDGLTP gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDMEX3A gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
50 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.01298701 1/8 1/9703
59 PULMONARY HYPERTENSION, PRIMARY, 4 0.01298701 1/8 1/9703
61 MENTAL RETARDATION, AUTOSOMAL DOMINANT 46 0.01298701 1/8 1/9703
62 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01298701 1/8 1/9703
26 Variegate Porphyria 0.01730977 1/8 2/9703
52 Porphyria, South African type 0.01730977 1/8 2/9703
56 HEMOCHROMATOSIS, TYPE 1 0.02224739 1/8 3/9703
27 Porphyria Cutanea Tarda 0.02306304 1/8 4/9703
28 Pilomatrixoma 0.02306304 1/8 4/9703
12 Lead Poisoning 0.02419000 1/8 7/9703
47 Hyperphosphatasia with Mental Retardation 0.02419000 1/8 7/9703
48 PULMONARY HYPERTENSION, PRIMARY, DEXFENFLURAMINE-ASSOCIATED 0.02419000 1/8 7/9703
49 Pulmonary Hypertension, Primary, Fenfluramine-Associated 0.02419000 1/8 7/9703
58 Pulmonary Hypertension, Primary, 1, With Hereditary Hemorrhagic Telangiectasia 0.02419000 1/8 7/9703
63 Pulmonary Hypertension, Primary, 1 0.02419000 1/8 7/9703
46 Familial pulmonary arterial hypertension 0.02590851 1/8 8/9703
25 Idiopathic pulmonary hypertension 0.02742265 1/8 9/9703
53 Pulmonary arterial hypertension 0.02876647 1/8 10/9703
3 Birth Weight 0.02926782 1/8 14/9703
8 Gliosis 0.02926782 1/8 17/9703
9 Hemochromatosis 0.02926782 1/8 12/9703
10 Hepatitis C 0.02926782 1/8 15/9703
15 Myopia 0.02926782 1/8 15/9703
33 Familial primary pulmonary hypertension 0.02926782 1/8 17/9703
34 Erythrocyte Mean Corpuscular Hemoglobin Test 0.02926782 1/8 13/9703
35 Hereditary hemochromatosis 0.02926782 1/8 12/9703
41 2-oxo-hept-3-ene-1,7-dioate hydratase activity 0.02926782 1/8 14/9703
42 Finding of Mean Corpuscular Hemoglobin 0.02926782 1/8 13/9703
54 Idiopathic pulmonary arterial hypertension 0.02926782 1/8 16/9703
60 Astrocytosis 0.02926782 1/8 17/9703
30 Congenital hernia of foramen of Morgagni 0.03064455 1/8 19/9703
31 Congenital hernia of foramen of Bochdalek 0.03064455 1/8 19/9703
17 Osteoarthritis of hip 0.03126865 1/8 20/9703
29 Congenital diaphragmatic hernia 0.03185494 1/8 21/9703
51 Hematopoetic Myelodysplasia 0.04261026 1/8 29/9703
7 Dermatitis, Atopic 0.04991026 1/8 36/9703
23 Eczema, Infantile 0.04991026 1/8 36/9703
Colon_Transverse
Number of cTWAS Genes in Tissue: 9SOX13 gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDTMEM179B gene(s) from the input list not found in DisGeNET CURATEDCPXM1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
9 Hypokinesia 0.004204018 1/4 3/9703
12 Bradykinesia 0.004204018 1/4 3/9703
13 Hypodynamia 0.004204018 1/4 3/9703
15 Hypokinesia, Antiorthostatic 0.004204018 1/4 3/9703
17 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.004204018 1/4 1/9703
11 Pilomatrixoma 0.004670409 1/4 4/9703
5 Morphine Dependence 0.005446283 1/4 7/9703
14 Morphine Abuse 0.005446283 1/4 7/9703
16 Hyperphosphatasia with Mental Retardation 0.005446283 1/4 7/9703
2 Cardiac Arrhythmia 0.017457239 1/4 25/9703
3 Dermatitis, Atopic 0.020913066 1/4 36/9703
8 Eczema, Infantile 0.020913066 1/4 36/9703
6 Substance Withdrawal Syndrome 0.026863005 1/4 58/9703
7 Drug Withdrawal Symptoms 0.026863005 1/4 58/9703
10 Withdrawal Symptoms 0.026863005 1/4 58/9703
Artery_Coronary
Number of cTWAS Genes in Tissue: 10RP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
13 Coronary Arteriosclerosis 0.01495779 2/7 65/9703
54 Alcohol abuse 0.01495779 2/7 67/9703
109 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.01495779 1/7 1/9703
110 Coronary Artery Disease 0.01495779 2/7 65/9703
112 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.01495779 1/7 1/9703
119 Glomerulopathy with fibronectin deposits 0.01495779 1/7 1/9703
120 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01495779 1/7 1/9703
124 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 70 0.01495779 1/7 1/9703
60 Variegate Porphyria 0.01626153 1/7 2/9703
81 Spondylometaphyseal dysplasia, 'corner fracture' type 0.01626153 1/7 2/9703
114 Porphyria, South African type 0.01626153 1/7 2/9703
107 Familial medullary thyroid carcinoma 0.02063325 1/7 3/9703
116 HEMOCHROMATOSIS, TYPE 1 0.02063325 1/7 3/9703
26 HSAN Type IV 0.02103132 1/7 4/9703
39 Hereditary Sensory and Autonomic Neuropathies 0.02103132 1/7 4/9703
55 Hereditary Sensory Radicular Neuropathy 0.02103132 1/7 4/9703
61 Porphyria Cutanea Tarda 0.02103132 1/7 4/9703
25 Hereditary Sensory Autonomic Neuropathy, Type 2 0.02482097 1/7 5/9703
4 Congenital Pain Insensitivity 0.02604590 1/7 7/9703
27 Hereditary Sensory Autonomic Neuropathy, Type 5 0.02604590 1/7 7/9703
30 Lead Poisoning 0.02604590 1/7 7/9703
74 Common Migraine 0.02604590 1/7 7/9703
89 Sensory Neuropathy, Hereditary 0.02604590 1/7 6/9703
108 Hyperphosphatasia with Mental Retardation 0.02604590 1/7 7/9703
24 Hereditary Sensory Autonomic Neuropathy, Type 1 0.02856724 1/7 8/9703
5 Aortic Valve Insufficiency 0.03238257 1/7 11/9703
8 Birth Weight 0.03238257 1/7 14/9703
22 Hemochromatosis 0.03238257 1/7 12/9703
23 Hepatitis C 0.03238257 1/7 15/9703
37 Neuralgia 0.03238257 1/7 16/9703
42 Psychosis, Brief Reactive 0.03238257 1/7 14/9703
47 Schizophreniform Disorders 0.03238257 1/7 14/9703
50 Neuralgia, Supraorbital 0.03238257 1/7 16/9703
53 Neuralgia, Vidian 0.03238257 1/7 16/9703
66 Neuralgia, Atypical 0.03238257 1/7 16/9703
67 Neuralgia, Stump 0.03238257 1/7 16/9703
77 Erythrocyte Mean Corpuscular Hemoglobin Test 0.03238257 1/7 13/9703
78 Hereditary hemochromatosis 0.03238257 1/7 12/9703
79 Neuralgia, Perineal 0.03238257 1/7 16/9703
80 Neuralgia, Iliohypogastric Nerve 0.03238257 1/7 16/9703
92 Neuralgia, Ilioinguinal 0.03238257 1/7 16/9703
93 Nerve Pain 0.03238257 1/7 16/9703
94 Paroxysmal Nerve Pain 0.03238257 1/7 16/9703
99 Finding of Mean Corpuscular Hemoglobin 0.03238257 1/7 13/9703
40 Osteoarthritis of hip 0.03952979 1/7 20/9703
71 Lung Injury 0.03972778 1/7 21/9703
111 Chronic Lung Injury 0.03972778 1/7 21/9703
95 Lewy Body Disease 0.04073991 1/7 22/9703
102 Ovarian Serous Adenocarcinoma 0.04170961 1/7 23/9703
49 West Syndrome 0.04180336 1/7 24/9703
52 Ureteral obstruction 0.04180336 1/7 24/9703
57 Left Ventricular Hypertrophy 0.04269457 1/7 25/9703
45 Schizoaffective Disorder 0.04676645 1/7 29/9703
56 Hyalinosis, Segmental Glomerular 0.04676645 1/7 28/9703
113 Hematopoetic Myelodysplasia 0.04676645 1/7 29/9703
101 Ovarian Mucinous Adenocarcinoma 0.04906868 1/7 31/9703
16 Diabetes Mellitus 0.04974755 1/7 32/9703Gene sets curated by Macarthur Lab
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_tissue=ctwas_genes_tissue)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_tissue]})
##########
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_tissue)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
} Artery_Aorta
Number of cTWAS Genes in Tissue: 26
gene_set nset ngenes percent padj
1 mgi_essential 1443 7 0.004851005 0.1410141
2 fda_approved_drug_targets 203 2 0.009852217 0.1735835
3 gwascatalog 3879 11 0.002835782 0.2271601
4 clinvar_path_likelypath 1814 6 0.003307607 0.2271601
5 core_essentials_hart 180 1 0.005555556 0.3218188
Artery_Tibial
Number of cTWAS Genes in Tissue: 23
gene_set nset ngenes percent padj
1 gwascatalog 3922 14 0.003569607 0.01597461
2 fda_approved_drug_targets 206 3 0.014563107 0.01597461
3 clinvar_path_likelypath 1865 7 0.003753351 0.08059937
4 mgi_essential 1468 5 0.003405995 0.16337587
5 core_essentials_hart 193 1 0.005181347 0.30495981
Skin_Sun_Exposed_Lower_leg
Number of cTWAS Genes in Tissue: 15
gene_set nset ngenes percent padj
1 gwascatalog 4062 11 0.002708026 0.005374155
2 mgi_essential 1511 3 0.001985440 0.423713125
3 fda_approved_drug_targets 218 1 0.004587156 0.423713125
4 clinvar_path_likelypath 1912 3 0.001569038 0.486566185
5 core_essentials_hart 199 0 0.000000000 1.000000000
