Weight - all weights
wesleycrouse
2022-02-28
Last updated: 2022-03-03
Checks: 6 1
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 380982d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Unstaged changes:
Modified: analysis/ukb-a-232_allweights.Rmd
Modified: analysis/ukb-a-249_allweights.Rmd
Modified: analysis/ukb-a-389_allweights.Rmd
Modified: analysis/ukb-a-66_allweights.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ukb-a-249_allweights.Rmd) and HTML (docs/ukb-a-249_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 380982d | wesleycrouse | 2022-03-01 | fixing typo in all weight reports |
| Rmd | 76fa2cd | wesleycrouse | 2022-03-01 | cleaning up all weight reports |
| html | 76fa2cd | wesleycrouse | 2022-03-01 | cleaning up all weight reports |
| html | 2509c32 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
| Rmd | 962fd16 | wesleycrouse | 2022-03-01 | additional traits for all weight analysis |
trait_id <- "ukb-a-249"
trait_name <- "Weight"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# #print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
# twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z) > sig_thresh]
#
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes )
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)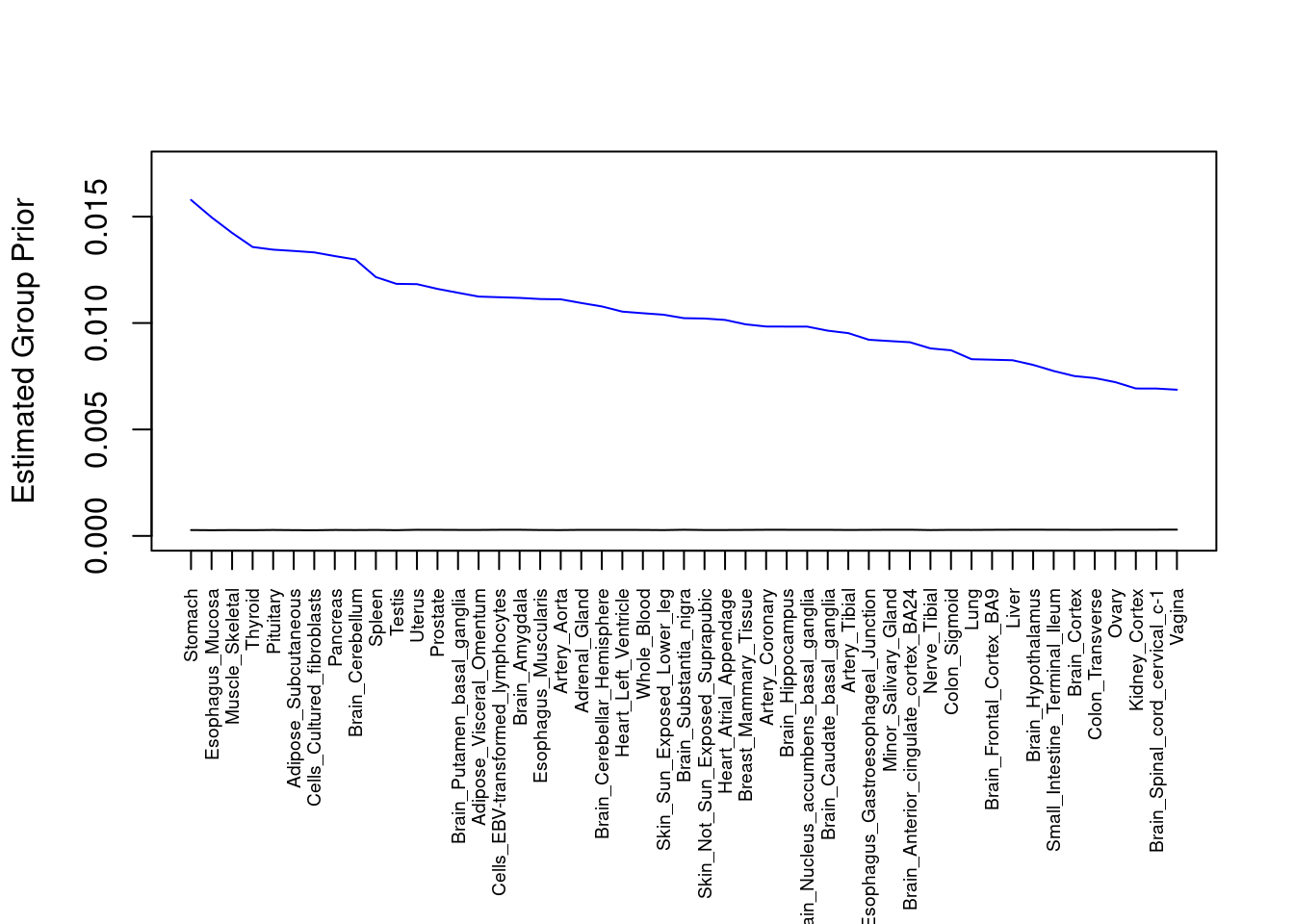
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)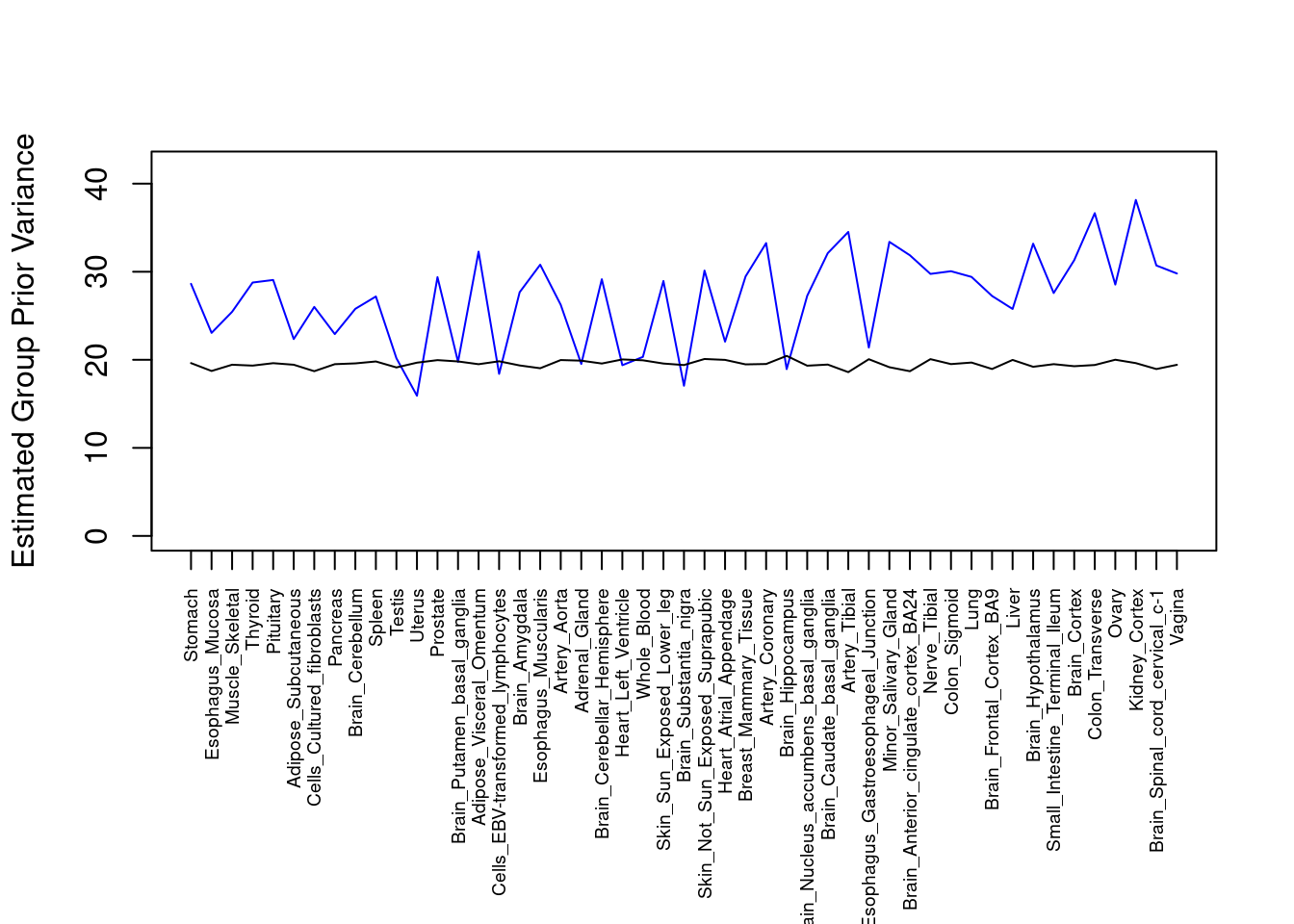
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
#plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g, output$pve_s)*1.1),
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)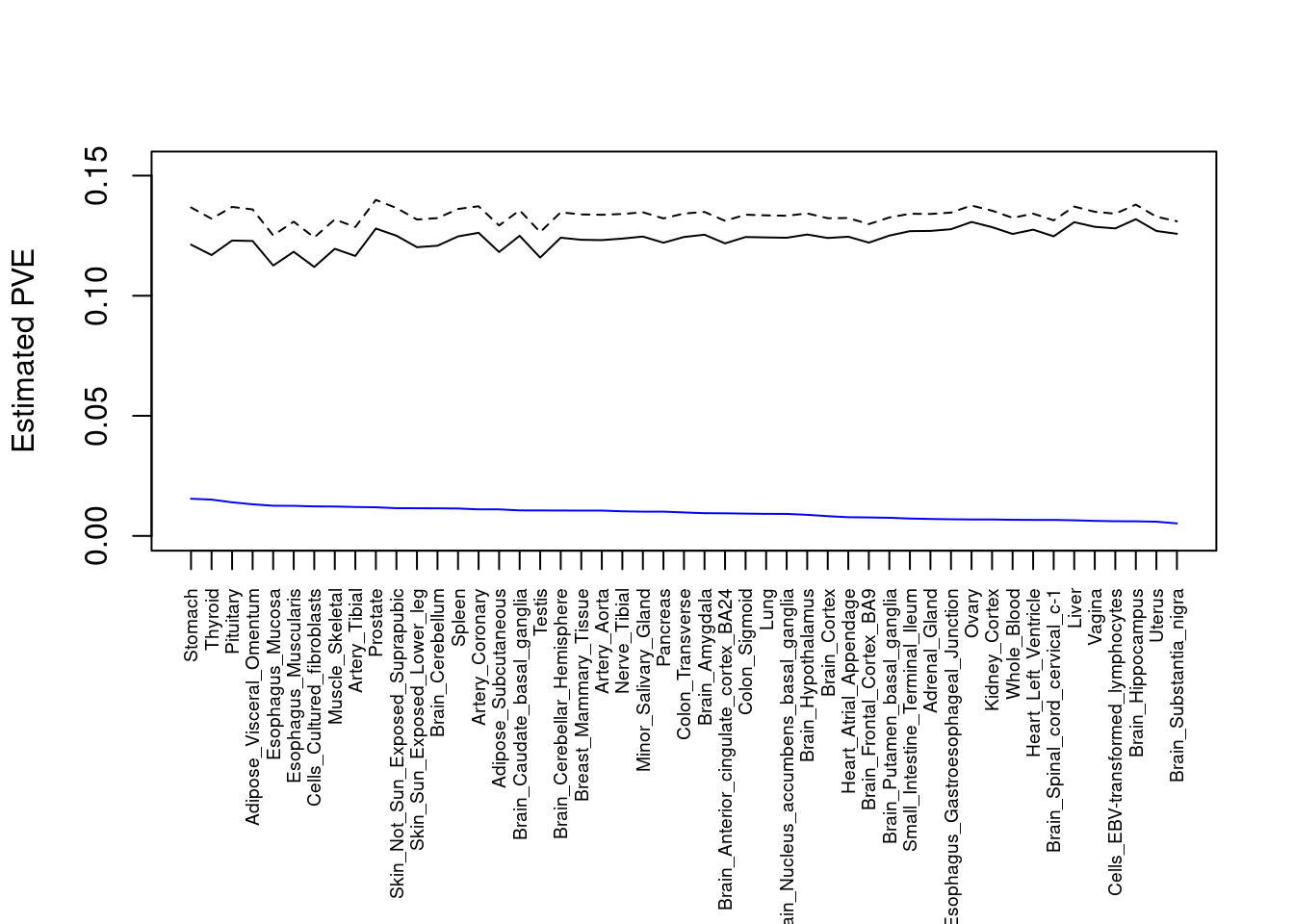
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")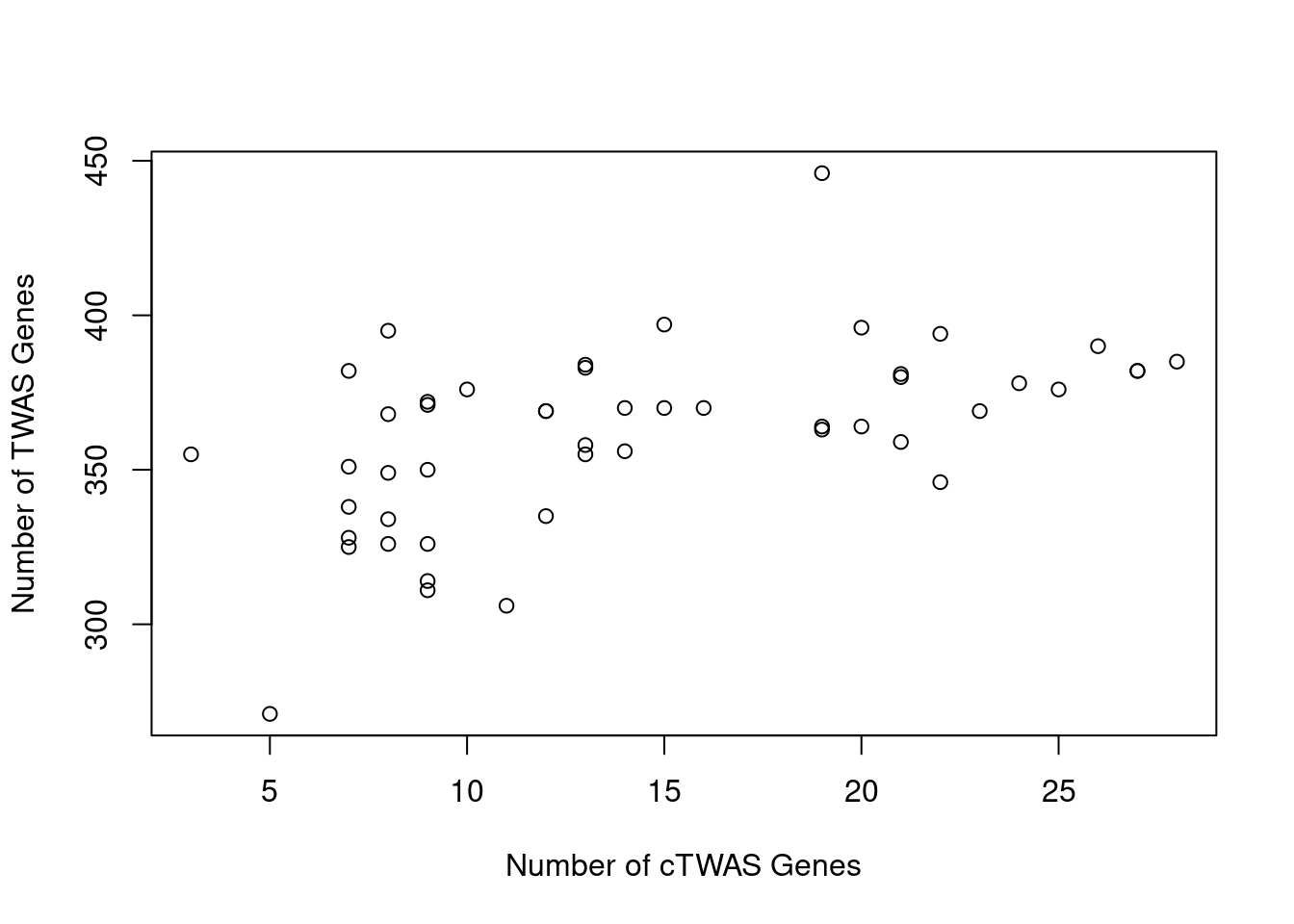
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 210#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 1749Enrichment analysis for cTWAS genes
#enrichment for cTWAS genes
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}[1] "GO_Biological_Process_2021"
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"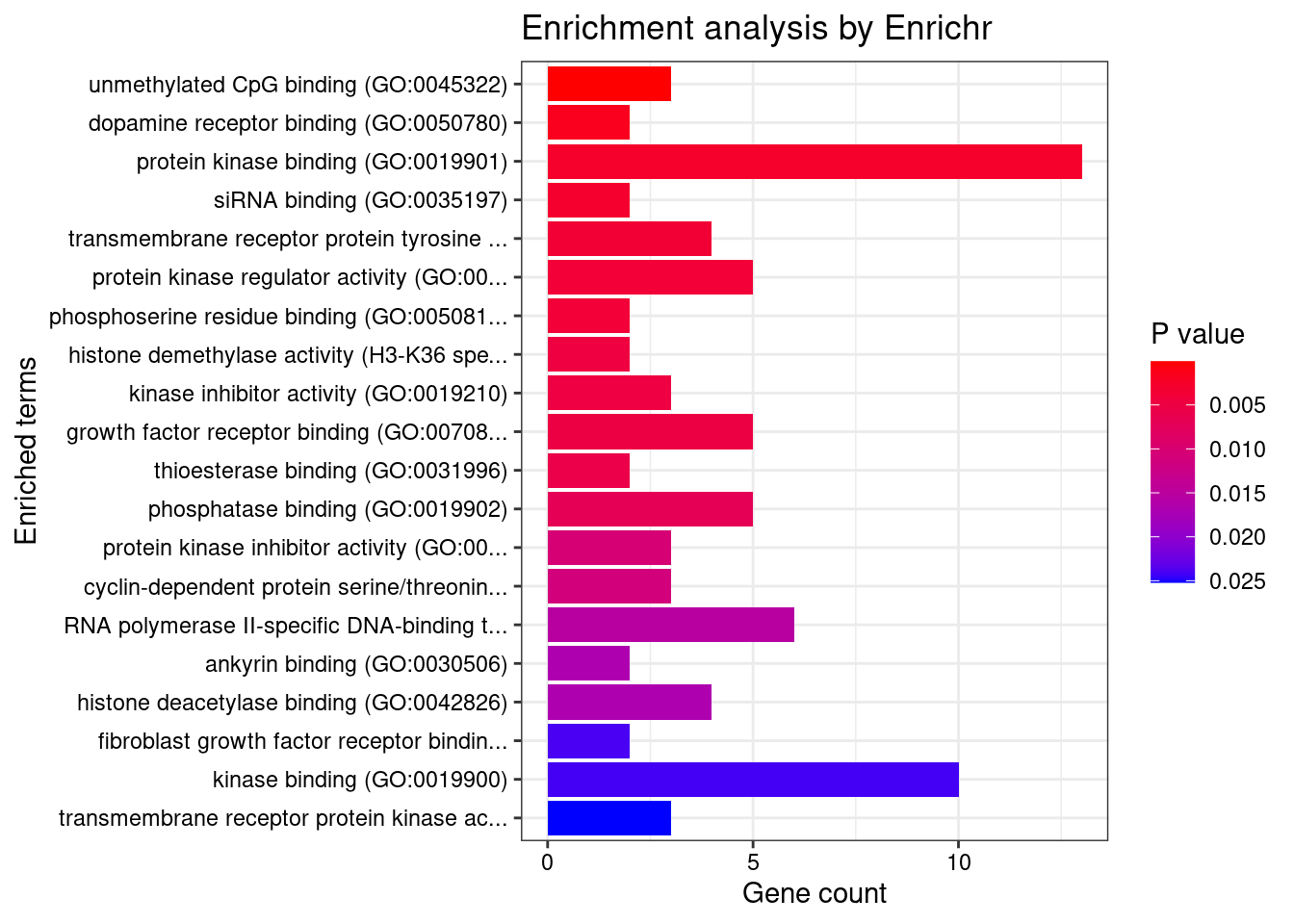
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term Overlap Adjusted.P.value
1 unmethylated CpG binding (GO:0045322) 3/8 0.01880987
Genes
1 KDM2A;CXXC1;TLR9Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}[1] "GO_Biological_Process_2021"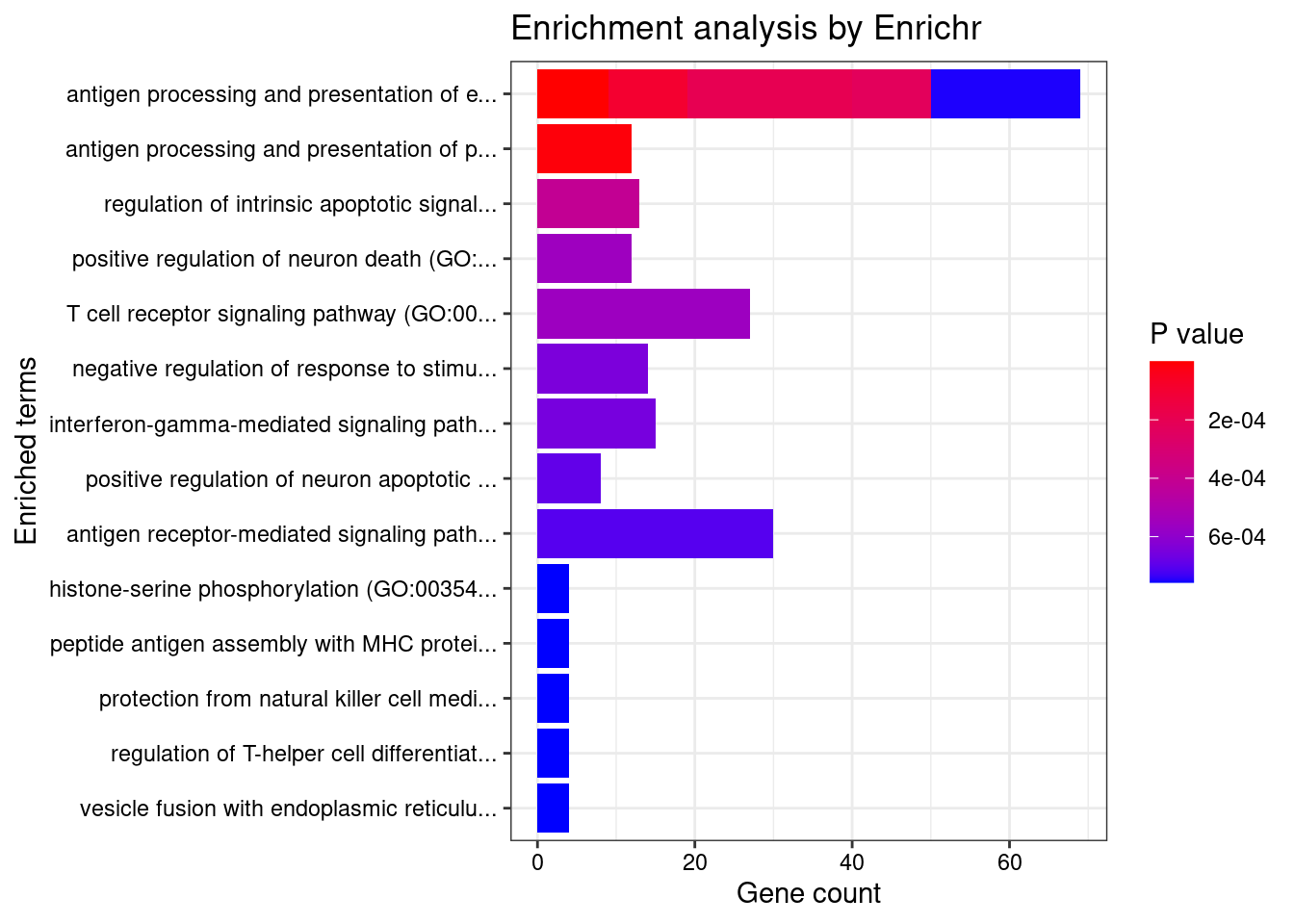
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term
1 antigen processing and presentation of endogenous peptide antigen (GO:0002483)
2 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474)
Overlap Adjusted.P.value
1 9/14 0.001722268
2 12/33 0.026325171
Genes
1 ERAP1;TAP2;TAP1;HLA-DRA;ABCB9;HLA-A;HLA-G;HLA-DRB1;HLA-E
2 HFE;ERAP1;HLA-B;TAP2;HLA-C;TAP1;ABCB9;HLA-A;HLA-G;SEC24C;HLA-E;TAPBP
[1] "GO_Cellular_Component_2021"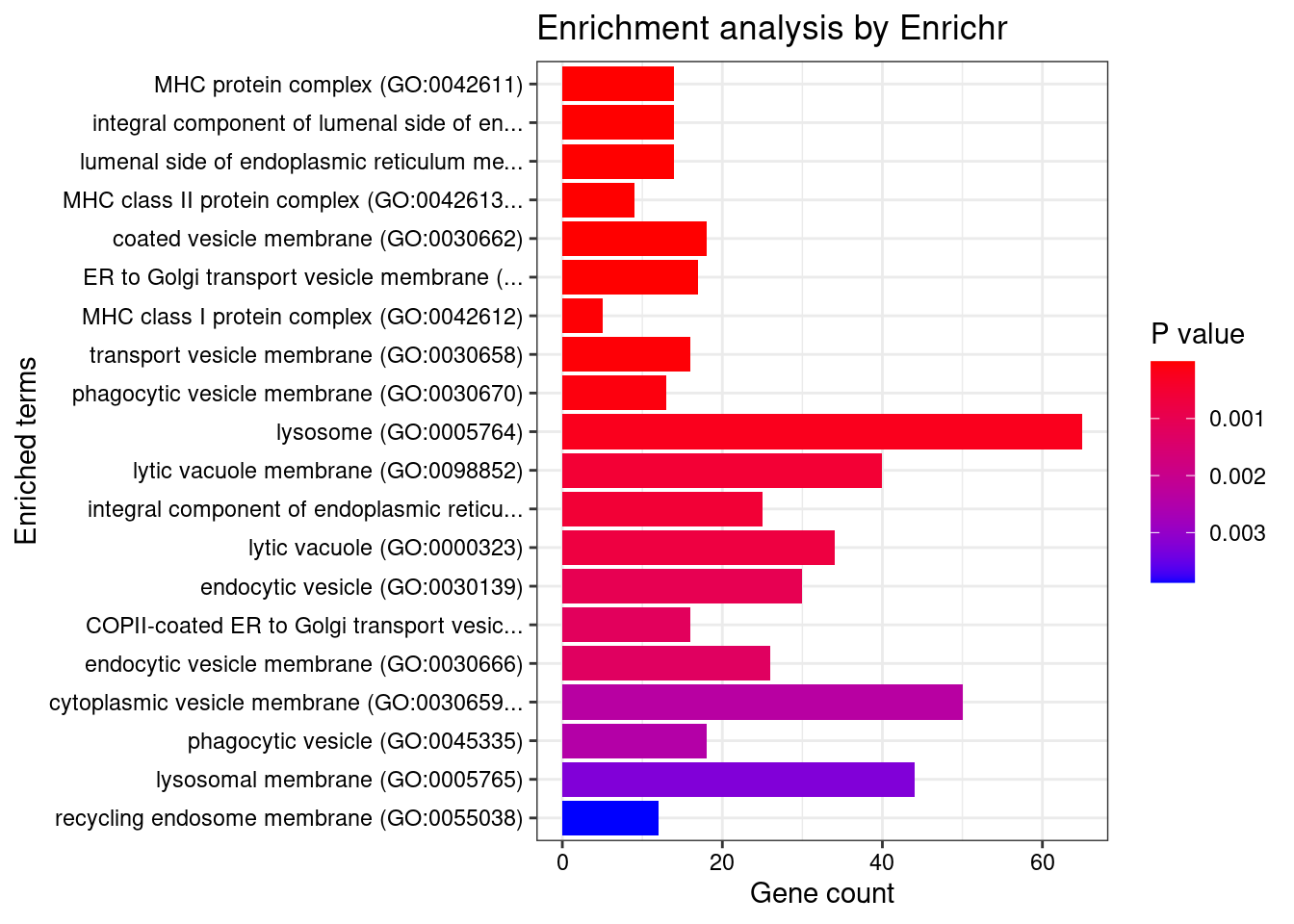
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term
1 MHC protein complex (GO:0042611)
2 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556)
3 lumenal side of endoplasmic reticulum membrane (GO:0098553)
4 MHC class II protein complex (GO:0042613)
5 coated vesicle membrane (GO:0030662)
6 ER to Golgi transport vesicle membrane (GO:0012507)
7 MHC class I protein complex (GO:0042612)
8 transport vesicle membrane (GO:0030658)
9 phagocytic vesicle membrane (GO:0030670)
10 lysosome (GO:0005764)
11 lytic vacuole membrane (GO:0098852)
12 integral component of endoplasmic reticulum membrane (GO:0030176)
13 lytic vacuole (GO:0000323)
14 endocytic vesicle (GO:0030139)
15 COPII-coated ER to Golgi transport vesicle (GO:0030134)
16 endocytic vesicle membrane (GO:0030666)
Overlap Adjusted.P.value
1 14/20 1.297702e-08
2 14/28 2.283210e-06
3 14/28 2.283210e-06
4 9/13 1.442975e-05
5 18/55 3.865885e-05
6 17/54 1.229098e-04
7 5/6 1.540683e-03