Colon_Transverse
Number of cTWAS Genes in Tissue: 9
gene_set nset ngenes percent padj
1 gwascatalog 3778 7 0.0018528322 0.0276595
2 mgi_essential 1392 3 0.0021551724 0.1880489
3 core_essentials_hart 186 0 0.0000000000 1.0000000
4 clinvar_path_likelypath 1782 1 0.0005611672 1.0000000
5 fda_approved_drug_targets 197 0 0.0000000000 1.0000000
Artery_Coronary
Number of cTWAS Genes in Tissue: 10
gene_set nset ngenes percent padj
1 fda_approved_drug_targets 190 2 0.010526316 0.05732035
2 gwascatalog 3581 6 0.001675510 0.09903238
3 clinvar_path_likelypath 1684 4 0.002375297 0.09903238
4 mgi_essential 1317 3 0.002277904 0.12516755
5 core_essentials_hart 176 1 0.005681818 0.14461588Summary of results across tissues
weight_groups <- as.data.frame(matrix(c("Adipose_Subcutaneous", "Adipose",
"Adipose_Visceral_Omentum", "Adipose",
"Adrenal_Gland", "Endocrine",
"Artery_Aorta", "Cardiovascular",
"Artery_Coronary", "Cardiovascular",
"Artery_Tibial", "Cardiovascular",
"Brain_Amygdala", "CNS",
"Brain_Anterior_cingulate_cortex_BA24", "CNS",
"Brain_Caudate_basal_ganglia", "CNS",
"Brain_Cerebellar_Hemisphere", "CNS",
"Brain_Cerebellum", "CNS",
"Brain_Cortex", "CNS",
"Brain_Frontal_Cortex_BA9", "CNS",
"Brain_Hippocampus", "CNS",
"Brain_Hypothalamus", "CNS",
"Brain_Nucleus_accumbens_basal_ganglia", "CNS",
"Brain_Putamen_basal_ganglia", "CNS",
"Brain_Spinal_cord_cervical_c-1", "CNS",
"Brain_Substantia_nigra", "CNS",
"Breast_Mammary_Tissue", "None",
"Cells_Cultured_fibroblasts", "Skin",
"Cells_EBV-transformed_lymphocytes", "Blood or Immune",
"Colon_Sigmoid", "Digestive",
"Colon_Transverse", "Digestive",
"Esophagus_Gastroesophageal_Junction", "Digestive",
"Esophagus_Mucosa", "Digestive",
"Esophagus_Muscularis", "Digestive",
"Heart_Atrial_Appendage", "Cardiovascular",
"Heart_Left_Ventricle", "Cardiovascular",
"Kidney_Cortex", "None",
"Liver", "None",
"Lung", "None",
"Minor_Salivary_Gland", "None",
"Muscle_Skeletal", "None",
"Nerve_Tibial", "None",
"Ovary", "None",
"Pancreas", "None",
"Pituitary", "Endocrine",
"Prostate", "None",
"Skin_Not_Sun_Exposed_Suprapubic", "Skin",
"Skin_Sun_Exposed_Lower_leg", "Skin",
"Small_Intestine_Terminal_Ileum", "Digestive",
"Spleen", "Blood or Immune",
"Stomach", "Digestive",
"Testis", "Endocrine",
"Thyroid", "Endocrine",
"Uterus", "None",
"Vagina", "None",
"Whole_Blood", "Blood or Immune"),
nrow=49, ncol=2, byrow=T), stringsAsFactors=F)
colnames(weight_groups) <- c("weight", "group")
#display tissue groups
print(weight_groups) weight group
1 Adipose_Subcutaneous Adipose
2 Adipose_Visceral_Omentum Adipose
3 Adrenal_Gland Endocrine
4 Artery_Aorta Cardiovascular
5 Artery_Coronary Cardiovascular
6 Artery_Tibial Cardiovascular
7 Brain_Amygdala CNS
8 Brain_Anterior_cingulate_cortex_BA24 CNS
9 Brain_Caudate_basal_ganglia CNS
10 Brain_Cerebellar_Hemisphere CNS
11 Brain_Cerebellum CNS
12 Brain_Cortex CNS
13 Brain_Frontal_Cortex_BA9 CNS
14 Brain_Hippocampus CNS
15 Brain_Hypothalamus CNS
16 Brain_Nucleus_accumbens_basal_ganglia CNS
17 Brain_Putamen_basal_ganglia CNS
18 Brain_Spinal_cord_cervical_c-1 CNS
19 Brain_Substantia_nigra CNS
20 Breast_Mammary_Tissue None
21 Cells_Cultured_fibroblasts Skin
22 Cells_EBV-transformed_lymphocytes Blood or Immune
23 Colon_Sigmoid Digestive
24 Colon_Transverse Digestive
25 Esophagus_Gastroesophageal_Junction Digestive
26 Esophagus_Mucosa Digestive
27 Esophagus_Muscularis Digestive
28 Heart_Atrial_Appendage Cardiovascular
29 Heart_Left_Ventricle Cardiovascular
30 Kidney_Cortex None
31 Liver None
32 Lung None
33 Minor_Salivary_Gland None
34 Muscle_Skeletal None
35 Nerve_Tibial None
36 Ovary None
37 Pancreas None
38 Pituitary Endocrine
39 Prostate None
40 Skin_Not_Sun_Exposed_Suprapubic Skin
41 Skin_Sun_Exposed_Lower_leg Skin
42 Small_Intestine_Terminal_Ileum Digestive
43 Spleen Blood or Immune
44 Stomach Digestive
45 Testis Endocrine
46 Thyroid Endocrine
47 Uterus None
48 Vagina None
49 Whole_Blood Blood or Immunegroups <- unique(weight_groups$group)
df_group <- list()
for (i in 1:length(groups)){
group <- groups[i]
weights <- weight_groups$weight[weight_groups$group==group]
df_group[[group]] <- list(ctwas=unique(unlist(lapply(df[weights], function(x){x$ctwas}))),
background=unique(unlist(lapply(df[weights], function(x){x$gene_pips$genename}))))
}
output <- output[sapply(weight_groups$weight, match, output$weight),,drop=F]
output$group <- weight_groups$group
output$n_ctwas_group <- sapply(output$group, function(x){length(df_group[[x]][["ctwas"]])})
output$n_ctwas_group[output$group=="None"] <- 0
#barplot of number of cTWAS genes in each tissue
output <- output[order(-output$n_ctwas),,drop=F]
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(output$n_ctwas, names.arg=output$weight, las=2, ylab="Number of cTWAS Genes", cex.names=0.6, main="Number of cTWAS Genes by Tissue")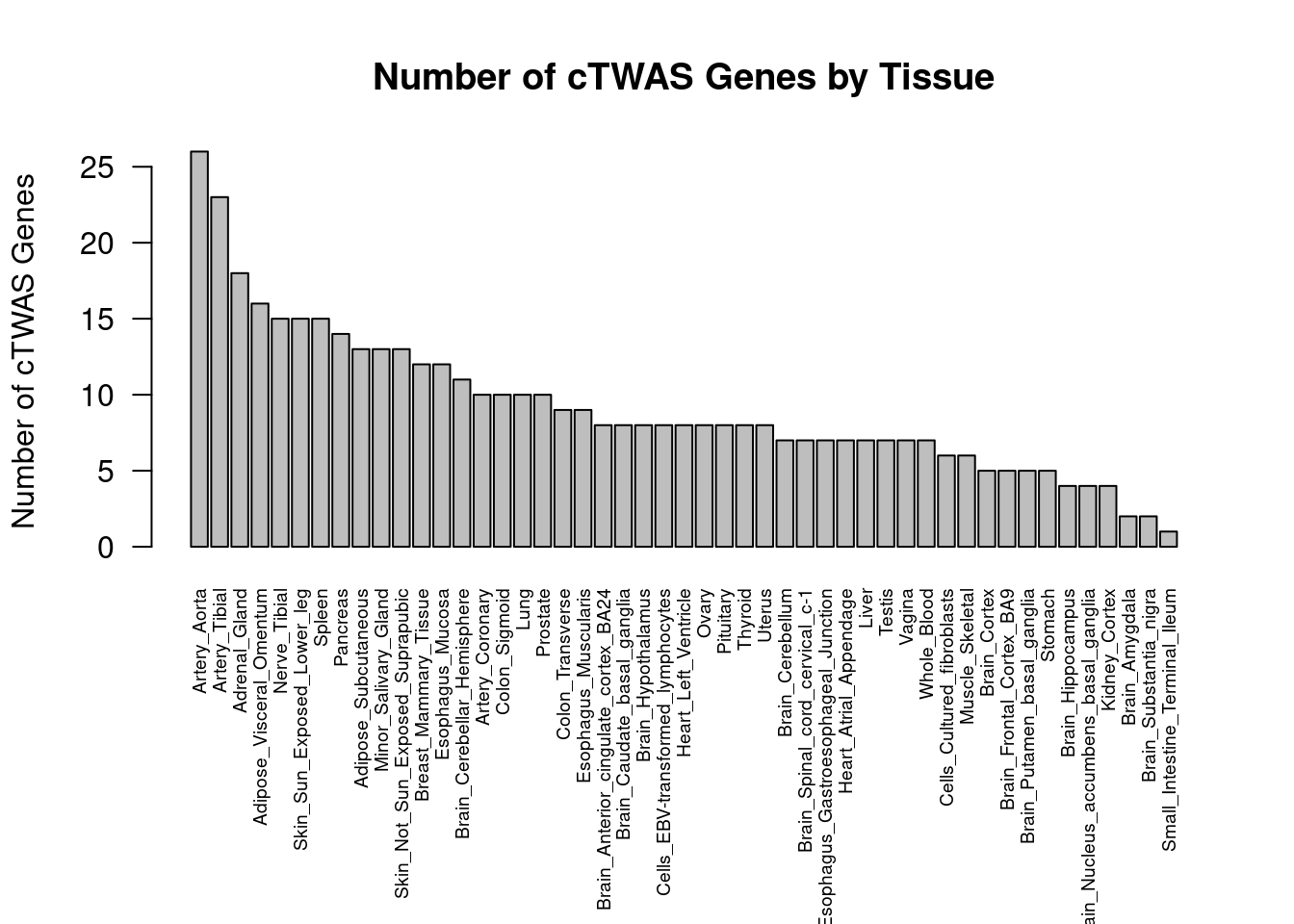
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
#barplot of number of cTWAS genes in each tissue
df_plot <- -sort(-sapply(groups[groups!="None"], function(x){length(df_group[[x]][["ctwas"]])}))
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(df_plot, las=2, ylab="Number of cTWAS Genes", main="Number of cTWAS Genes by Tissue Group")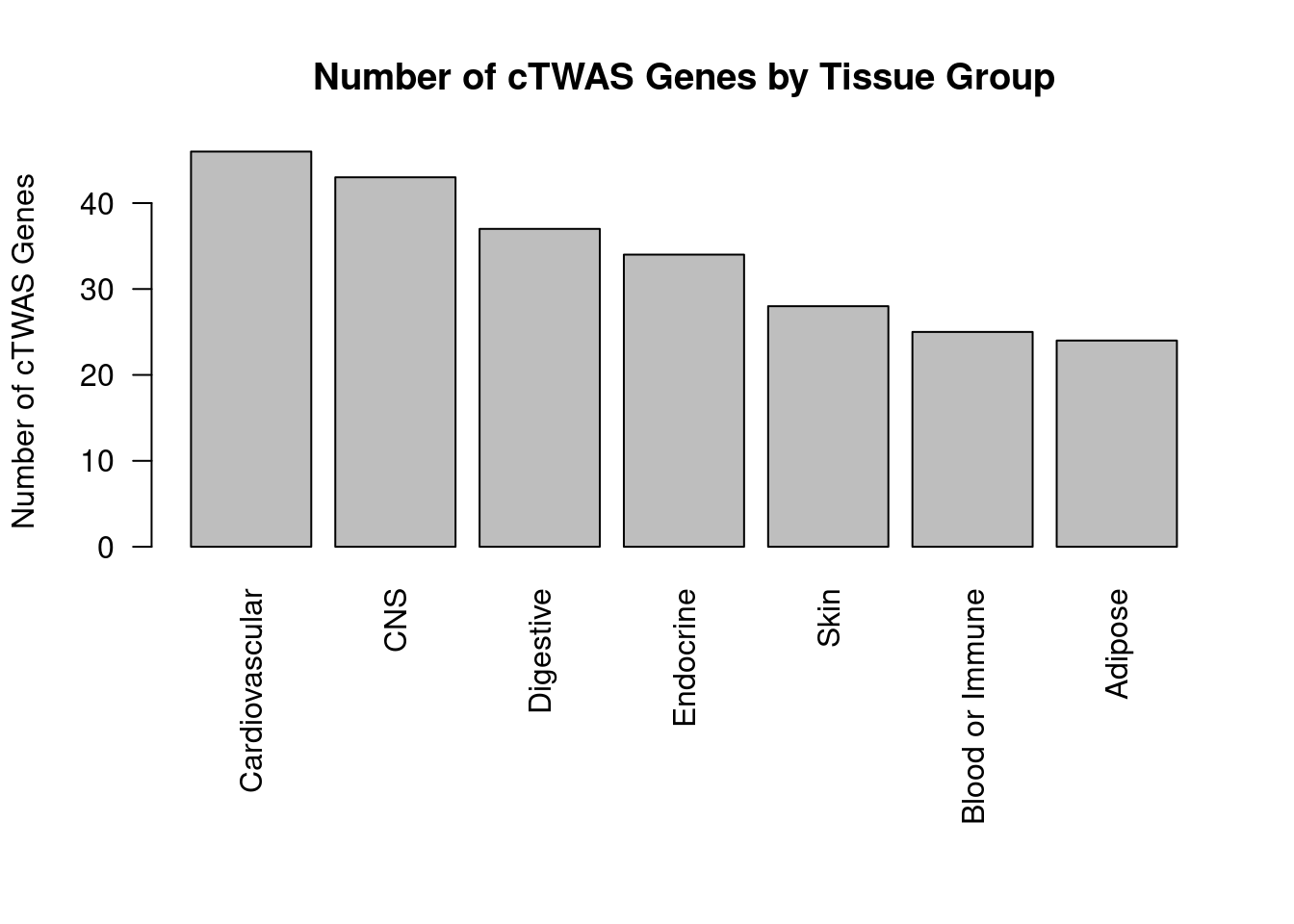
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
Enrichment analysis for cTWAS genes in each tissue group
GO
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_group, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Adipose
Number of cTWAS Genes in Tissue Group: 24
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
| Version | Author | Date |
|---|---|---|
| d14af05 | wesleycrouse | 2022-04-04 |
Endocrine
Number of cTWAS Genes in Tissue Group: 34
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Cardiovascular
Number of cTWAS Genes in Tissue Group: 46
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 positive regulation of phosphatidylinositol 3-kinase signaling (GO:0014068) 4/77 0.01934406 NTRK1;GPER1;FN1;SIRT1
2 regulation of phosphatidylinositol 3-kinase signaling (GO:0014066) 4/106 0.03381938 NTRK1;GPER1;FN1;SIRT1
3 positive regulation of protein phosphorylation (GO:0001934) 6/371 0.03797663 NTRK1;HFE;GPER1;NGF;SIRT1;GDF7
4 nerve growth factor signaling pathway (GO:0038180) 2/10 0.03797663 NTRK1;NGF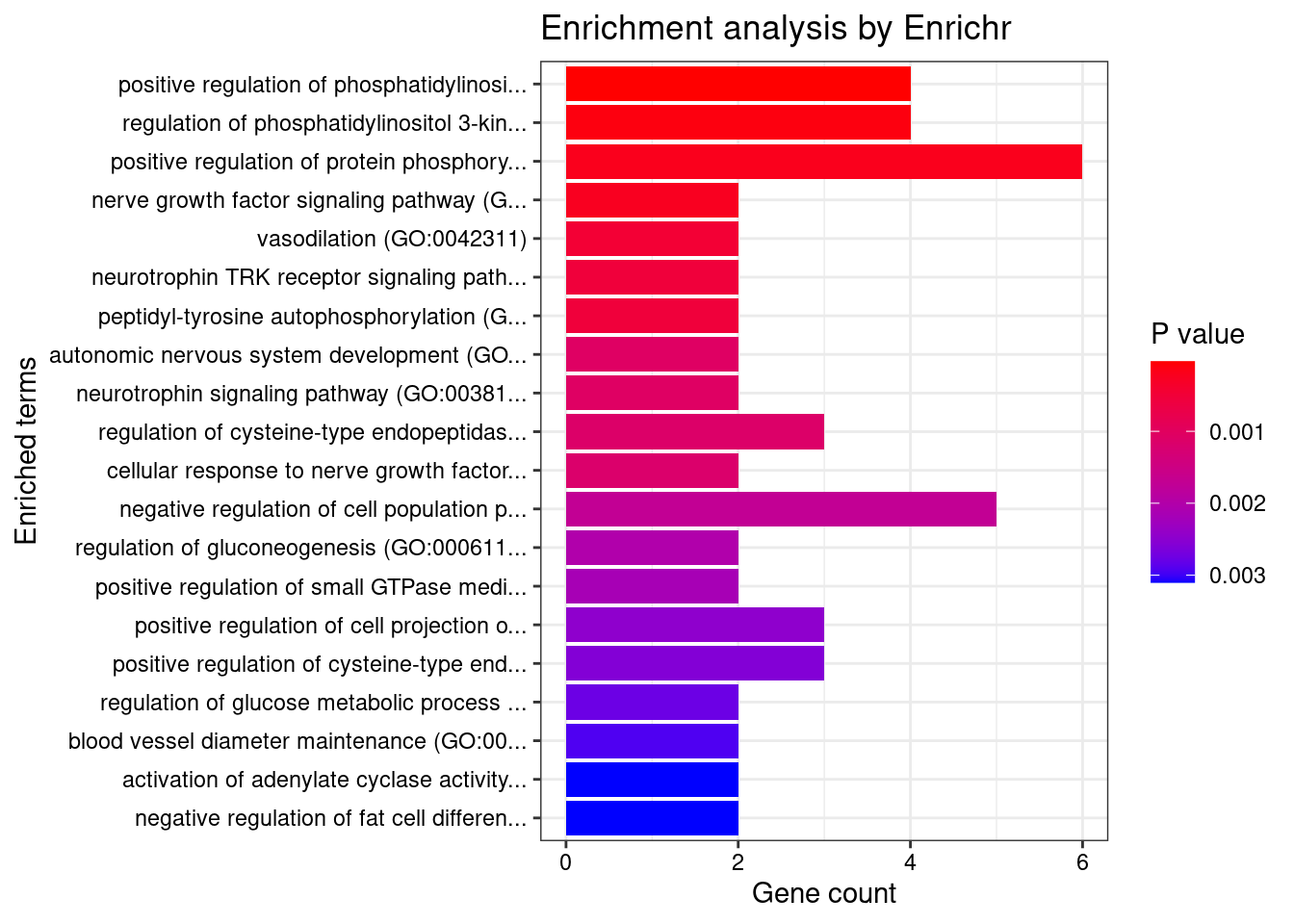
CNS
Number of cTWAS Genes in Tissue Group: 43
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)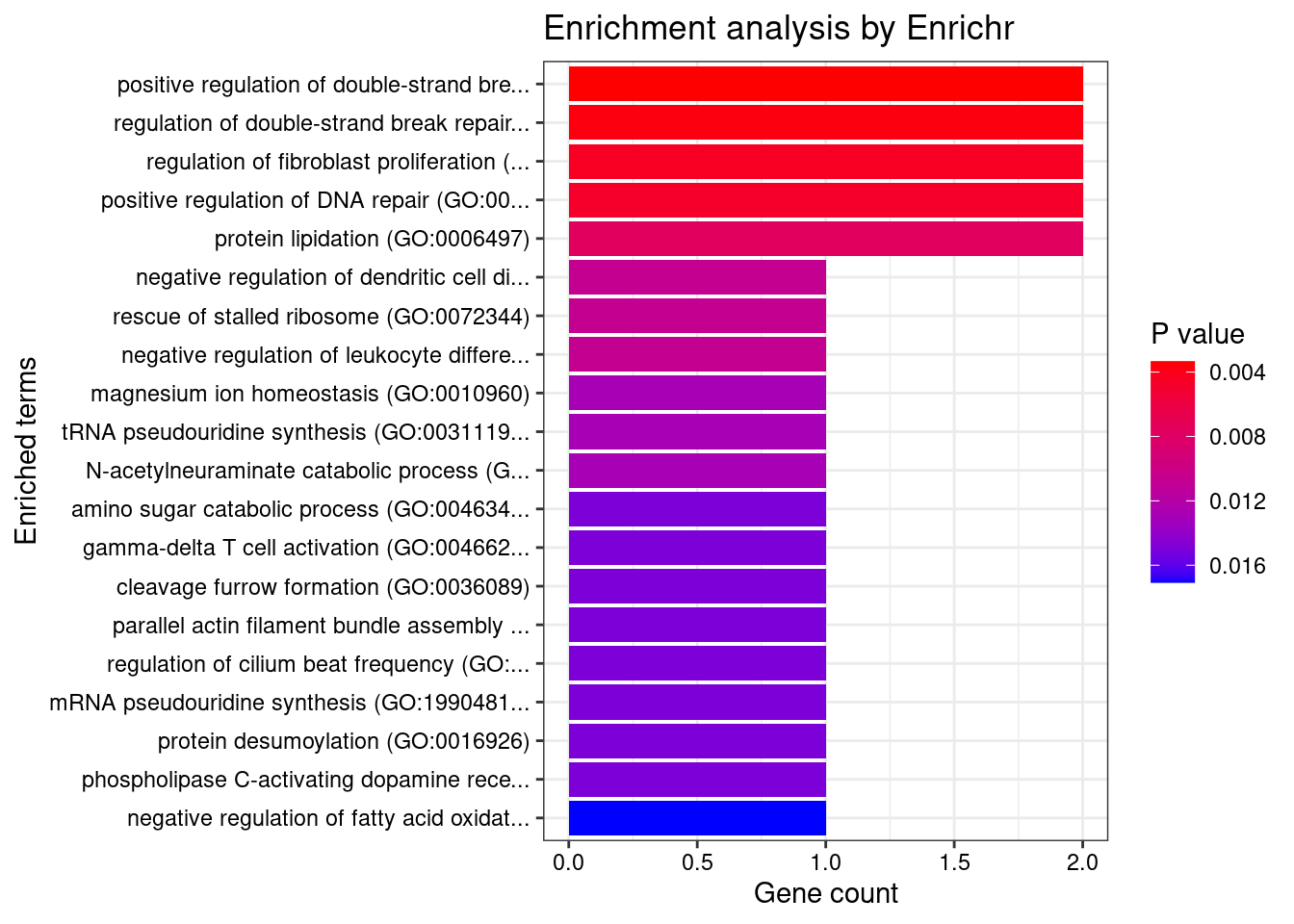
None
Number of cTWAS Genes in Tissue Group: 60
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)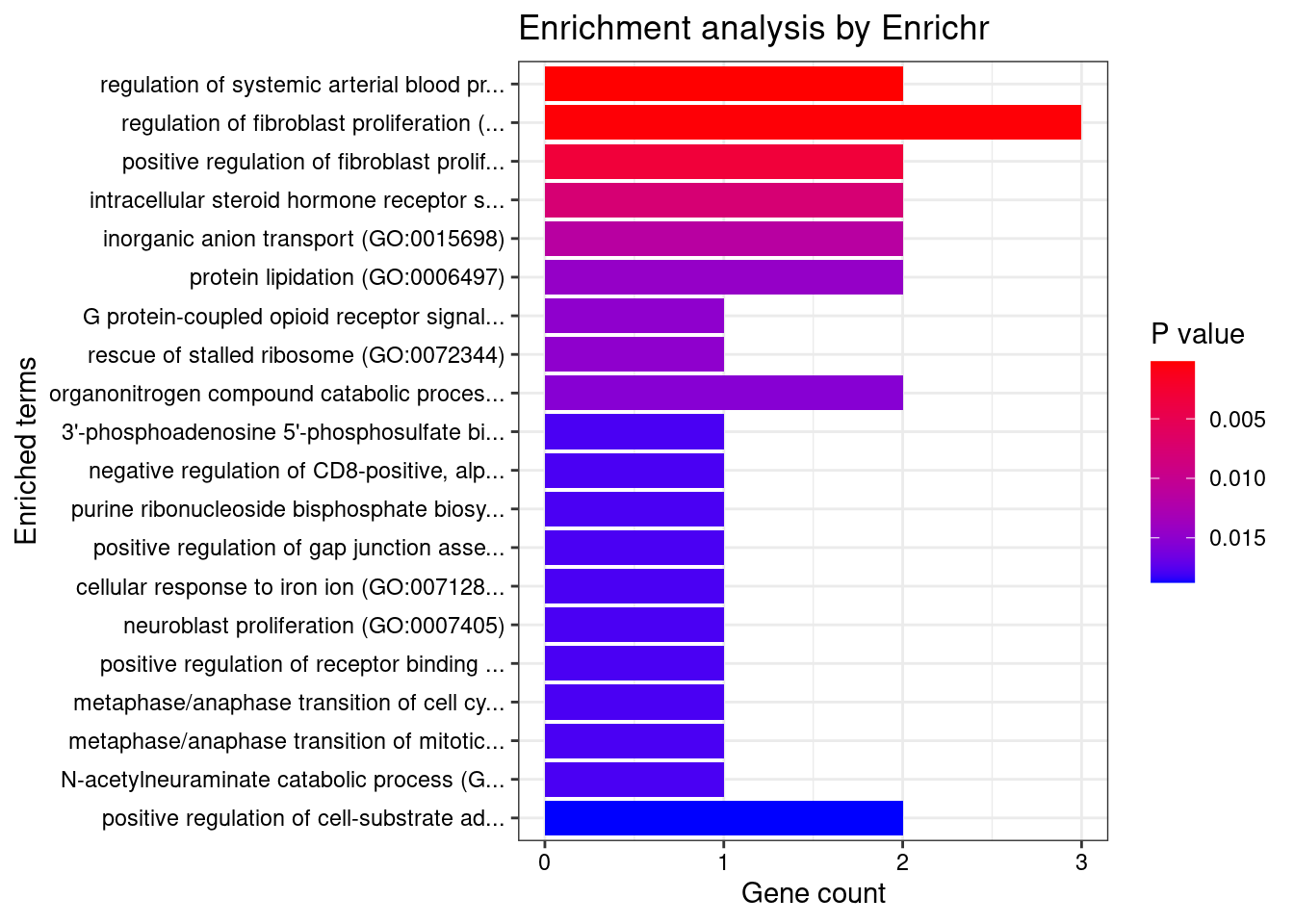
Skin
Number of cTWAS Genes in Tissue Group: 28
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of lipase activity (GO:0060191) 2/14 0.04391487 FURIN;RHOC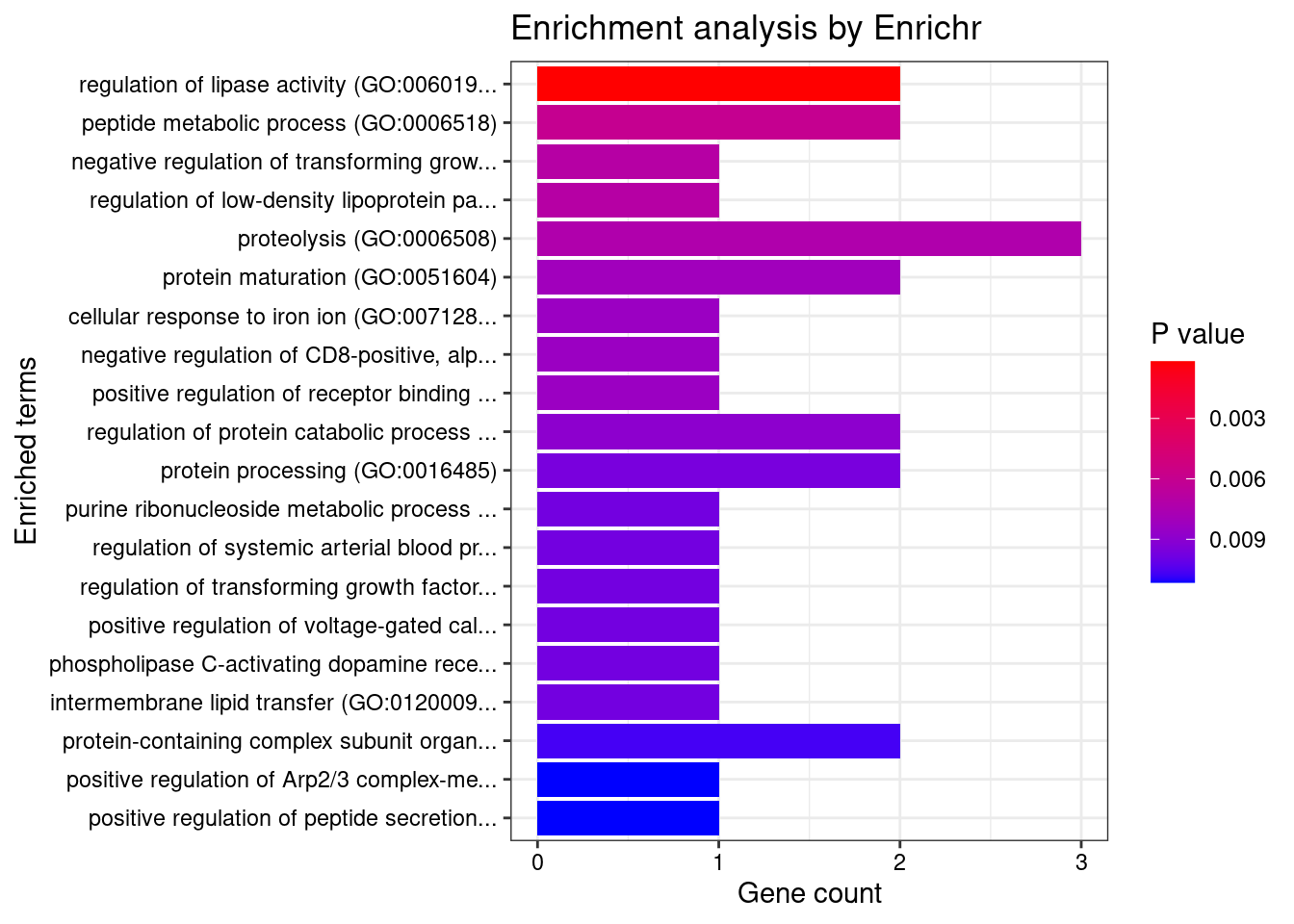
Blood or Immune
Number of cTWAS Genes in Tissue Group: 25
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of fibroblast proliferation (GO:0048145) 3/46 0.007683618 MORC3;FN1;AGT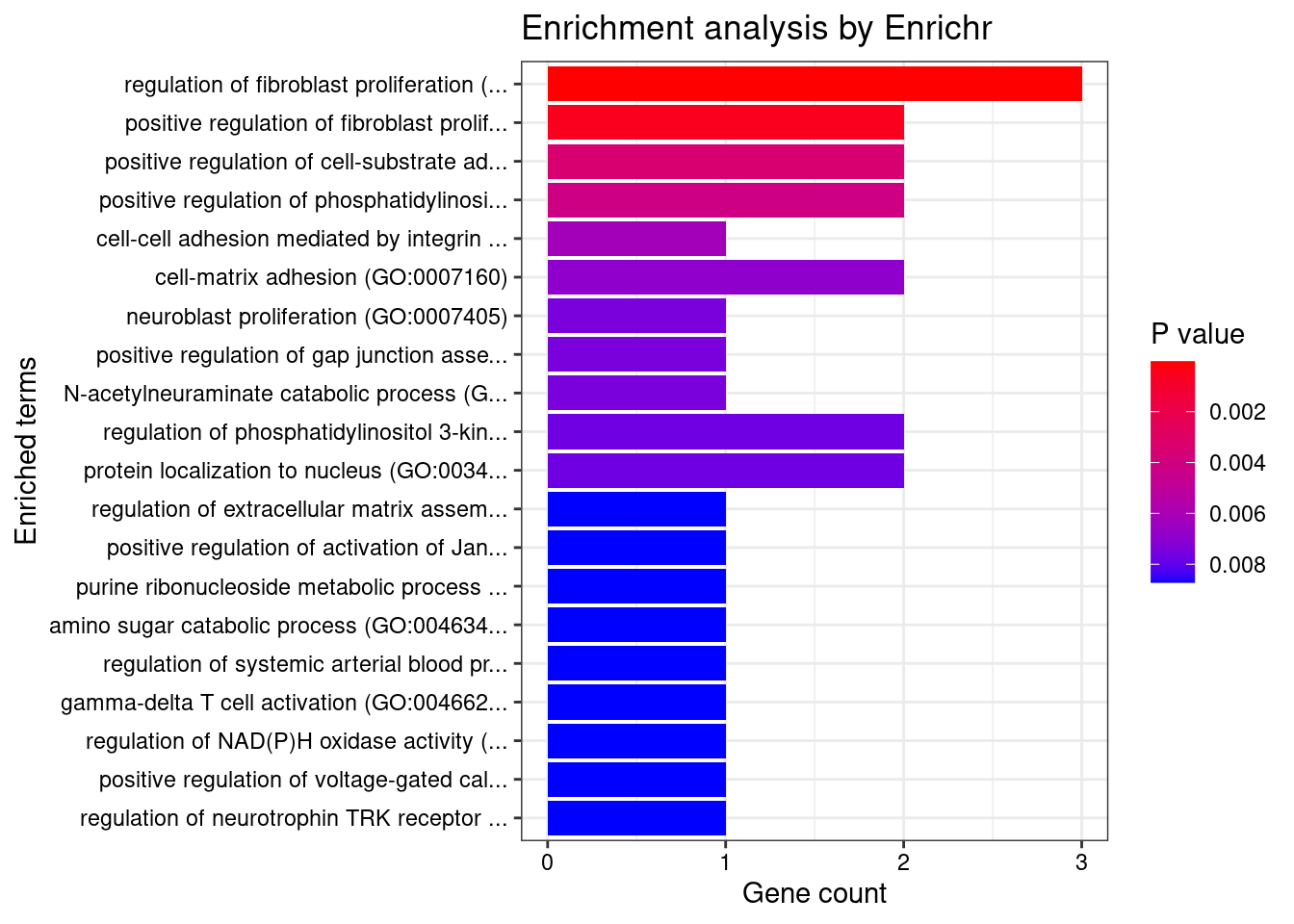
Digestive
Number of cTWAS Genes in Tissue Group: 37
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)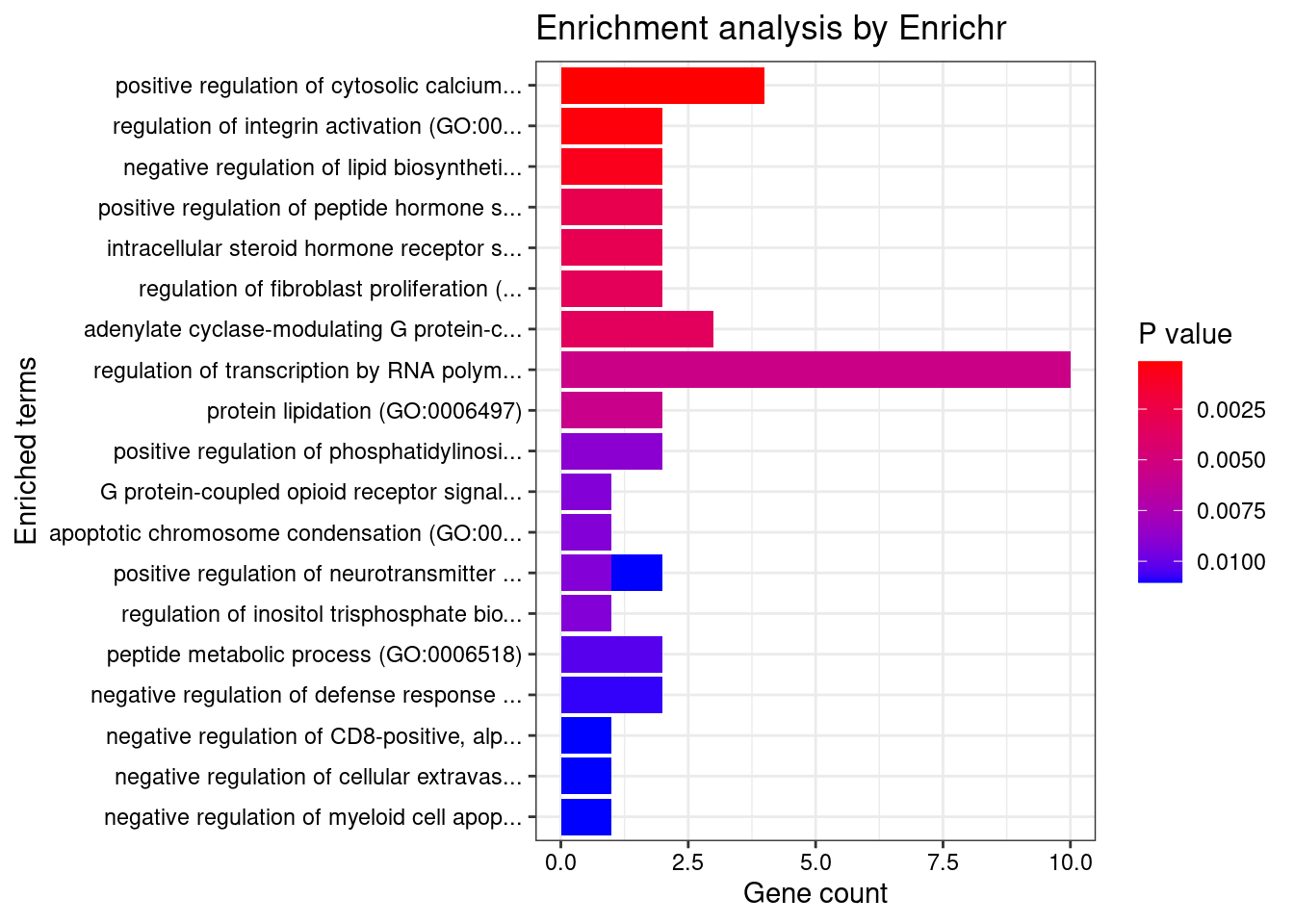
KEGG
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_group, referenceGene=background_group,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 24
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Endocrine
Number of cTWAS Genes in Tissue Group: 34
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Cardiovascular
Number of cTWAS Genes in Tissue Group: 46
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
CNS
Number of cTWAS Genes in Tissue Group: 43
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
None
Number of cTWAS Genes in Tissue Group: 60
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Skin