8 16/60 1.895911e-03
9 13/45 3.590180e-03
10 65/477 8.465994e-03
11 40/267 1.782614e-02
12 25/142 1.782614e-02
13 34/219 2.132486e-02
14 30/189 2.746241e-02
15 16/79 2.967855e-02
16 26/158 2.967855e-02
Genes
1 HLA-DRB5;HFE;HLA-B;HLA-C;HLA-A;HLA-E;HLA-DMA;HLA-DMB;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1
2 HLA-DRB5;SPPL2B;HLA-B;HLA-C;HLA-A;SPPL3;HLA-G;HLA-E;TAPBP;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1
3 HLA-DRB5;SPPL2B;HLA-B;HLA-C;HLA-A;SPPL3;HLA-G;HLA-E;TAPBP;HLA-DRA;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1
4 HLA-DRB5;HLA-DMA;HLA-DMB;HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB2;HLA-DRB1
5 SEC16B;HLA-DRB5;DENND1A;HLA-B;HLA-C;HLA-A;HLA-G;HLA-E;AP1G1;HLA-DRA;KDELR2;CNIH2;HLA-DQA2;SEC24C;HLA-DQB2;HLA-DQA1;SEC31B;HLA-DRB1
6 SEC16B;HLA-DRB5;PEF1;PDCD6;HLA-B;HLA-C;HLA-A;HLA-G;HLA-E;HLA-DRA;CNIH2;HLA-DQA2;SEC24C;HLA-DQB2;HLA-DQA1;SEC31B;HLA-DRB1
7 HFE;HLA-B;HLA-C;HLA-A;HLA-E
8 SEC16B;HLA-DRB5;RAB3A;HLA-B;HLA-C;ECE2;HLA-A;HLA-G;HLA-E;HLA-DRA;CNIH2;HLA-DQA2;SEC24C;HLA-DQB2;HLA-DQA1;HLA-DRB1
9 HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G;HLA-E;TAPBP;VAMP8;STX4;PIK3C3;ATP6V0A1;VAMP3
10 SLC35F6;TINAGL1;SPPL2B;CTSW;RPTOR;NAGLU;AP1G1;HYAL1;HYAL3;FLOT1;AP1S1;PIP4K2A;HLA-DOA;HLA-DOB;AP2M1;UBXN6;VPS33A;PRSS16;TSC2;AP3B1;AP5B1;TMEM175;VAMP8;PLA2G15;HCK;NPC1;TLR9;SPNS1;DAGLB;RAB7B;HLA-DQB2;RAMP2;SRC;ABCB9;BCL10;WDR24;TMEM165;CLN3;HLA-DMA;HLA-DMB;GPC1;MYO6;NEU1;VPS11;GPC5;FNIP1;HLA-DQA2;SLC17A2;HLA-DQA1;SLC17A4;ATP6V0A1;CD164;HLA-DRB5;AP3D1;CYBRD1;RAB27A;MTOR;TPCN2;P2RX4;GNB1;OCIAD1;HLA-DRA;PPT2;OCIAD2;HLA-DRB1
11 SLC35F6;SPPL2B;ABCB9;WDR24;TMEM165;RPTOR;CLN3;HLA-DMA;HLA-DMB;AP1G1;MYO6;VPS11;FLOT1;AP1S1;HLA-DOA;FNIP1;HLA-DQA2;HLA-DOB;HLA-DQA1;UBXN6;AP2M1;ATP6V0A1;HLA-DRB5;VPS33A;AP3D1;CYBRD1;AP3B1;AP5B1;TMEM175;MTOR;TPCN2;VAMP8;P2RX4;NPC1;GNB1;SPNS1;HLA-DRA;DAGLB;HLA-DRB1;HLA-DQB2
12 ANKLE2;ATF6B;SPPL2B;CCDC47;ABCB9;SPPL3;CLN3;EMC3;SLC37A4;HLA-DQA2;HLA-DQA1;HLA-DRB5;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G;HLA-E;TAPBP;TBL2;HLA-DRA;ERGIC3;HLA-DRB1;HLA-DQB2
13 RAMP2;TINAGL1;SRC;CTSW;ABCB9;BCL10;RPTOR;CLN3;NAGLU;HYAL1;NEU1;HYAL3;VPS11;PIP4K2A;SLC17A2;HLA-DOB;SLC17A4;CD164;VPS33A;RAB27A;PRSS16;TSC2;TMEM175;MTOR;TPCN2;PLA2G15;HCK;NPC1;TLR9;OCIAD1;HLA-DRA;PPT2;OCIAD2;RAB7B
14 CEMIP;RAB5B;CORO1A;MYO6;RAB24;DVL2;VPS11;HLA-DQA2;RAB11FIP3;AP2M1;HLA-DQA1;RAB11FIP4;HLA-DRB5;AMN;NOS3;PTCH1;SFTPC;SCIMP;MTOR;DNM2;RABEP1;DLG4;RAB35;NOSTRIN;HLA-DRA;HYOU1;RAB7B;RAPGEF6;HLA-DRB1;HLA-DQB2
15 SEC16B;HLA-DRB5;HLA-B;HLA-C;HLA-A;HLA-G;HLA-E;HLA-DRA;ERGIC3;CNIH2;HLA-DQA2;SEC24C;HLA-DQB2;HLA-DQA1;CNIH4;HLA-DRB1