Number of cTWAS Genes in Tissue Group: 28
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Blood or Immune
Number of cTWAS Genes in Tissue Group: 25
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Digestive
Number of cTWAS Genes in Tissue Group: 37
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_group, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 24PAQR5 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDASCC2 gene(s) from the input list not found in DisGeNET CURATEDLINC00930 gene(s) from the input list not found in DisGeNET CURATEDMAP6D1 gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDRGS19 gene(s) from the input list not found in DisGeNET CURATEDSSPO gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
139 Sleep Apnea, Obstructive 0.02573353 1/14 1/9703
153 Upper Airway Resistance Sleep Apnea Syndrome 0.02573353 1/14 1/9703
174 Band Heterotopia of Brain 0.02573353 1/14 1/9703
178 Charcot-Marie-Tooth disease, Type 4B2 0.02573353 1/14 1/9703
179 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.02573353 1/14 1/9703
187 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.02573353 1/14 1/9703
199 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.02573353 1/14 1/9703
202 PULMONARY HYPERTENSION, PRIMARY, 4 0.02573353 1/14 1/9703
206 Glomerulopathy with fibronectin deposits 0.02573353 1/14 1/9703
207 MANDIBULOFACIAL DYSOSTOSIS WITH ALOPECIA 0.02573353 1/14 1/9703
208 BAND HETEROTOPIA 0.02573353 1/14 1/9703
209 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.02573353 1/14 1/9703
74 Cerebrovascular accident 0.03700664 2/14 62/9703
81 Alcohol abuse 0.03700664 2/14 67/9703
84 Hypokinesia 0.03700664 1/14 3/9703
91 Variegate Porphyria 0.03700664 1/14 2/9703
111 Bradykinesia 0.03700664 1/14 3/9703
117 Hypodynamia 0.03700664 1/14 3/9703
133 Spondylometaphyseal dysplasia, 'corner fracture' type 0.03700664 1/14 2/9703
152 Hypokinesia, Antiorthostatic 0.03700664 1/14 3/9703
154 Acute Cerebrovascular Accidents 0.03700664 2/14 54/9703
191 Porphyria, South African type 0.03700664 1/14 2/9703
196 HEMOCHROMATOSIS, TYPE 1 0.03700664 1/14 3/9703
198 Ovarian clear cell carcinoma 0.03700664 1/14 3/9703
204 MIGRAINE WITH OR WITHOUT AURA, SUSCEPTIBILITY TO, 1 0.03700664 1/14 3/9703
24 Cleft Palate 0.04332742 2/14 81/9703
92 Porphyria Cutanea Tarda 0.04332742 1/14 4/9703
116 Mandibulofacial Dysostosis 0.04332742 1/14 4/9703
176 Subcortical Band Heterotopia 0.04332742 1/14 4/9703
Endocrine
Number of cTWAS Genes in Tissue Group: 34TMEM175 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDUSP36 gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDC20orf187 gene(s) from the input list not found in DisGeNET CURATEDSLC2A4RG gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDRP11-286N22.10 gene(s) from the input list not found in DisGeNET CURATEDKIAA1614 gene(s) from the input list not found in DisGeNET CURATEDRP4-758J18.13 gene(s) from the input list not found in DisGeNET CURATEDSHISA8 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDARHGEF25 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDRGS19 gene(s) from the input list not found in DisGeNET CURATEDCTD-2349P21.5 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
72 Lead Poisoning 0.01258463 2/15 7/9703
162 Androgenetic Alopecia 0.01258463 2/15 15/9703
183 Female pattern alopecia (disorder) 0.01258463 2/15 15/9703
256 Microsatellite Instability 0.01258463 2/15 15/9703
276 Replication Error Phenotype 0.01258463 2/15 15/9703
317 Alopecia, Male Pattern 0.01258463 2/15 15/9703
8 Alopecia 0.01531639 2/15 19/9703
154 Pseudopelade 0.01531639 2/15 19/9703
40 Hearing Loss, Sudden 0.01800071 1/15 1/9703
155 Left Ventricular Hypertrophy 0.01800071 2/15 25/9703
176 Microvascular Angina 0.01800071 1/15 1/9703
200 Vitamin D-Dependent Rickets, Type 2A 0.01800071 1/15 1/9703
221 Maxillofacial Abnormalities 0.01800071 1/15 1/9703
248 Blood Coagulation Disorders, Inherited 0.01800071 1/15 1/9703
261 Deafness, Sudden 0.01800071 1/15 1/9703
277 Disproportionate tall stature 0.01800071 1/15 1/9703
278 Band Heterotopia of Brain 0.01800071 1/15 1/9703
281 HOMOCYSTINURIA DUE TO DEFICIENCY OF N(5,10)-METHYLENETETRAHYDROFOLATE REDUCTASE ACTIVITY 0.01800071 1/15 1/9703
282 Mthfr Deficiency, Thermolabile Type 0.01800071 1/15 1/9703
283 Methylenetetrahydrofolate reductase deficiency 0.01800071 1/15 1/9703
285 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.01800071 1/15 1/9703
296 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.01800071 1/15 1/9703
307 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.01800071 1/15 1/9703
312 MACROCEPHALY AND EPILEPTIC ENCEPHALOPATHY 0.01800071 1/15 1/9703
314 Glomerulopathy with fibronectin deposits 0.01800071 1/15 1/9703
315 HOMOCYSTINURIA DUE TO MTHFR DEFICIENCY 0.01800071 1/15 1/9703
320 BAND HETEROTOPIA 0.01800071 1/15 1/9703
321 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01800071 1/15 1/9703
25 Carcinoma, Transitional Cell 0.01986704 2/15 41/9703
161 Crohn's disease of large bowel 0.02008776 2/15 44/9703
187 Crohn's disease of the ileum 0.02008776 2/15 44/9703
227 Regional enteritis 0.02008776 2/15 44/9703
259 IIeocolitis 0.02008776 2/15 44/9703
38 Crohn Disease 0.02289346 2/15 50/9703
119 Sinus Thrombosis, Intracranial 0.02289346 1/15 2/9703
127 Thyrotoxicosis 0.02289346 1/15 2/9703
134 Vitamin D Deficiency 0.02289346 1/15 2/9703
164 Variegate Porphyria 0.02289346 1/15 2/9703
179 Cervical Intraepithelial Neoplasia 0.02289346 1/15 2/9703
214 Spondylometaphyseal dysplasia, 'corner fracture' type 0.02289346 1/15 2/9703
240 Petrous Sinus Thrombophlebitis 0.02289346 1/15 2/9703
241 Intracranial Sinus Thrombophlebitis 0.02289346 1/15 2/9703
242 Petrous Sinus Thrombosis 0.02289346 1/15 2/9703
300 Porphyria, South African type 0.02289346 1/15 2/9703
73 Chronic Lymphocytic Leukemia 0.02289837 2/15 55/9703
52 Endometrial Neoplasms 0.02486814 2/15 58/9703
71 Langer-Giedion Syndrome 0.02810843 1/15 3/9703
121 Spinal Cord Diseases 0.02810843 1/15 3/9703
147 Alcohol abuse 0.02810843 2/15 67/9703
163 Malnutrition 0.02810843 1/15 3/9703
249 Acute schizophrenia 0.02810843 1/15 3/9703
304 HEMOCHROMATOSIS, TYPE 1 0.02810843 1/15 3/9703
305 Familial Hypophosphatemic Rickets 0.02810843 1/15 3/9703
306 Ovarian clear cell carcinoma 0.02810843 1/15 3/9703
15 Bipolar Disorder 0.03029392 4/15 477/9703
11 Anencephaly 0.03094952 1/15 4/9703
91 Meningomyelocele 0.03094952 1/15 4/9703
115 Rickets 0.03094952 1/15 4/9703
152 Myelocele 0.03094952 1/15 4/9703
165 Porphyria Cutanea Tarda 0.03094952 1/15 4/9703
217 Endometrial Carcinoma 0.03094952 2/15 72/9703
237 Acquired Meningomyelocele 0.03094952 1/15 4/9703
279 Subcortical Band Heterotopia 0.03094952 1/15 4/9703
286 Neural tube defect, folate-sensitive 0.03094952 1/15 4/9703
301 THROMBOPHILIA DUE TO THROMBIN DEFECT 0.03094952 1/15 4/9703
4 Affective Disorders, Psychotic 0.03539204 1/15 5/9703
32 Congenital clubfoot 0.03539204 1/15 5/9703
110 Parkinson Disease 0.03539204 2/15 85/9703
258 Coronary Restenosis 0.03539204 1/15 5/9703
269 Autism Spectrum Disorders 0.03539204 2/15 85/9703
302 MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 0.03539204 1/15 5/9703
228 Breast Carcinoma 0.03543697 4/15 538/9703
41 Presenile dementia 0.03881984 2/15 99/9703
59 Graft-vs-Host Disease 0.03881984 1/15 6/9703
86 Malignant neoplasm of stomach 0.03881984 3/15 300/9703
124 Stomach Neoplasms 0.03881984 3/15 297/9703
181 Lennox-Gastaut syndrome 0.03881984 1/15 6/9703
191 Familial Alzheimer Disease (FAD) 0.03881984 2/15 100/9703
218 Alzheimer Disease, Late Onset 0.03881984 2/15 99/9703
224 Acute Confusional Senile Dementia 0.03881984 2/15 99/9703
230 Alzheimer's Disease, Focal Onset 0.03881984 2/15 99/9703
231 Alzheimer Disease, Early Onset 0.03881984 2/15 99/9703
247 Carcinoma in situ of uterine cervix 0.03881984 1/15 6/9703
275 Hereditary Diffuse Gastric Cancer 0.03881984 3/15 293/9703
292 Spina bifida aperta of cervical spine 0.03881984 1/15 6/9703
9 Alzheimer's Disease 0.03910871 2/15 101/9703
180 Depression, Postpartum 0.03991929 1/15 7/9703
280 Hyperphosphatasia with Mental Retardation 0.03991929 1/15 7/9703
61 Hematological Disease 0.04313813 1/15 8/9703
77 Leukopenia 0.04313813 1/15 8/9703
166 Williams Syndrome 0.04313813 1/15 8/9703
177 Embryonal Rhabdomyosarcoma 0.04313813 1/15 8/9703
196 Inflammatory disease of mucous membrane 0.04313813 1/15 8/9703
3 Adenocarcinoma 0.04427622 2/15 116/9703
167 Adenocarcinoma, Basal Cell 0.04427622 2/15 116/9703
168 Adenocarcinoma, Oxyphilic 0.04427622 2/15 116/9703
169 Carcinoma, Cribriform 0.04427622 2/15 116/9703
170 Carcinoma, Granular Cell 0.04427622 2/15 116/9703
171 Adenocarcinoma, Tubular 0.04427622 2/15 116/9703
212 Thrombophilia 0.04510074 1/15 9/9703
Cardiovascular
Number of cTWAS Genes in Tissue Group: 46LRRC10B gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDZNF692 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDRP11-1055B8.3 gene(s) from the input list not found in DisGeNET CURATEDZBTB46 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDKIAA1462 gene(s) from the input list not found in DisGeNET CURATEDSHB gene(s) from the input list not found in DisGeNET CURATEDRP11-373D23.3 gene(s) from the input list not found in DisGeNET CURATEDRP4-758J18.13 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDKIF13B gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDGDF7 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDZNF827 gene(s) from the input list not found in DisGeNET CURATEDDDI2 gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDCAMK1D gene(s) from the input list not found in DisGeNET CURATEDRP11-405A12.2 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
75 HSAN Type IV 0.002444182 2/22 4/9703
106 Hereditary Sensory and Autonomic Neuropathies 0.002444182 2/22 4/9703
143 Hereditary Sensory Radicular Neuropathy 0.002444182 2/22 4/9703
205 Lung Injury 0.002444182 3/22 21/9703
359 Chronic Lung Injury 0.002444182 3/22 21/9703
46 Diabetes Mellitus 0.002905741 3/22 32/9703
74 Hereditary Sensory Autonomic Neuropathy, Type 2 0.002905741 2/22 5/9703
262 Sensory Neuropathy, Hereditary 0.003808543 2/22 6/9703
9 Congenital Pain Insensitivity 0.004259707 2/22 7/9703
76 Hereditary Sensory Autonomic Neuropathy, Type 5 0.004259707 2/22 7/9703
73 Hereditary Sensory Autonomic Neuropathy, Type 1 0.005156187 2/22 8/9703
17 Birth Weight 0.014130852 2/22 14/9703
142 Alcohol abuse 0.014130852 3/22 67/9703
66 Heart failure 0.026951733 3/22 110/9703
67 Congestive heart failure 0.026951733 3/22 110/9703
88 Left-Sided Heart Failure 0.026951733 3/22 110/9703
179 Heart Failure, Right-Sided 0.026951733 3/22 110/9703
245 Sleep Apnea, Obstructive 0.026951733 1/22 1/9703
295 Upper Airway Resistance Sleep Apnea Syndrome 0.026951733 1/22 1/9703
312 Lewy Body Disease 0.026951733 2/22 22/9703
346 Band Heterotopia of Brain 0.026951733 1/22 1/9703
347 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 0.026951733 1/22 1/9703
350 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.026951733 1/22 1/9703
353 Myocardial Failure 0.026951733 3/22 110/9703
354 Heart Decompensation 0.026951733 3/22 110/9703
362 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.026951733 1/22 1/9703
379 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.026951733 1/22 1/9703
385 PULMONARY HYPERTENSION, PRIMARY, 4 0.026951733 1/22 1/9703
386 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 0.026951733 1/22 1/9703
389 Phakomatosis cesiomarmorata 0.026951733 1/22 1/9703
393 Glomerulopathy with fibronectin deposits 0.026951733 1/22 1/9703
396 MANDIBULOFACIAL DYSOSTOSIS WITH ALOPECIA 0.026951733 1/22 1/9703
397 BAND HETEROTOPIA 0.026951733 1/22 1/9703
404 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.026951733 1/22 1/9703
416 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 70 0.026951733 1/22 1/9703
199 Degenerative Diseases, Central Nervous System 0.037854244 2/22 39/9703
249 Neurodegenerative Disorders 0.037854244 2/22 39/9703
294 Degenerative Diseases, Spinal Cord 0.037854244 2/22 39/9703
35 Corneal Ulcer 0.038460802 1/22 2/9703
64 Congenital Heart Defects 0.038460802 2/22 44/9703
156 Variegate Porphyria 0.038460802 1/22 2/9703
180 Congenital hemangioma 0.038460802 1/22 2/9703
238 Spondylometaphyseal dysplasia, 'corner fracture' type 0.038460802 1/22 2/9703
327 Port-wine stain with oculocutaneous melanosis 0.038460802 1/22 2/9703
369 Porphyria, South African type 0.038460802 1/22 2/9703
383 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1 0.038460802 1/22 2/9703
388 Phakomatosis cesioflammea 0.038460802 1/22 2/9703
395 Autosomal dominant hypocalcemia 0.038460802 1/22 2/9703
407 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1, WITH BARTTER SYNDROME 0.038460802 1/22 2/9703
105 Neuroblastoma 0.041512116 2/22 47/9703
132 Thrombosis 0.044153661 2/22 49/9703
331 Cardiomyopathy, Familial Idiopathic 0.045047180 2/22 50/9703
305 Acute Cerebrovascular Accidents 0.048686382 2/22 54/9703
345 Familial medullary thyroid carcinoma 0.048686382 1/22 3/9703
376 HEMOCHROMATOSIS, TYPE 1 0.048686382 1/22 3/9703
378 Ovarian clear cell carcinoma 0.048686382 1/22 3/9703
390 MIGRAINE WITH OR WITHOUT AURA, SUSCEPTIBILITY TO, 1 0.048686382 1/22 3/9703
405 Idiopathic basal ganglia calcification 1 0.048686382 1/22 3/9703
CNS
Number of cTWAS Genes in Tissue Group: 43ARHGEF25 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDASCC2 gene(s) from the input list not found in DisGeNET CURATEDLINC00930 gene(s) from the input list not found in DisGeNET CURATEDRP11-757G1.6 gene(s) from the input list not found in DisGeNET CURATEDSLC2A4RG gene(s) from the input list not found in DisGeNET CURATEDTMEM175 gene(s) from the input list not found in DisGeNET CURATEDSF3B3 gene(s) from the input list not found in DisGeNET CURATEDTMEM176B gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDSOX13 gene(s) from the input list not found in DisGeNET CURATEDRP11-286N22.10 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDPGBD2 gene(s) from the input list not found in DisGeNET CURATEDC22orf31 gene(s) from the input list not found in DisGeNET CURATEDSTK38L gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDSPIRE1 gene(s) from the input list not found in DisGeNET CURATEDRP5-965G21.3 gene(s) from the input list not found in DisGeNET CURATEDSENP3 gene(s) from the input list not found in DisGeNET CURATEDZNF598 gene(s) from the input list not found in DisGeNET CURATEDZBTB46 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
6 Alopecia 0.01704428 2/17 19/9703
65 Spasmophilia 0.01704428 1/17 1/9703
68 Tetany 0.01704428 1/17 1/9703
105 Age related macular degeneration 0.01704428 2/17 30/9703
110 Tetany, Neonatal 0.01704428 1/17 1/9703
154 Tetanilla 0.01704428 1/17 1/9703
167 Cerebral Autosomal Recessive Arteriopathy with Subcortical Infarcts and Leukoencephalopathy 0.01704428 1/17 1/9703
168 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 0.01704428 1/17 1/9703
170 Macular Degeneration, Age-Related, 7 0.01704428 1/17 1/9703
172 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.01704428 1/17 1/9703
175 CILIARY DYSKINESIA, PRIMARY, 7 (disorder) 0.01704428 1/17 1/9703
180 MACULAR DEGENERATION, AGE-RELATED, 8 0.01704428 1/17 1/9703
181 HYPOMAGNESEMIA 6, RENAL 0.01704428 1/17 1/9703
186 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.01704428 1/17 1/9703
190 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 0.01704428 1/17 1/9703
193 Phakomatosis cesiomarmorata 0.01704428 1/17 1/9703
194 Very long chain acyl-CoA dehydrogenase deficiency 0.01704428 1/17 1/9703