16 STX4;HLA-DQA2;AP2M1;HLA-DQA1;ATP6V0A1;HLA-DRB5;NOS3;PTCH1;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G;HLA-E;DNM2;TAPBP;VAMP8;DLG4;RAB35;NOSTRIN;HLA-DRA;PIK3C3;HLA-DRB1;HLA-DQB2;VAMP3
[1] "GO_Molecular_Function_2021"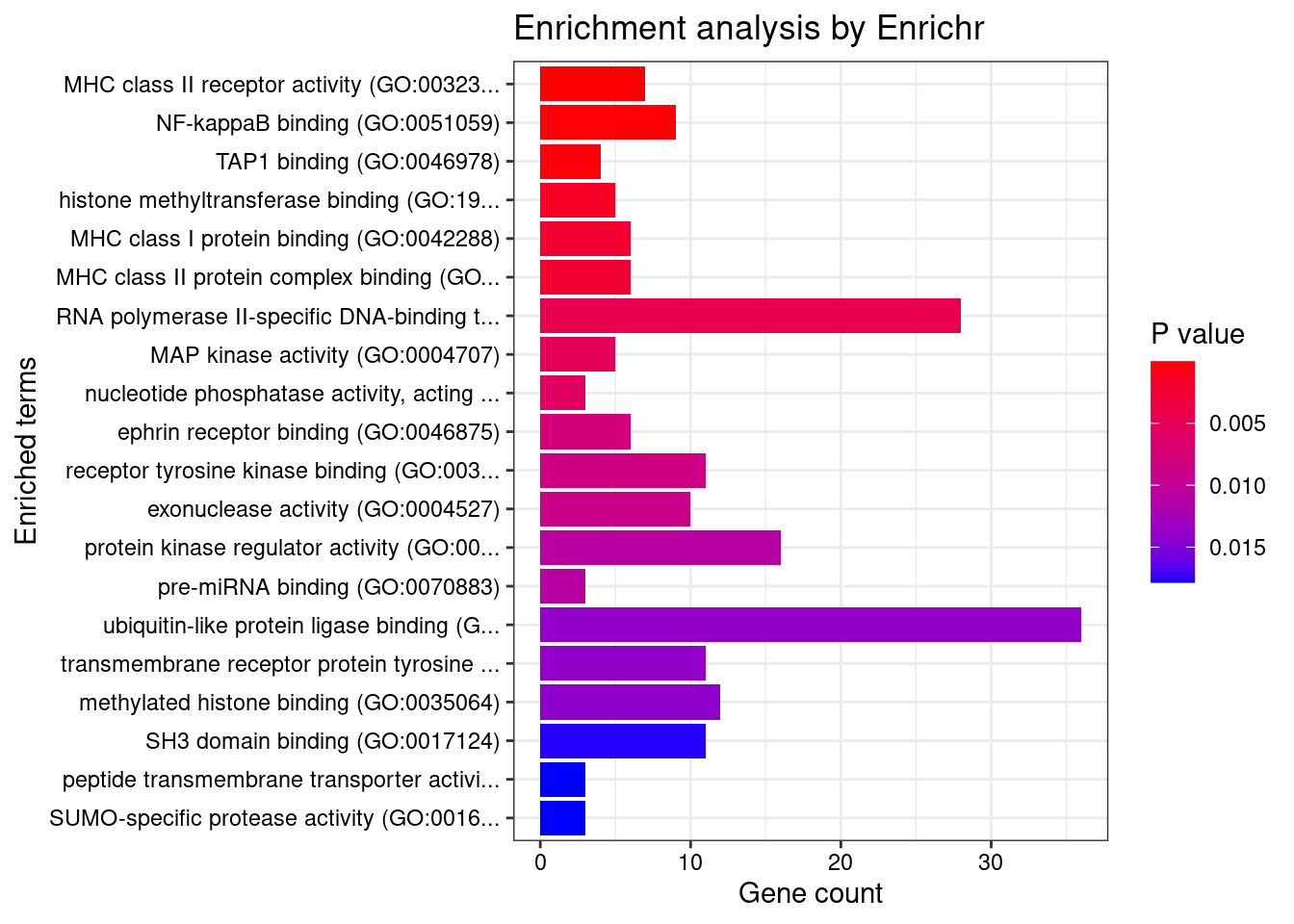
| Version | Author | Date |
|---|---|---|
| 2509c32 | wesleycrouse | 2022-03-01 |
Term Overlap Adjusted.P.value
1 MHC class II receptor activity (GO:0032395) 7/10 0.002968506
Genes
1 HLA-DRA;HLA-DOA;HLA-DOB;HLA-DQA2;HLA-DQA1;HLA-DQB2;HLA-DRB1Enrichment analysis for cTWAS genes in top tissues separately
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
ctwas_genes_tissue <- df[[tissue]]$ctwas
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
print(db)
enrich_results <- GO_enrichment[[db]]
print(plotEnrich(GO_enrichment[[db]]))
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
}
}Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"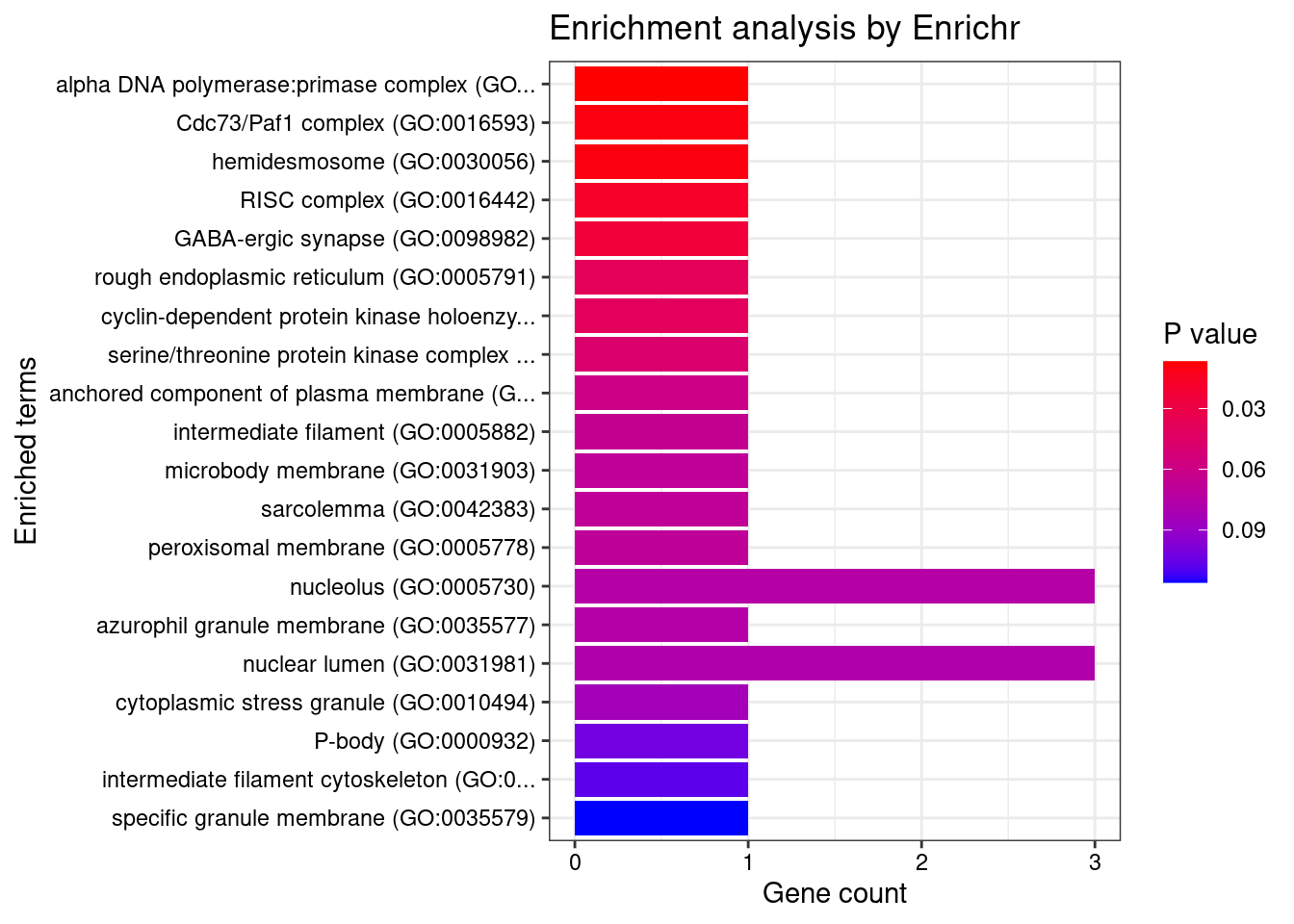
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"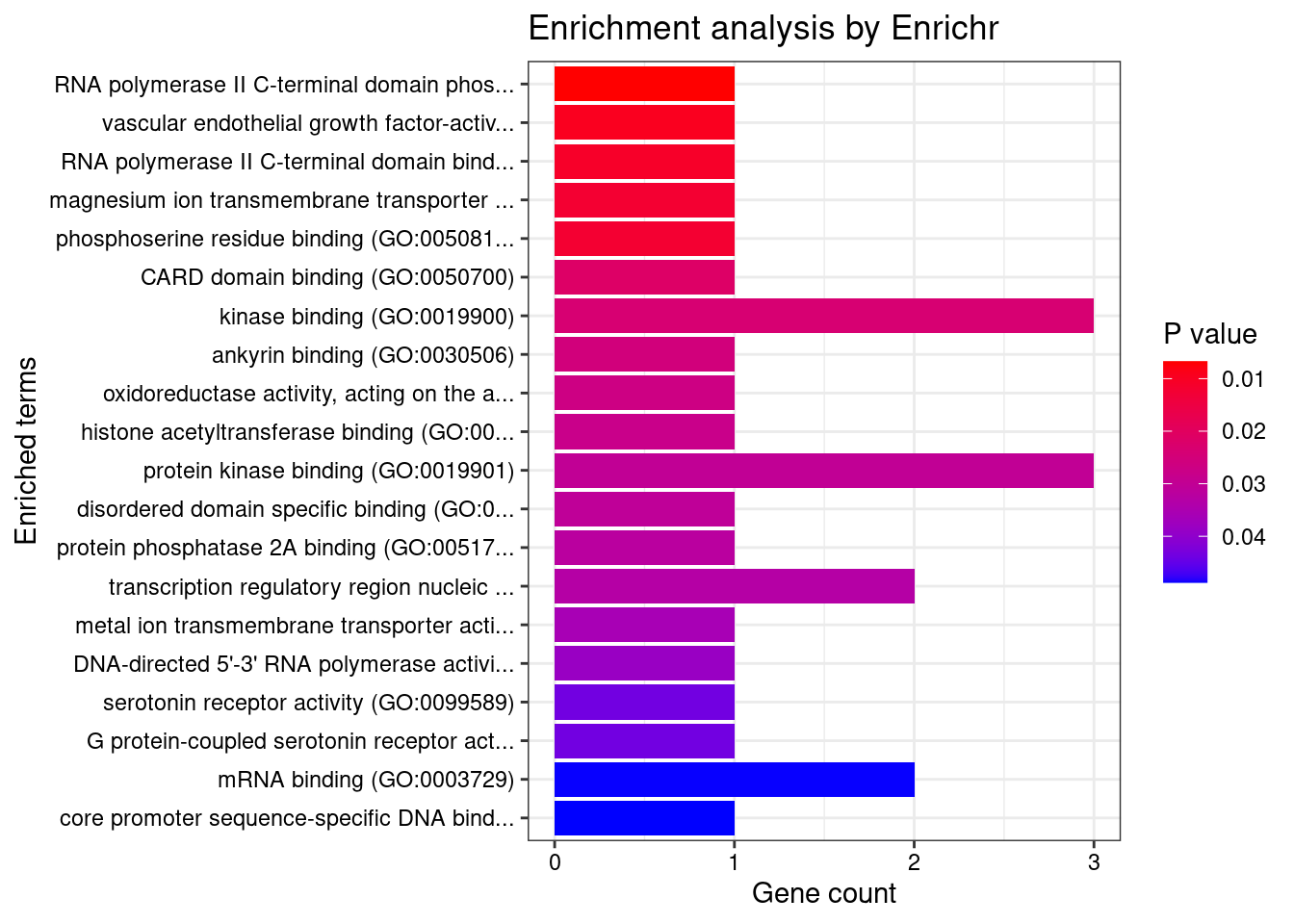
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"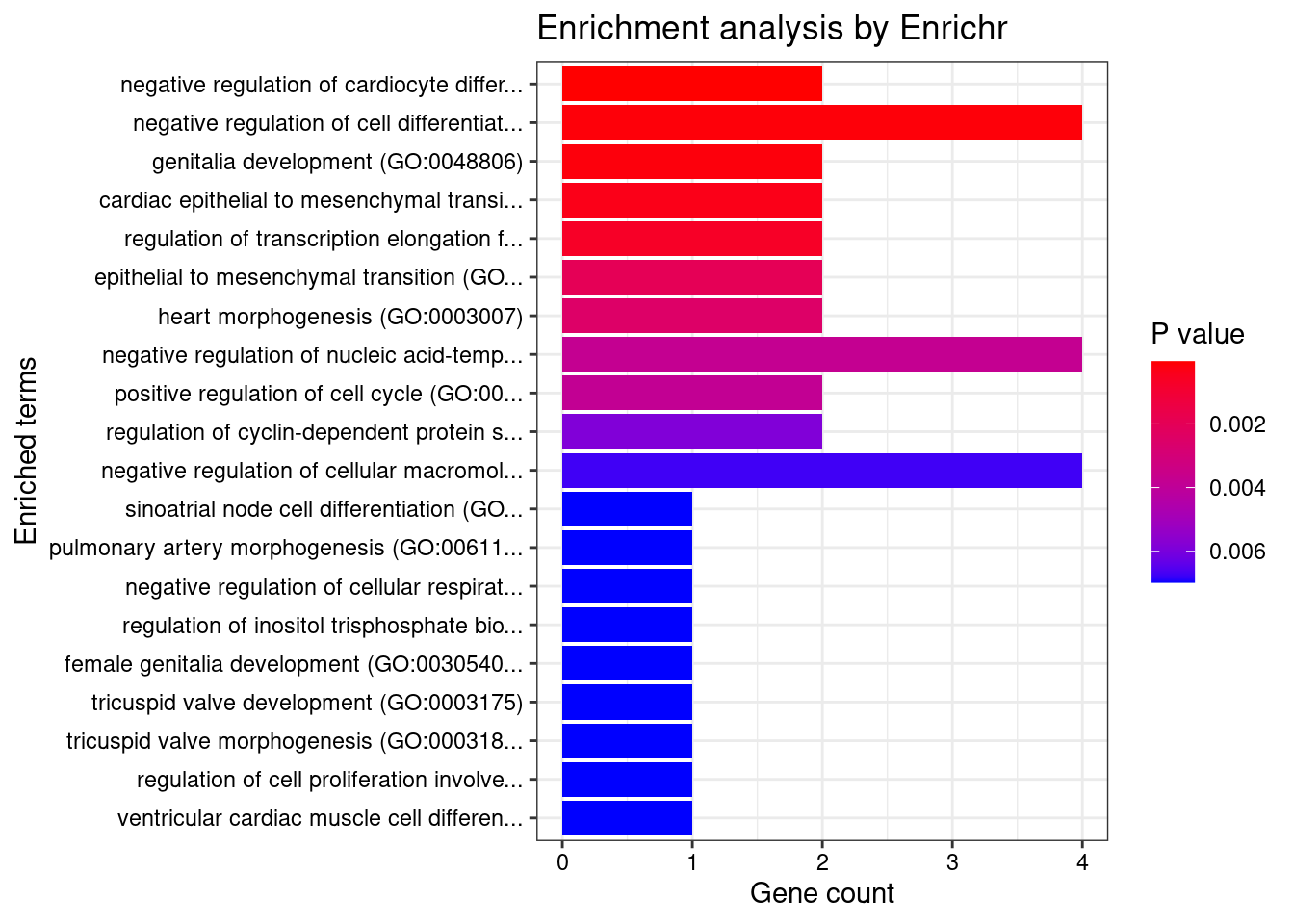
Term
1 negative regulation of cardiocyte differentiation (GO:1905208)