195 Glomerulopathy with fibronectin deposits 0.01704428 1/17 1/9703
199 CEREBRAL ARTERIOPATHY, AUTOSOMAL DOMINANT, WITH SUBCORTICAL INFARCTS AND LEUKOENCEPHALOPATHY, TYPE 2 0.01704428 1/17 1/9703
200 HYPOMAGNESEMIA, SEIZURES, AND MENTAL RETARDATION 0.01704428 1/17 1/9703
207 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01704428 1/17 1/9703
209 Ciliary Dyskinesia, Primary, 7, With Or Without Situs Inversus 0.01704428 1/17 1/9703
39 Hydatidiform Mole 0.02140943 1/17 2/9703
101 Congenital hemangioma 0.02140943 1/17 2/9703
114 Metabolic myopathy 0.02140943 1/17 2/9703
117 Hydatidiform Mole, Partial 0.02140943 1/17 2/9703
134 Spondylometaphyseal dysplasia, 'corner fracture' type 0.02140943 1/17 2/9703
151 CADASIL Syndrome 0.02140943 1/17 2/9703
160 Port-wine stain with oculocutaneous melanosis 0.02140943 1/17 2/9703
162 CADASILM 0.02140943 1/17 2/9703
188 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1 0.02140943 1/17 2/9703
192 Phakomatosis cesioflammea 0.02140943 1/17 2/9703
197 Autosomal dominant hypocalcemia 0.02140943 1/17 2/9703
206 Familial primary hypomagnesemia with normocalciuria and normocalcemia 0.02140943 1/17 2/9703
210 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1, WITH BARTTER SYNDROME 0.02140943 1/17 2/9703
42 Intestinal Obstruction 0.02807671 1/17 3/9703
82 Intervertebral Disc Degeneration 0.02807671 1/17 3/9703
95 Neurofibrosarcoma 0.02807671 1/17 3/9703
109 Congenital secretory diarrhea, sodium type (disorder) 0.02807671 1/17 3/9703
185 Ovarian clear cell carcinoma 0.02807671 1/17 3/9703
141 Complete hydatidiform mole 0.03479512 1/17 4/9703
152 Malignant Peripheral Nerve Sheath Tumor 0.03479512 1/17 4/9703
171 CYSTIC FIBROSIS MODIFIER 1 0.03479512 1/17 4/9703
123 Malignant melanoma of iris 0.03975949 1/17 5/9703
124 Malignant melanoma of choroid 0.03975949 1/17 5/9703
176 Cerebral Small Vessel Diseases 0.03975949 1/17 5/9703
182 MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 0.03975949 1/17 5/9703
None
Number of cTWAS Genes in Tissue Group: 60TMEM175 gene(s) from the input list not found in DisGeNET CURATEDEFR3B gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDNPW gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDCIB4 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDYEATS2 gene(s) from the input list not found in DisGeNET CURATEDARL4A gene(s) from the input list not found in DisGeNET CURATEDRP4-758J18.13 gene(s) from the input list not found in DisGeNET CURATEDRP5-965G21.3 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDPGBD2 gene(s) from the input list not found in DisGeNET CURATEDLINC01169 gene(s) from the input list not found in DisGeNET CURATEDCDC16 gene(s) from the input list not found in DisGeNET CURATEDBAHCC1 gene(s) from the input list not found in DisGeNET CURATEDFAM212A gene(s) from the input list not found in DisGeNET CURATEDZNF598 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDRGS19 gene(s) from the input list not found in DisGeNET CURATEDDDI2 gene(s) from the input list not found in DisGeNET CURATEDRP11-757G1.6 gene(s) from the input list not found in DisGeNET CURATEDCTDSPL gene(s) from the input list not found in DisGeNET CURATEDNUDT16L1 gene(s) from the input list not found in DisGeNET CURATEDSF3B3 gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDSNX11 gene(s) from the input list not found in DisGeNET CURATEDPAQR5 gene(s) from the input list not found in DisGeNET CURATEDNDUFAF8 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
8 Alkalosis 0.03748852 1/28 1/9703
18 Cardiac Arrhythmia 0.03748852 2/28 25/9703
32 Cardiomyopathy, Alcoholic 0.03748852 1/28 1/9703
86 Hypokalemia 0.03748852 2/28 7/9703
148 Ureteral obstruction 0.03748852 2/28 24/9703
158 Hyalinosis, Segmental Glomerular 0.03748852 2/28 28/9703
163 Left Ventricular Hypertrophy 0.03748852 2/28 25/9703
196 Female Pseudohermaphroditism 0.03748852 1/28 1/9703
273 Trichomegaly 0.03748852 1/28 1/9703
283 Glucose Metabolism Disorders 0.03748852 1/28 1/9703
303 NEPHROLITHIASIS, CALCIUM OXALATE 0.03748852 1/28 1/9703
304 Band Heterotopia of Brain 0.03748852 1/28 1/9703
305 Glucocorticoid Receptor Deficiency 0.03748852 1/28 1/9703
306 Pseudohermaphroditism, Female, With Hypokalemia, Due To Glucocorticoid Resistance 0.03748852 1/28 1/9703
307 BODY COMPOSITION, BENEFICIAL 0.03748852 1/28 1/9703
310 Deafness, Autosomal Recessive 28 0.03748852 1/28 1/9703
312 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.03748852 1/28 1/9703
322 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.03748852 1/28 1/9703
324 CILIARY DYSKINESIA, PRIMARY, 7 (disorder) 0.03748852 1/28 1/9703
326 Myopathy, Mitochondrial Progressive, With Congenital Cataract, Hearing Loss, And Developmental Delay 0.03748852 1/28 1/9703
331 46, XX Disorders of Sex Development 0.03748852 1/28 1/9703
342 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.03748852 1/28 1/9703
349 PULMONARY HYPERTENSION, PRIMARY, 4 0.03748852 1/28 1/9703
354 Glomerulopathy with fibronectin deposits 0.03748852 1/28 1/9703
355 SPASTIC PARAPLEGIA 45, AUTOSOMAL RECESSIVE 0.03748852 1/28 1/9703
362 BAND HETEROTOPIA 0.03748852 1/28 1/9703
367 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.03748852 1/28 1/9703
369 Ciliary Dyskinesia, Primary, 7, With Or Without Situs Inversus 0.03748852 1/28 1/9703
373 Familial isolated trichomegaly 0.03748852 1/28 1/9703
53 Diabetes Mellitus 0.04721394 2/28 32/9703
Skin
Number of cTWAS Genes in Tissue Group: 28RP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDARL4A gene(s) from the input list not found in DisGeNET CURATEDPGBD2 gene(s) from the input list not found in DisGeNET CURATEDGLTP gene(s) from the input list not found in DisGeNET CURATEDDDI2 gene(s) from the input list not found in DisGeNET CURATEDTTC33 gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDSF3B3 gene(s) from the input list not found in DisGeNET CURATEDLRRC10B gene(s) from the input list not found in DisGeNET CURATEDMEX3A gene(s) from the input list not found in DisGeNET CURATEDRP4-534N18.2 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDSSPO gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
106 HYPOCALCIURIC HYPERCALCEMIA, FAMILIAL, TYPE II (disorder) 0.02308802 1/14 1/9703
112 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.02308802 1/14 1/9703
124 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.02308802 1/14 1/9703
130 PULMONARY HYPERTENSION, PRIMARY, 4 0.02308802 1/14 1/9703
131 HYPOCALCEMIA, AUTOSOMAL DOMINANT 2 0.02308802 1/14 1/9703
133 Phakomatosis cesiomarmorata 0.02308802 1/14 1/9703
135 SPASTIC PARAPLEGIA 45, AUTOSOMAL RECESSIVE 0.02308802 1/14 1/9703
138 MENTAL RETARDATION, AUTOSOMAL DOMINANT 46 0.02308802 1/14 1/9703
139 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.02308802 1/14 1/9703
54 Variegate Porphyria 0.02442976 1/14 2/9703
69 Congenital hemangioma 0.02442976 1/14 2/9703
100 Port-wine stain with oculocutaneous melanosis 0.02442976 1/14 2/9703
116 Porphyria, South African type 0.02442976 1/14 2/9703
128 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1 0.02442976 1/14 2/9703
132 Phakomatosis cesioflammea 0.02442976 1/14 2/9703
136 Autosomal dominant hypocalcemia 0.02442976 1/14 2/9703
141 HYPOCALCEMIA, AUTOSOMAL DOMINANT 1, WITH BARTTER SYNDROME 0.02442976 1/14 2/9703
121 HEMOCHROMATOSIS, TYPE 1 0.03276535 1/14 3/9703
123 Ovarian clear cell carcinoma 0.03276535 1/14 3/9703
55 Porphyria Cutanea Tarda 0.03949997 1/14 4/9703
67 Pilomatrixoma 0.03949997 1/14 4/9703
80 Malignant melanoma of iris 0.04144720 1/14 5/9703
81 Malignant melanoma of choroid 0.04144720 1/14 5/9703
119 MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 0.04144720 1/14 5/9703
137 Cone-Rod Dystrophies 0.04144720 1/14 5/9703
24 Lead Poisoning 0.04673258 1/14 7/9703
107 Hyperphosphatasia with Mental Retardation 0.04673258 1/14 7/9703
109 PULMONARY HYPERTENSION, PRIMARY, DEXFENFLURAMINE-ASSOCIATED 0.04673258 1/14 7/9703
110 Pulmonary Hypertension, Primary, Fenfluramine-Associated 0.04673258 1/14 7/9703
127 Pulmonary Hypertension, Primary, 1, With Hereditary Hemorrhagic Telangiectasia 0.04673258 1/14 7/9703
140 Pulmonary Hypertension, Primary, 1 0.04673258 1/14 7/9703
65 Embryonal Rhabdomyosarcoma 0.04866353 1/14 8/9703
68 Uveal melanoma 0.04866353 1/14 8/9703
103 Familial pulmonary arterial hypertension 0.04866353 1/14 8/9703
Blood or Immune
Number of cTWAS Genes in Tissue Group: 25PAQR5 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDCERS5 gene(s) from the input list not found in DisGeNET CURATEDNPNT gene(s) from the input list not found in DisGeNET CURATEDCTDNEP1 gene(s) from the input list not found in DisGeNET CURATEDGDF7 gene(s) from the input list not found in DisGeNET CURATEDGIT2 gene(s) from the input list not found in DisGeNET CURATEDCLCN6 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDHSCB gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDLINC01451 gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATEDRP5-965G21.3 gene(s) from the input list not found in DisGeNET CURATEDSOX13 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
4 Alkalosis 0.01281952 1/10 1/9703
17 Cardiomyopathy, Alcoholic 0.01281952 1/10 1/9703
32 Diabetic Nephropathy 0.01281952 2/10 44/9703
39 Nodular glomerulosclerosis 0.01281952 2/10 41/9703
40 Focal glomerulosclerosis 0.01281952 2/10 36/9703
88 Ureteral obstruction 0.01281952 2/10 24/9703
92 Hyalinosis, Segmental Glomerular 0.01281952 2/10 28/9703
94 Left Ventricular Hypertrophy 0.01281952 2/10 25/9703
165 Band Heterotopia of Brain 0.01281952 1/10 1/9703
168 SPINAL MUSCULAR ATROPHY WITH RESPIRATORY DISTRESS 1 0.01281952 1/10 1/9703
169 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.01281952 1/10 1/9703
184 Glomerulopathy with fibronectin deposits 0.01281952 1/10 1/9703
185 SPASTIC PARAPLEGIA 45, AUTOSOMAL RECESSIVE 0.01281952 1/10 1/9703
186 CHARCOT-MARIE-TOOTH DISEASE, AXONAL, TYPE 2S 0.01281952 1/10 1/9703
191 BAND HETEROTOPIA 0.01281952 1/10 1/9703
196 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.01281952 1/10 1/9703
72 Drug Overdose 0.02158076 1/10 2/9703
73 Peptic Ulcer Hemorrhage 0.02158076 1/10 2/9703
128 Spondylometaphyseal dysplasia, 'corner fracture' type 0.02158076 1/10 2/9703
8 Anuria 0.02382543 1/10 3/9703
50 Malignant Hypertension 0.02382543 1/10 3/9703
51 Renal hypertension 0.02382543 1/10 3/9703
82 Retinopathy of Prematurity 0.02382543 1/10 3/9703
182 Vascular Remodeling 0.02382543 1/10 3/9703
183 Pulmonary Arterial Remodeling 0.02382543 1/10 3/9703
199 Alveolitis, Fibrosing 0.02382543 2/10 83/9703
79 Pulmonary Fibrosis 0.02404244 2/10 85/9703
111 Allanson Pantzar McLeod syndrome 0.02731028 1/10 4/9703
166 Subcortical Band Heterotopia 0.02731028 1/10 4/9703
173 Renal Tubular Dysgenesis With Choanal Atresia And Athelia 0.02731028 1/10 4/9703
7 Aneurysm, Dissecting 0.03010766 1/10 5/9703
27 Sudden death 0.03010766 1/10 5/9703
119 Dissection of aorta 0.03010766 1/10 5/9703
188 Dissection, Blood Vessel 0.03010766 1/10 5/9703
174 Neointima 0.03410619 1/10 6/9703
175 Neointima Formation 0.03410619 1/10 6/9703
9 Aortic Aneurysm 0.03671271 1/10 7/9703
53 Hypokalemia 0.03671271 1/10 7/9703
167 Hyperphosphatasia with Mental Retardation 0.03671271 1/10 7/9703
10 Aortic Valve Insufficiency 0.04583257 1/10 11/9703
19 Carotid Artery Diseases 0.04583257 1/10 11/9703
33 Diabetic Retinopathy 0.04583257 1/10 11/9703
98 Aortic Aneurysm, Abdominal 0.04583257 1/10 10/9703
131 Endomyocardial Fibrosis 0.04583257 1/10 9/9703
132 Carotid Atherosclerosis 0.04583257 1/10 11/9703
133 External Carotid Artery Diseases 0.04583257 1/10 11/9703
138 Internal Carotid Artery Diseases 0.04583257 1/10 11/9703
139 Arterial Diseases, Common Carotid 0.04583257 1/10 11/9703
150 Diabetic Cardiomyopathies 0.04583257 1/10 11/9703
90 Essential Hypertension 0.04897647 1/10 12/9703
48 Hyperemia 0.04910486 1/10 13/9703
89 Venous Engorgement 0.04910486 1/10 13/9703
99 Reactive Hyperemia 0.04910486 1/10 13/9703
117 Active Hyperemia 0.04910486 1/10 13/9703
Digestive
Number of cTWAS Genes in Tissue Group: 37PAQR5 gene(s) from the input list not found in DisGeNET CURATEDZNF467 gene(s) from the input list not found in DisGeNET CURATEDASCC2 gene(s) from the input list not found in DisGeNET CURATEDSLC2A4RG gene(s) from the input list not found in DisGeNET CURATEDRP11-757G1.6 gene(s) from the input list not found in DisGeNET CURATEDC17orf82 gene(s) from the input list not found in DisGeNET CURATEDMORC3 gene(s) from the input list not found in DisGeNET CURATEDCCNT2 gene(s) from the input list not found in DisGeNET CURATEDZNF415 gene(s) from the input list not found in DisGeNET CURATEDSOX13 gene(s) from the input list not found in DisGeNET CURATEDSSBP3 gene(s) from the input list not found in DisGeNET CURATEDCPXM1 gene(s) from the input list not found in DisGeNET CURATEDRP11-84A19.4 gene(s) from the input list not found in DisGeNET CURATEDSTK38L gene(s) from the input list not found in DisGeNET CURATEDLIMA1 gene(s) from the input list not found in DisGeNET CURATEDTMEM179B gene(s) from the input list not found in DisGeNET CURATEDLRRC10B gene(s) from the input list not found in DisGeNET CURATEDMRPL21 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
29 Corpus Luteum Cyst 0.02509888 2/19 35/9703
62 Ovarian Cysts 0.02509888 2/19 35/9703
83 Left Ventricular Hypertrophy 0.02509888 2/19 25/9703
121 Erythrocyte Mean Corpuscular Hemoglobin Test 0.02509888 2/19 13/9703
147 Finding of Mean Corpuscular Hemoglobin 0.02509888 2/19 13/9703
157 Band Heterotopia of Brain 0.02509888 1/19 1/9703
160 GLOMERULOPATHY WITH FIBRONECTIN DEPOSITS 2 (disorder) 0.02509888 1/19 1/9703
163 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 7 (finding) 0.02509888 1/19 1/9703
164 CILIARY DYSKINESIA, PRIMARY, 7 (disorder) 0.02509888 1/19 1/9703
166 Myopathy, Mitochondrial Progressive, With Congenital Cataract, Hearing Loss, And Developmental Delay 0.02509888 1/19 1/9703
175 MENTAL RETARDATION, AUTOSOMAL DOMINANT 14 0.02509888 1/19 1/9703
179 Very long chain acyl-CoA dehydrogenase deficiency 0.02509888 1/19 1/9703
180 Glomerulopathy with fibronectin deposits 0.02509888 1/19 1/9703
183 BAND HETEROTOPIA 0.02509888 1/19 1/9703
184 HYPERPHOSPHATASIA WITH MENTAL RETARDATION SYNDROME 1 0.02509888 1/19 1/9703
186 Ciliary Dyskinesia, Primary, 7, With Or Without Situs Inversus 0.02509888 1/19 1/9703
87 Variegate Porphyria 0.03596311 1/19 2/9703
112 Metabolic myopathy 0.03596311 1/19 2/9703
125 Toxic effect of carbon tetrachloride 0.03596311 1/19 2/9703
126 Spondylometaphyseal dysplasia, 'corner fracture' type 0.03596311 1/19 2/9703
170 Porphyria, South African type 0.03596311 1/19 2/9703
80 Hypokinesia 0.04191805 1/19 3/9703
102 Bradykinesia 0.04191805 1/19 3/9703
106 Hypodynamia 0.04191805 1/19 3/9703
139 Hypokinesia, Antiorthostatic 0.04191805 1/19 3/9703
173 HEMOCHROMATOSIS, TYPE 1 0.04191805 1/19 3/9703
174 Ovarian clear cell carcinoma 0.04191805 1/19 3/9703
76 Alcohol abuse 0.04568637 2/19 67/9703
89 Porphyria Cutanea Tarda 0.04568637 1/19 4/9703
101 Pilomatrixoma 0.04568637 1/19 4/9703
103 Physiological Sexual Disorders 0.04568637 1/19 4/9703
158 Subcortical Band Heterotopia 0.04568637 1/19 4/9703
168 Chromosome 2q37 deletion syndrome 0.04568637 1/19 4/9703Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_group=ctwas_genes_group)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_group]})
#hypergeometric test
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_group)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
#multiple testing correction
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 24