2 negative regulation of cell differentiation (GO:0045596)
3 genitalia development (GO:0048806)
4 cardiac epithelial to mesenchymal transition (GO:0060317)
5 regulation of transcription elongation from RNA polymerase II promoter (GO:0034243)
Overlap Adjusted.P.value Genes
1 2/7 0.01193454 HEY2;FRS2
2 4/191 0.01713474 ZADH2;LEO1;GDF5;TBX3
3 2/14 0.01713474 LHCGR;TBX3
4 2/20 0.02669293 HEY2;TBX3
5 2/27 0.03921114 HEXIM1;LEO1
[1] "GO_Cellular_Component_2021"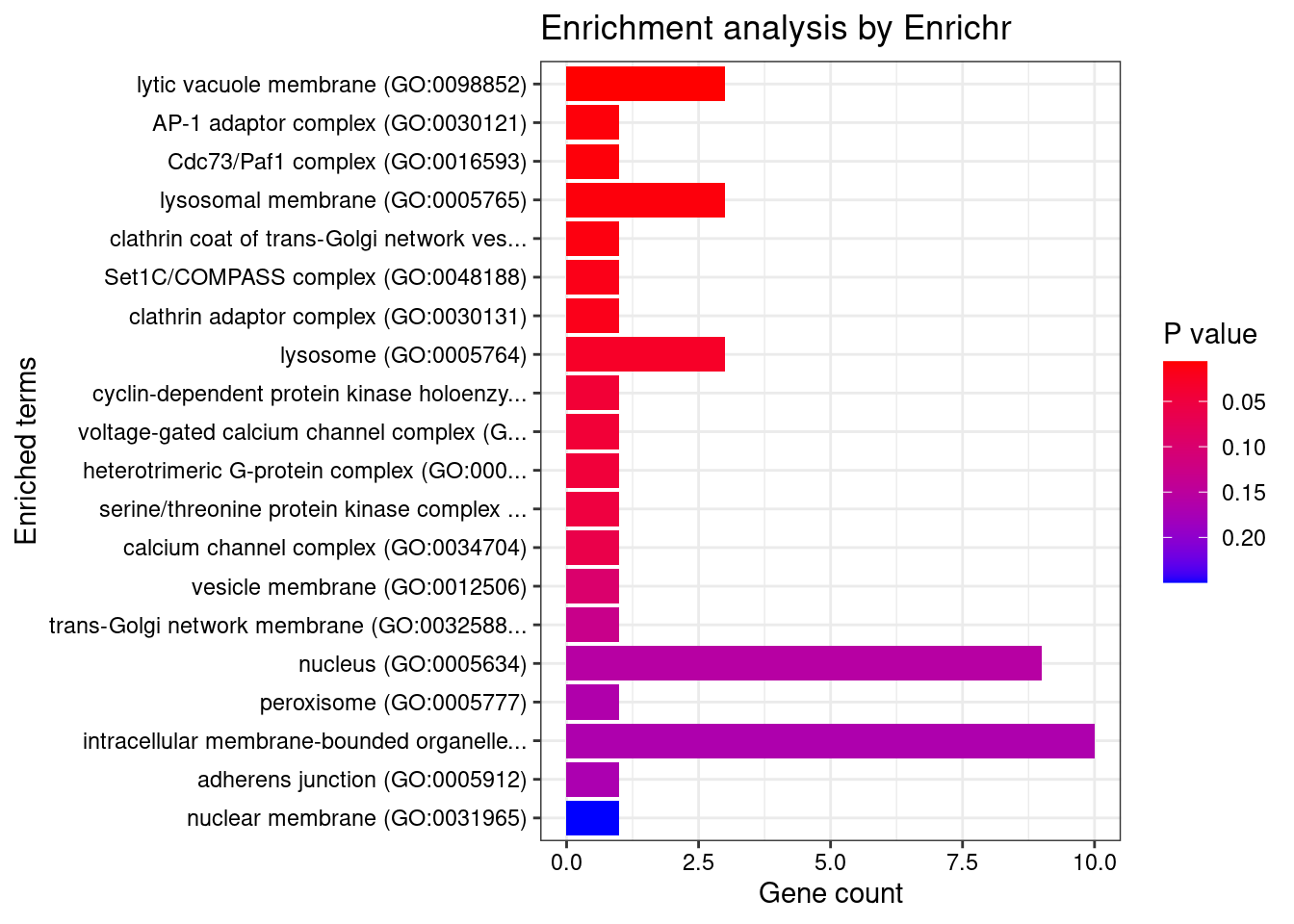
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"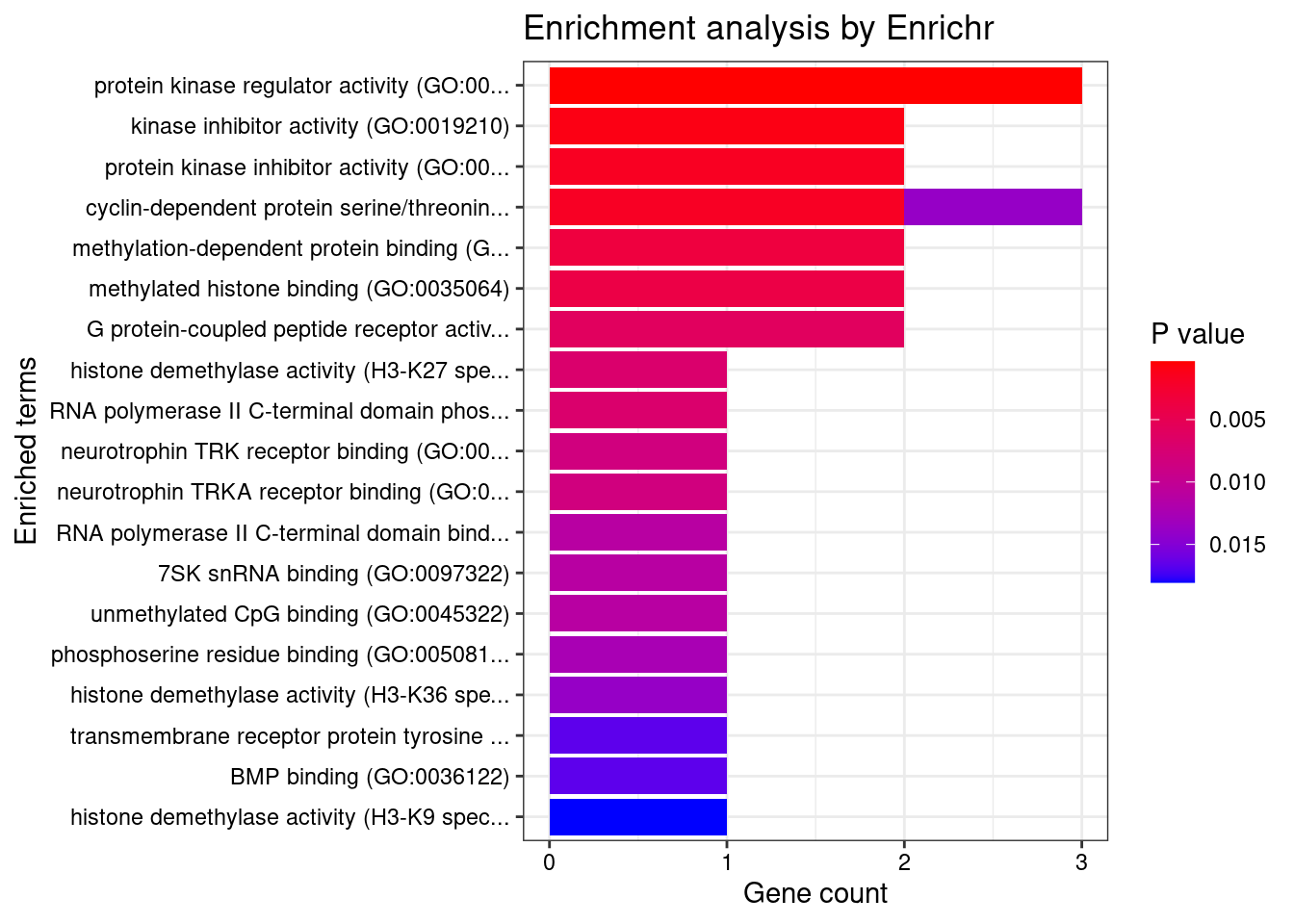
Term
1 protein kinase regulator activity (GO:0019887)
2 kinase inhibitor activity (GO:0019210)
3 protein kinase inhibitor activity (GO:0004860)
4 cyclin-dependent protein serine/threonine kinase regulator activity (GO:0016538)
5 methylation-dependent protein binding (GO:0140034)
6 methylated histone binding (GO:0035064)
7 G protein-coupled peptide receptor activity (GO:0008528)
8 histone demethylase activity (H3-K27 specific) (GO:0071558)
9 RNA polymerase II C-terminal domain phosphoserine binding (GO:1990269)
10 neurotrophin TRK receptor binding (GO:0005167)
11 neurotrophin TRKA receptor binding (GO:0005168)
12 RNA polymerase II C-terminal domain binding (GO:0099122)