gene_set nset ngenes percent padj
1 gwascatalog 4611 16 0.003469963 0.002070824
2 fda_approved_drug_targets 260 3 0.011538462 0.021228616
3 mgi_essential 1729 6 0.003470214 0.069573451
4 clinvar_path_likelypath 2144 7 0.003264925 0.069573451
5 core_essentials_hart 212 1 0.004716981 0.295412111
Endocrine
Number of cTWAS Genes in Tissue Group: 34
gene_set nset ngenes percent padj
1 gwascatalog 5415 17 0.003139428 0.03353774
2 mgi_essential 2030 6 0.002955665 0.25103220
3 fda_approved_drug_targets 302 2 0.006622517 0.25103220
4 core_essentials_hart 239 1 0.004184100 0.35085689
5 clinvar_path_likelypath 2493 6 0.002406739 0.35085689
Cardiovascular
Number of cTWAS Genes in Tissue Group: 46
gene_set nset ngenes percent padj
1 fda_approved_drug_targets 286 5 0.017482517 0.00595081
2 mgi_essential 1969 12 0.006094464 0.01692767
3 gwascatalog 5199 22 0.004231583 0.02690972
4 clinvar_path_likelypath 2407 11 0.004570004 0.08172504
5 core_essentials_hart 243 1 0.004115226 0.49727949
CNS
Number of cTWAS Genes in Tissue Group: 43
gene_set nset ngenes percent padj
1 gwascatalog 5430 18 0.003314917 0.3652811
2 clinvar_path_likelypath 2528 9 0.003560127 0.3652811
3 mgi_essential 2086 5 0.002396932 0.5756643
4 core_essentials_hart 245 1 0.004081633 0.5756643
5 fda_approved_drug_targets 316 1 0.003164557 0.5756643
None
Number of cTWAS Genes in Tissue Group: 60
gene_set nset ngenes percent padj
1 gwascatalog 5644 30 0.005315379 0.005497656
2 mgi_essential 2139 11 0.005142590 0.140689190
3 fda_approved_drug_targets 320 3 0.009375000 0.140689190
4 core_essentials_hart 256 2 0.007812500 0.213087487
5 clinvar_path_likelypath 2608 11 0.004217791 0.213087487
Skin
Number of cTWAS Genes in Tissue Group: 28
gene_set nset ngenes percent padj
1 gwascatalog 5122 16 0.003123780 0.02125153
2 mgi_essential 1930 5 0.002590674 0.37920993
3 core_essentials_hart 232 1 0.004310345 0.37920993
4 clinvar_path_likelypath 2345 6 0.002558635 0.37920993
5 fda_approved_drug_targets 275 1 0.003636364 0.37920993
Blood or Immune
Number of cTWAS Genes in Tissue Group: 25
gene_set nset ngenes percent padj
1 gwascatalog 4798 12 0.002501042 0.1665561
2 core_essentials_hart 222 2 0.009009009 0.1665561
3 mgi_essential 1797 4 0.002225932 0.4339454
4 fda_approved_drug_targets 255 1 0.003921569 0.4339454
5 clinvar_path_likelypath 2208 4 0.001811594 0.5081103
Digestive
Number of cTWAS Genes in Tissue Group: 37
gene_set nset ngenes percent padj
1 gwascatalog 5418 20 0.003691399 0.01519409
2 mgi_essential 2055 10 0.004866180 0.02198368
3 clinvar_path_likelypath 2501 7 0.002798880 0.45812527
4 core_essentials_hart 246 1 0.004065041 0.48542112
5 fda_approved_drug_targets 310 1 0.003225806 0.48542112Analysis of TWAS False Positives by Region
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_tag, function(x){unlist(region_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from region total to get combined pip for other genes in region
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip < 0.5)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip < 0.5)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
| Version | Author | Date |
|---|---|---|
| 95e0f8e | wesleycrouse | 2022-04-07 |
Analysis of TWAS False Positives by Credible Set
cTWAS is using susie settings that mask credible sets consisting of variables with minimum pairwise correlations below a specified threshold. The default threshold is 0.5. I think this is intended to mask credible sets with “diffuse” support. As a consequence, many of the genes considered here (TWAS false positives; significant z score but low PIP) are not assigned to a credible set (have cs_index=0). For this reason, the first figure is not really appropriate for answering the question “are TWAS false positives due to SNPs or genes”.
The second figure includes only TWAS genes that are assigned to a reported causal set (i.e. they are in a “pure” causal set with high pairwise correlations). I think that this figure is closer to the intended analysis. However, it may be biased in some way because we have excluded many TWAS false positive genes that are in “impure” credible sets.
Some alternatives to these figures include the region-based analysis in the previous section; or re-analysis with lower/no minimum pairwise correlation threshold (“min_abs_corr” option in susie_get_cs) for reporting credible sets.
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
| Version | Author | Date |
|---|---|---|
| 95e0f8e | wesleycrouse | 2022-04-07 |
####################
#using only genes assigned to a credible set
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
#exclude genes that are not assigned to a credible set, cs_index==0
gene_pips <- gene_pips[as.numeric(sapply(gene_pips$region_cs_tag, function(x){rev(unlist(strsplit(x, "_")))[1]}))!=0,]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
cTWAS genes without genome-wide significant SNP nearby
novel_genes <- data.frame(genename=as.character(), weight=as.character(), susie_pip=as.numeric(), snp_maxz=as.numeric())
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$ctwas,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, sapply(gene_pips$region_tag, function(x){region_pips[x,"snp_maxz"]}))
names(gene_pips)[ncol(gene_pips)] <- "snp_maxz"
if (nrow(gene_pips)>0){
gene_pips$weight <- names(df)[i]
gene_pips <- gene_pips[gene_pips$snp_maxz < qnorm(1-(5E-8/2), lower=T),c("genename", "weight", "susie_pip", "snp_maxz")]
novel_genes <- rbind(novel_genes, gene_pips)
}
}
novel_genes_summary <- data.frame(genename=unique(novel_genes$genename))
novel_genes_summary$nweights <- sapply(novel_genes_summary$genename, function(x){length(novel_genes$weight[novel_genes$genename==x])})
novel_genes_summary$weights <- sapply(novel_genes_summary$genename, function(x){paste(novel_genes$weight[novel_genes$genename==x],collapse=", ")})
novel_genes_summary <- novel_genes_summary[order(-novel_genes_summary$nweights),]
novel_genes_summary[,c("genename","nweights")] genename nweights
1 PIGV 21
2 RP11-84A19.4 20
8 ARID1A 16
13 RINT1 13
4 ENPEP 10
14 ZNF467 8
44 MORC3 8
46 HSCB 8
5 EML1 7
10 BAHCC1 7
6 ZNF415 6
34 DNAH11 5
39 ZDHHC13 5
25 AATK 4
30 DDI2 4
37 PGBD2 4
38 GNPDA1 4
47 TMEM175 4
7 ASCC2 3
16 HHIPL1 3
19 GPER1 3
24 LRRC10B 3
40 RP5-965G21.3 3
41 SOX13 3
48 RP11-286N22.10 3
54 SSBP3 3
9 LINC00930 2
21 SLC20A2 2
27 NTRK1 2
36 STK38L 2
50 ARHGEF25 2
52 RP4-534N18.2 2
65 ARL4A 2
67 GLTP 2
3 MRAS 1
11 SLC19A1 1
12 KIAA1614 1
15 VDR 1
17 CKB 1
18 SHISA8 1
20 KIF13B 1
22 SHB 1
23 KIAA1462 1
26 RP11-1055B8.3 1
28 RP11-373D23.3 1
29 PROCR 1
31 ZNF692 1
32 SLC9A3 1
33 KHDC3L 1
35 ARMS2 1
42 HTRA1 1
43 SPIRE1 1
45 PUS7 1
49 C22orf31 1
51 NUDT16L1 1
53 SSPO 1
55 MAEA 1
56 TMEM179B 1
57 CPXM1 1
58 PPP3R1 1
59 CAMK1D 1
60 NDUFAF8 1
61 LINC01169 1
62 SLC26A1 1
63 NR3C1 1
64 TRIOBP 1
66 TTC33 1
68 WASF3 1
69 MEX3A 1
70 KCNQ5 1
71 LINC01451 1
72 PTGER4 1
73 CTD-2349P21.5 1
74 MAPK10 1
75 MYO1F 1Tissue-specificity for cTWAS genes
gene_pips_by_weight <- data.frame(genename=as.character(ctwas_genes))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips
gene_pips <- gene_pips[match(ctwas_genes, gene_pips$genename),,drop=F]
#gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- -1
gene_pips$susie_pip[is.na(gene_pips$susie_pip)] <- 0 #missing values coded as PIP=0
gene_pips_by_weight <- cbind(gene_pips_by_weight, gene_pips$susie_pip)
names(gene_pips_by_weight)[ncol(gene_pips_by_weight)] <- names(df)[i]
}
gene_pips_by_weight <- as.matrix(gene_pips_by_weight[,-1])
rownames(gene_pips_by_weight) <- ctwas_genes
#number of tissues with PIP>0.5 for cTWAS genes
ctwas_frequency <- rowSums(gene_pips_by_weight>0.5)
hist(ctwas_frequency, col="grey", breaks=0:max(ctwas_frequency), xlim=c(0,ncol(gene_pips_by_weight)),
xlab="Number of Tissues with PIP>0.5",
ylab="Number of cTWAS Genes",
main="Tissue Specificity for cTWAS Genes")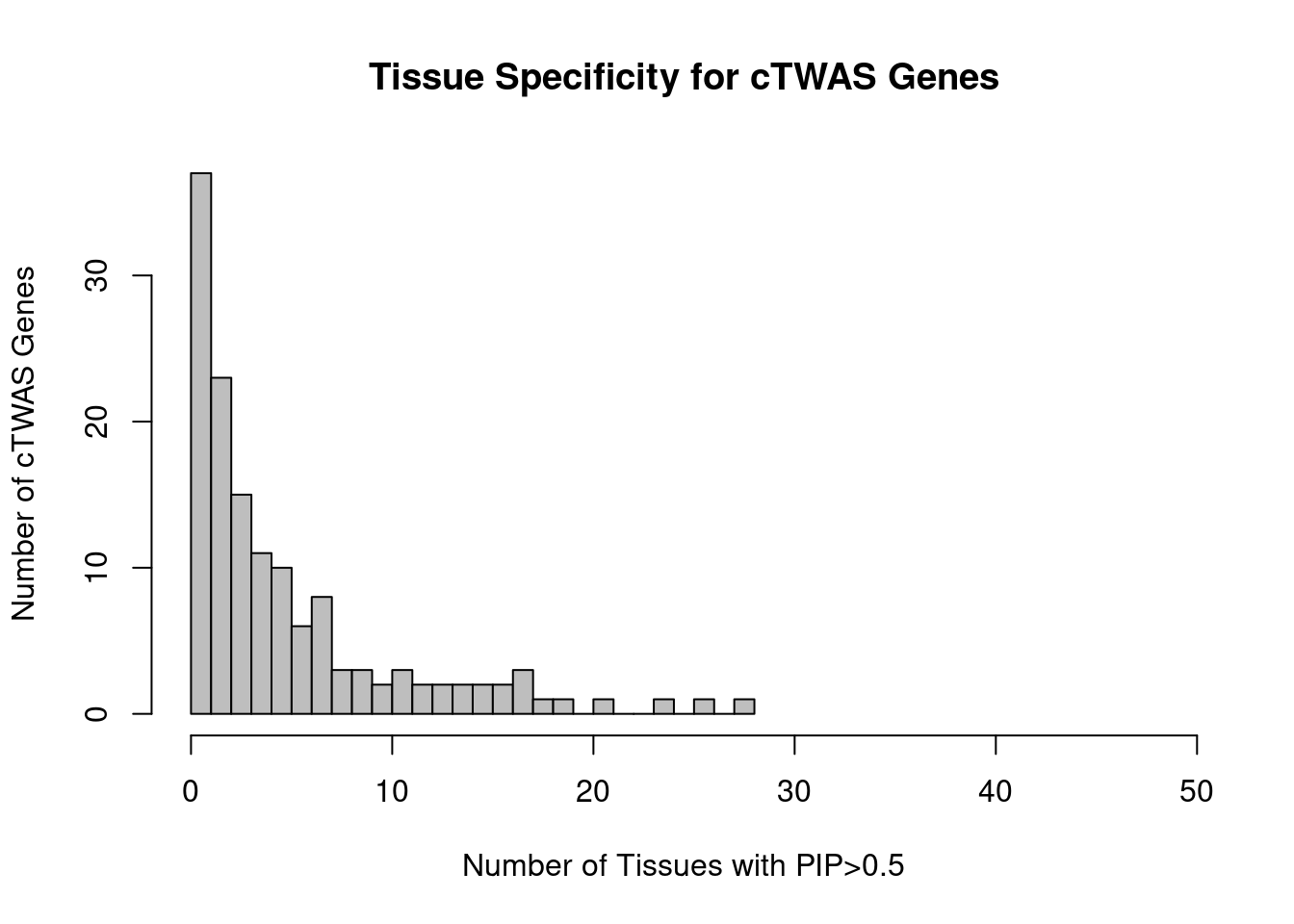
#heatmap of gene PIPs
cluster_ctwas_genes <- hclust(dist(gene_pips_by_weight))
cluster_ctwas_weights <- hclust(dist(t(gene_pips_by_weight)))
plot(cluster_ctwas_weights, cex=0.6)
plot(cluster_ctwas_genes, cex=0.6, labels=F)
par(mar=c(14.1, 4.1, 4.1, 2.1))
image(t(gene_pips_by_weight[rev(cluster_ctwas_genes$order),rev(cluster_ctwas_weights$order)]),
axes=F)
mtext(text=colnames(gene_pips_by_weight)[cluster_ctwas_weights$order], side=1, line=0.3, at=seq(0,1,1/(ncol(gene_pips_by_weight)-1)), las=2, cex=0.8)
mtext(text=rownames(gene_pips_by_weight)[cluster_ctwas_genes$order], side=2, line=0.3, at=seq(0,1,1/(nrow(gene_pips_by_weight)-1)), las=1, cex=0.4)
cTWAS genes with highest proportion of total PIP on a single tissue
#genes with highest porportion of PIP on a single tissue
gene_pips_proportion <- gene_pips_by_weight/rowSums(gene_pips_by_weight)
proportion_table <- data.frame(genename=as.character(rownames(gene_pips_proportion)))
proportion_table$max_pip_prop <- apply(gene_pips_proportion,1,max)
proportion_table$max_weight <- colnames(gene_pips_proportion)[apply(gene_pips_proportion,1,which.max)]
proportion_table[order(-proportion_table$max_pip_prop),] genename max_pip_prop max_weight
106 FGF5 1.00000000 Kidney_Cortex
129 MEX3A 1.00000000 Skin_Sun_Exposed_Lower_leg
138 C20orf187 1.00000000 Testis
97 MAEA 0.99970816 Colon_Transverse
108 LINC01169 0.86630869 Lung
130 KCNQ5 0.86055264 Skin_Sun_Exposed_Lower_leg
63 KHDC3L 0.85710895 Brain_Amygdala
89 CNNM2 0.85002654 Brain_Putamen_basal_ganglia
128 FURIN 0.79557650 Skin_Not_Sun_Exposed_Suprapubic
87 C22orf31 0.78860732 Brain_Hypothalamus
36 SHISA8 0.77821055 Adrenal_Gland
19 SBF2 0.75694260 Adipose_Visceral_Omentum
56 NGF 0.74119098 Artery_Tibial
131 NPNT 0.72162883 Spleen
82 PUS7 0.71458121 Brain_Cortex
121 CIB4 0.65665869 Pancreas
91 NUDT16L1 0.62934740 Breast_Mammary_Tissue
95 GIT2 0.55835346 Cells_EBV-transformed_lymphocytes
40 KIF13B 0.53258520 Artery_Aorta
134 CTDNEP1 0.51931412 Spleen
67 SENP3 0.51623809 Brain_Anterior_cingulate_cortex_BA24
26 KIAA1614 0.51389867 Adrenal_Gland
93 RP4-534N18.2 0.50346201 Heart_Atrial_Appendage
94 IGHMBP2 0.48885870 Cells_EBV-transformed_lymphocytes
103 C17orf82 0.47037555 Esophagus_Mucosa
115 NR3C1 0.46139808 Nerve_Tibial
54 PROCR 0.44178010 Artery_Coronary
43 KIAA1462 0.44010861 Artery_Aorta
78 HTRA1 0.42780857 Brain_Cerebellum
57 ZNF692 0.41807915 Artery_Tibial
68 MYOZ1 0.41279707 Heart_Atrial_Appendage
137 CTD-2349P21.5 0.39822338 Testis
113 YEATS2 0.39382047 Nerve_Tibial
140 MYO1F 0.39161869 Uterus
101 PPP3R1 0.37559664 Esophagus_Mucosa
29 VDR 0.37487015 Adrenal_Gland
46 RP11-405A12.2 0.36296813 Artery_Aorta
110 CTDSPL 0.36151872 Muscle_Skeletal
107 NDUFAF8 0.35147932 Liver
127 WASF3 0.33981282 Skin_Not_Sun_Exposed_Suprapubic
13 ASCC2 0.33333331 Adipose_Subcutaneous
122 THBS2 0.31744957 Pancreas
38 PHACTR1 0.30444883 Artery_Aorta
98 TMEM179B 0.29597329 Colon_Transverse
133 CERS5 0.27139479 Testis
69 STK38L 0.27046500 Brain_Caudate_basal_ganglia
118 ARL4A 0.26706013 Skin_Not_Sun_Exposed_Suprapubic
32 CRK 0.26382765 Adrenal_Gland
116 COL21A1 0.25642457 Nerve_Tibial
139 MAPK10 0.25091479 Thyroid
71 PGBD2 0.24657441 Brain_Cerebellum
60 LIMA1 0.24606523 Artery_Tibial
55 DDI2 0.24538355 Artery_Tibial
114 SLC26A1 0.24501308 Nerve_Tibial
126 GLTP 0.24205516 Skin_Sun_Exposed_Lower_leg
41 SLC20A2 0.24091134 Artery_Tibial
66 ARMS2 0.24026652 Brain_Anterior_cingulate_cortex_BA24
76 SLC2A4RG 0.23903783 Thyroid
136 MTHFR 0.23813957 Testis
37 GDF7 0.23205766 Artery_Aorta
99 OVOL1 0.22156620 Colon_Transverse
21 LINC00930 0.21574799 Adipose_Visceral_Omentum
104 GFER 0.21240645 Esophagus_Muscularis
24 SLC19A1 0.20090206 Adipose_Visceral_Omentum
102 FARP2 0.19964184 Esophagus_Muscularis
105 CAMK1D 0.19785381 Heart_Left_Ventricle
8 SSPO 0.18258522 Adipose_Subcutaneous
61 ZBTB46 0.17978029 Brain_Anterior_cingulate_cortex_BA24
59 ADRB1 0.17977725 Artery_Tibial
124 RHOC 0.17842155 Skin_Sun_Exposed_Lower_leg
74 RP11-757G1.6 0.17771990 Prostate
35 TCEA2 0.17585453 Adrenal_Gland
17 MAP6D1 0.17427304 Adipose_Visceral_Omentum
44 SIRT1 0.17086685 Artery_Aorta
77 SOX13 0.16617866 Brain_Cerebellum
123 NT5C2 0.16486535 Prostate
88 TMEM176B 0.16420632 Brain_Putamen_basal_ganglia
86 RP11-286N22.10 0.16078177 Brain_Nucleus_accumbens_basal_ganglia
80 SPIRE1 0.16037666 Brain_Cerebellum
70 SF3B3 0.15917279 Prostate
20 ADAMTS8 0.15901463 Artery_Aorta
83 ACADVL 0.15712579 Colon_Sigmoid
45 LRRC10B 0.15694033 Skin_Not_Sun_Exposed_Suprapubic
109 AGT 0.15642883 Muscle_Skeletal
5 MRAS 0.15221499 Adipose_Subcutaneous
48 AATK 0.15189669 Artery_Aorta
119 NPW 0.14976436 Ovary
42 SHB 0.14966207 Artery_Aorta
120 EFR3B 0.14872042 Pancreas
33 DLG4 0.14806604 Thyroid
39 GPER1 0.14801735 Esophagus_Muscularis
18 EDNRA 0.14596165 Adipose_Visceral_Omentum
53 MRPL21 0.14134652 Artery_Coronary
16 ULK4 0.14069820 Skin_Sun_Exposed_Lower_leg
125 TTC33 0.13974398 Skin_Not_Sun_Exposed_Suprapubic
58 ZNF827 0.13708465 Artery_Tibial
90 ARHGEF25 0.13589120 Brain_Substantia_nigra
135 PTGER4 0.12969263 Stomach
73 ZDHHC13 0.12864028 Brain_Hypothalamus
79 ZNF598 0.12566126 Brain_Cerebellum
132 LINC01451 0.12541195 Spleen
75 RP5-965G21.3 0.12509770 Brain_Cerebellar_Hemisphere
112 CDC16 0.11888537 Muscle_Skeletal
62 SLC9A3 0.11835951 Brain_Amygdala
84 HSCB 0.11535717 Heart_Left_Ventricle
50 GNA11 0.11376852 Artery_Aorta
64 CLCN6 0.10822737 Skin_Sun_Exposed_Lower_leg
34 USP36 0.10696715 Adrenal_Gland
51 NTRK1 0.10337033 Artery_Coronary
85 TMEM175 0.10262839 Pancreas
100 CPXM1 0.10217385 Colon_Transverse
52 RP11-373D23.3 0.10121264 Artery_Coronary
30 HHIPL1 0.09361330 Lung
3 KCNK3 0.09059667 Adipose_Subcutaneous
117 TRIOBP 0.08798104 Nerve_Tibial
22 BAHCC1 0.08595632 Ovary
31 CKB 0.08542344 Adrenal_Gland
96 SSBP3 0.08452710 Colon_Transverse
49 RP11-1055B8.3 0.08118196 Artery_Aorta
92 SNX11 0.07611229 Breast_Mammary_Tissue
81 MORC3 0.07528762 Pancreas
27 RINT1 0.07405352 Cells_EBV-transformed_lymphocytes
6 ENPEP 0.07263773 Lung
111 FAM212A 0.07203150 Muscle_Skeletal
65 DNAH11 0.06936665 Brain_Cortex
12 RGS19 0.06870624 Adipose_Subcutaneous
47 CACNB3 0.06630927 Artery_Tibial
9 EML1 0.06500174 Artery_Tibial
72 GNPDA1 0.06466171 Spleen
25 RP4-758J18.13 0.06437376 Kidney_Cortex
11 ZNF415 0.05952835 Adipose_Visceral_Omentum
14 ARID1A 0.05893765 Cells_Cultured_fibroblasts
10 PAQR5 0.05717632 Artery_Aorta
23 OPRL1 0.05659471 Prostate
7 HFE 0.05643812 Small_Intestine_Terminal_Ileum
15 FN1 0.05385348 Artery_Aorta
2 RP11-84A19.4 0.04861800 Adipose_Subcutaneous
4 CCNT2 0.03856262 Adipose_Subcutaneous
28 ZNF467 0.03779791 Artery_Aorta
1 PIGV 0.03574467 Brain_Caudate_basal_ganglia
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.5 disgenet2r_0.99.2 WebGestaltR_0.4.4 enrichR_3.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.8 reshape2_1.4.3 purrr_0.3.4 lattice_0.20-38 colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 utf8_1.2.1 rlang_0.4.11 later_0.8.0 pillar_1.6.1 withr_2.4.1 glue_1.4.2 DBI_1.1.1 gdtools_0.1.9 rngtools_1.5 doRNG_1.8.2 plyr_1.8.4 foreach_1.5.1 lifecycle_1.0.0 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0 workflowr_1.6.2 codetools_0.2-16 evaluate_0.14 labeling_0.3 knitr_1.23 doParallel_1.0.16 httpuv_1.5.1 curl_3.3 parallel_3.6.1 fansi_0.5.0 Rcpp_1.0.6 readr_1.4.0 promises_1.0.1 scales_1.1.0 jsonlite_1.6 apcluster_1.4.8 farver_2.1.0 fs_1.3.1 hms_1.1.0 rjson_0.2.20 digest_0.6.20 stringi_1.4.3 dplyr_1.0.7 grid_3.6.1 rprojroot_2.0.2 tools_3.6.1 magrittr_2.0.1 tibble_3.1.2 crayon_1.4.1
[56] whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2 Matrix_1.2-18 svglite_1.2.2 rmarkdown_1.13 httr_1.4.1 iterators_1.0.13 R6_2.5.0 igraph_1.2.4.1 git2r_0.26.1 compiler_3.6.1