13 7SK snRNA binding (GO:0097322)
14 unmethylated CpG binding (GO:0045322)
Overlap Adjusted.P.value Genes
1 3/98 0.02119850 HEXIM1;CCND2;SH3BP5L
2 2/33 0.02672416 HEXIM1;SH3BP5L
3 2/43 0.02672416 HEXIM1;SH3BP5L
4 2/44 0.02672416 HEXIM1;CCND2
5 2/63 0.04202174 CXXC1;KDM7A
6 2/68 0.04202174 CXXC1;KDM7A
7 2/82 0.04718541 LHCGR;CYSLTR2
8 1/5 0.04718541 KDM7A
9 1/5 0.04718541 LEO1
10 1/6 0.04718541 FRS2
11 1/6 0.04718541 FRS2
12 1/8 0.04936575 LEO1
13 1/8 0.04936575 HEXIM1
14 1/8 0.04936575 CXXC1
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"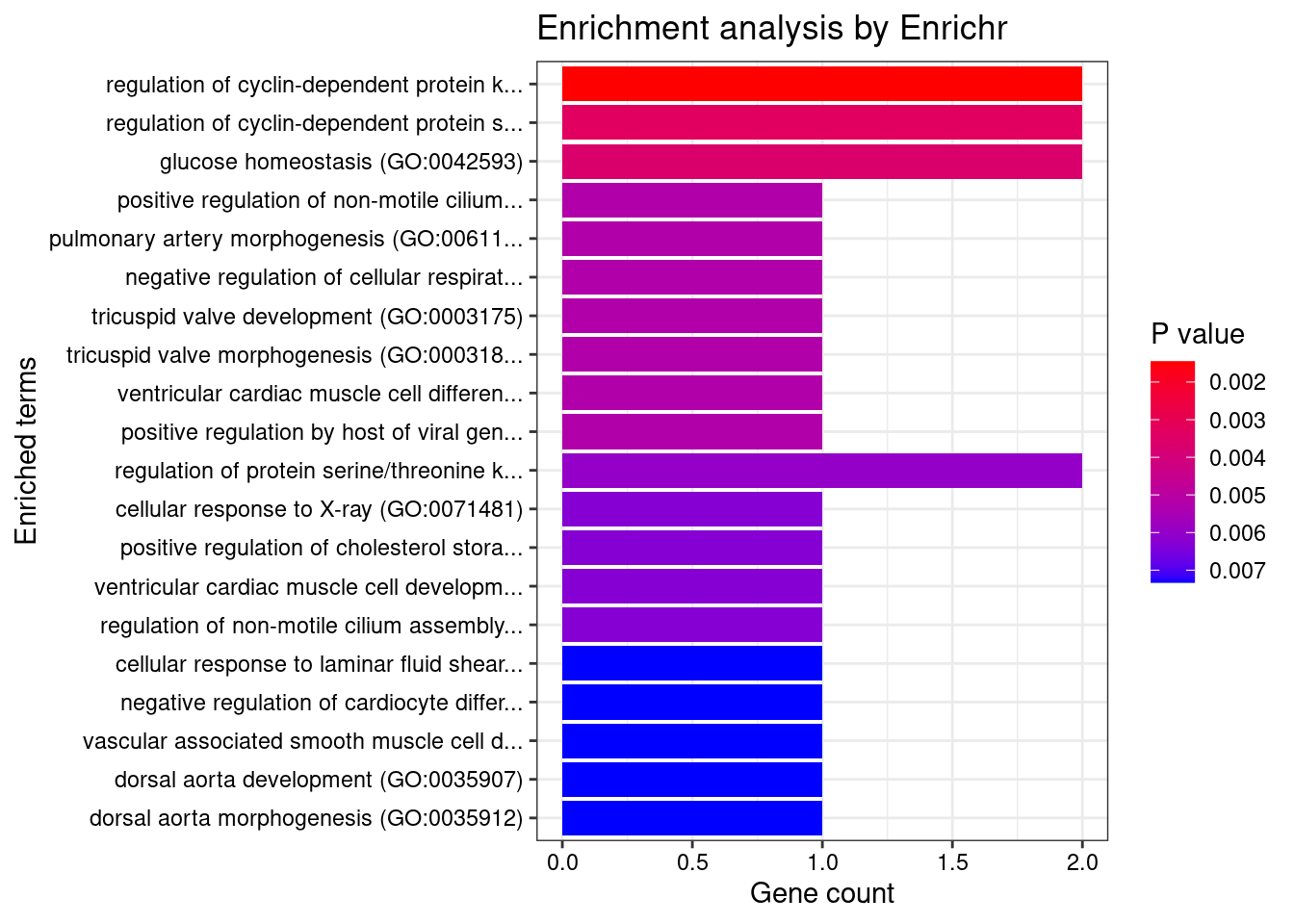
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"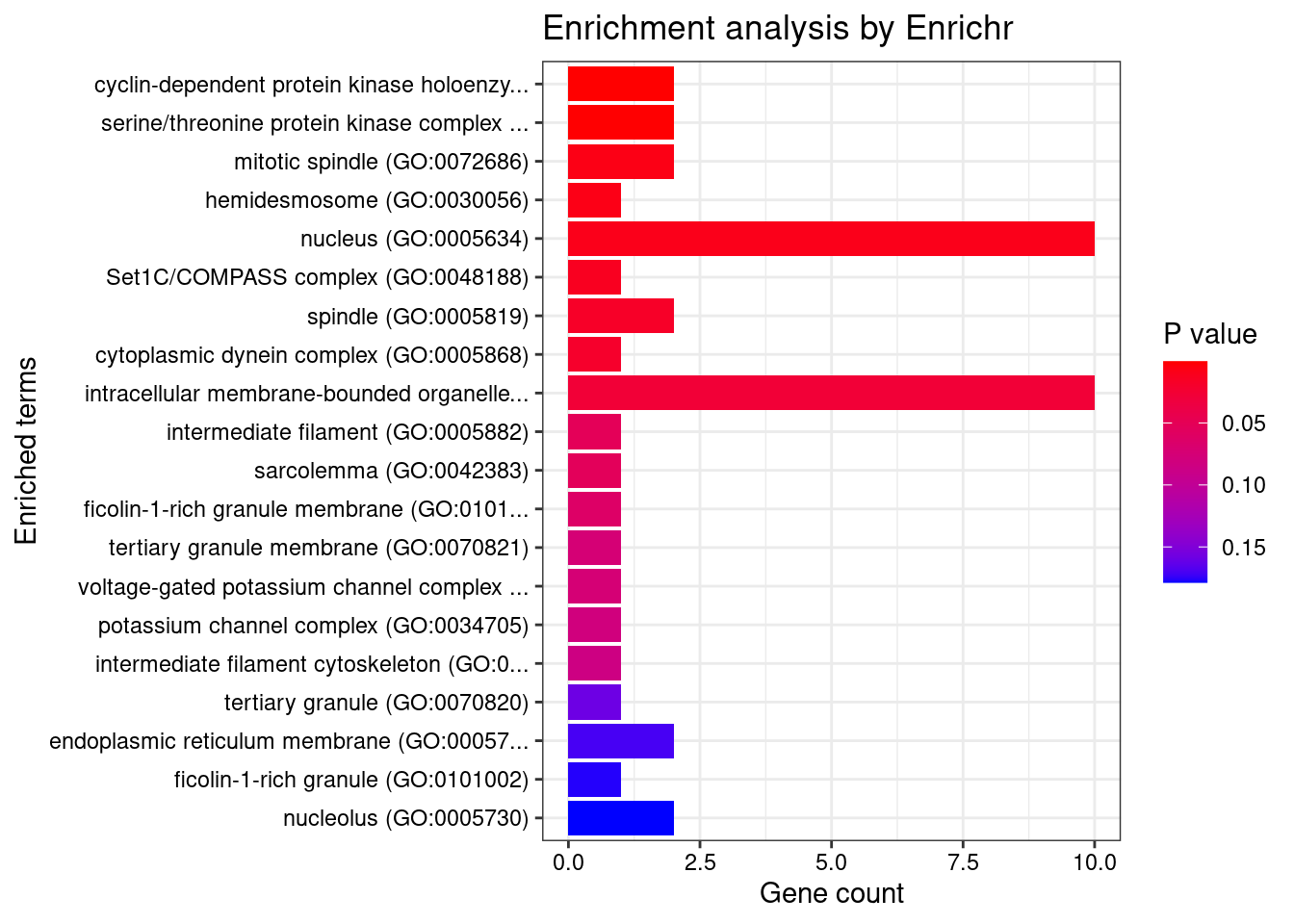
Term Overlap
1 cyclin-dependent protein kinase holoenzyme complex (GO:0000307) 2/30
2 serine/threonine protein kinase complex (GO:1902554) 2/37
Adjusted.P.value Genes
1 0.01196983 CCND2;CNPPD1
2 0.01196983 CCND2;CNPPD1
[1] "GO_Molecular_Function_2021"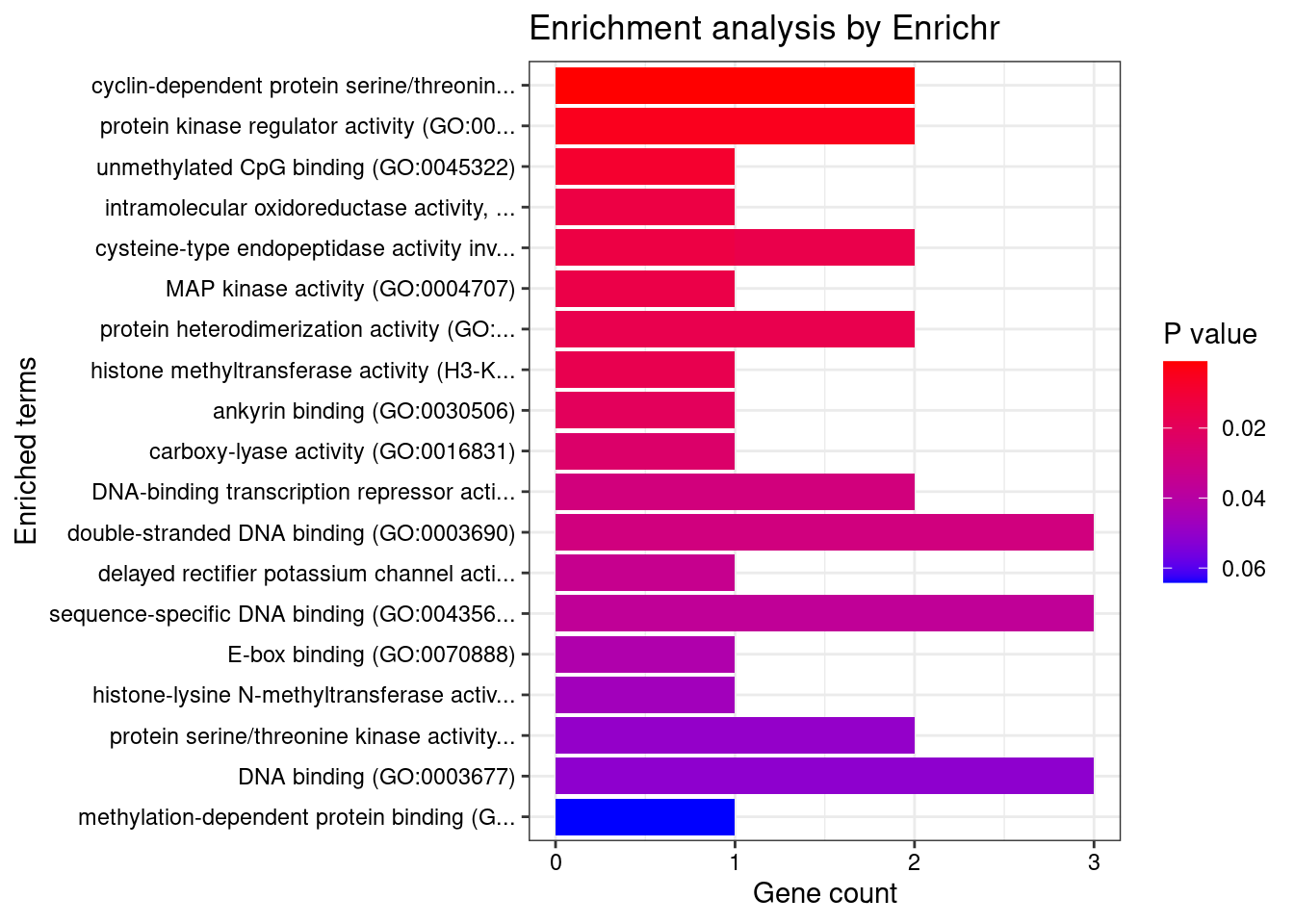
Term
1 cyclin-dependent protein serine/threonine kinase regulator activity (GO:0016538)
Overlap Adjusted.P.value Genes
1 2/44 0.03869054 CCND2;CNPPD1
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"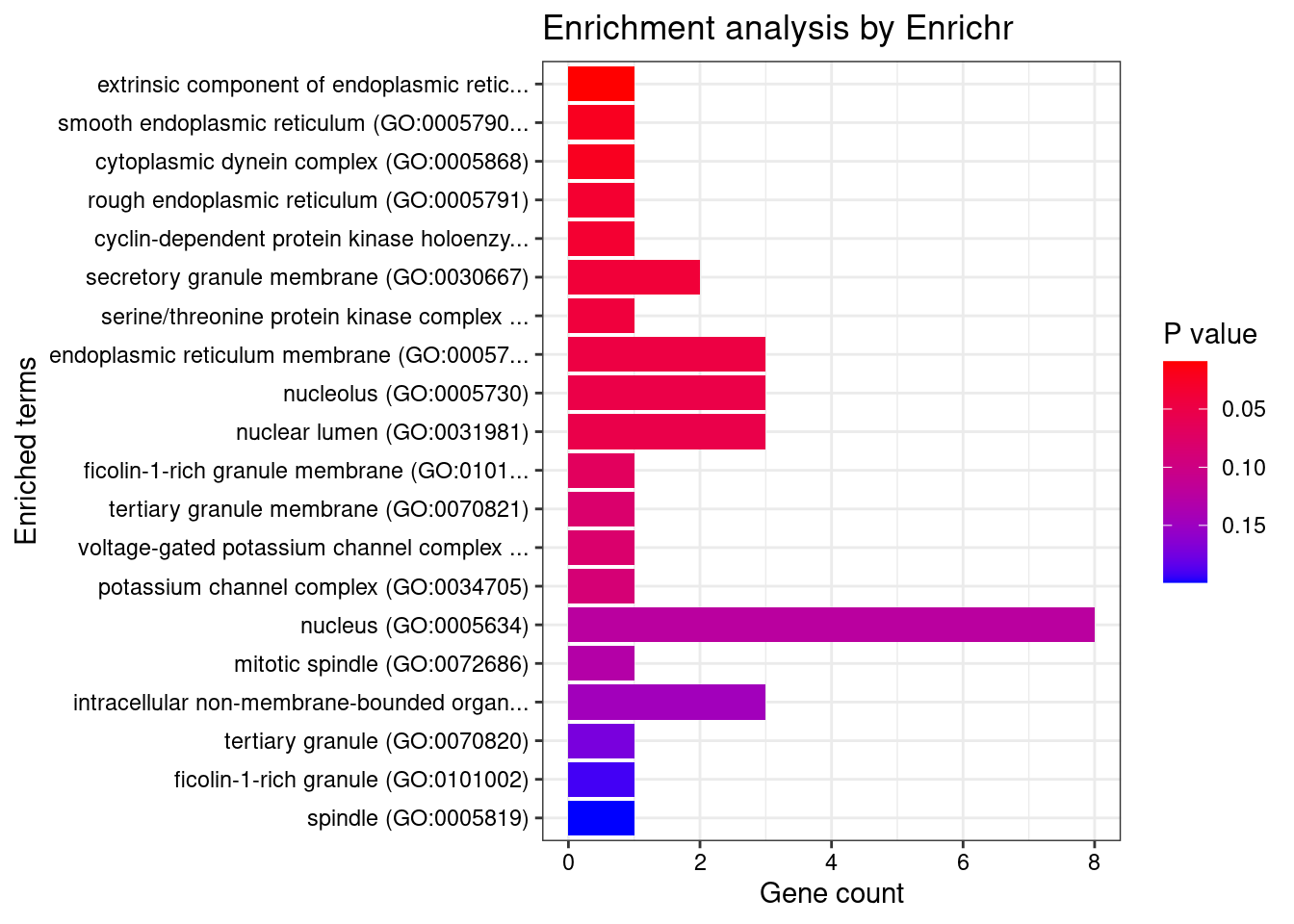
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"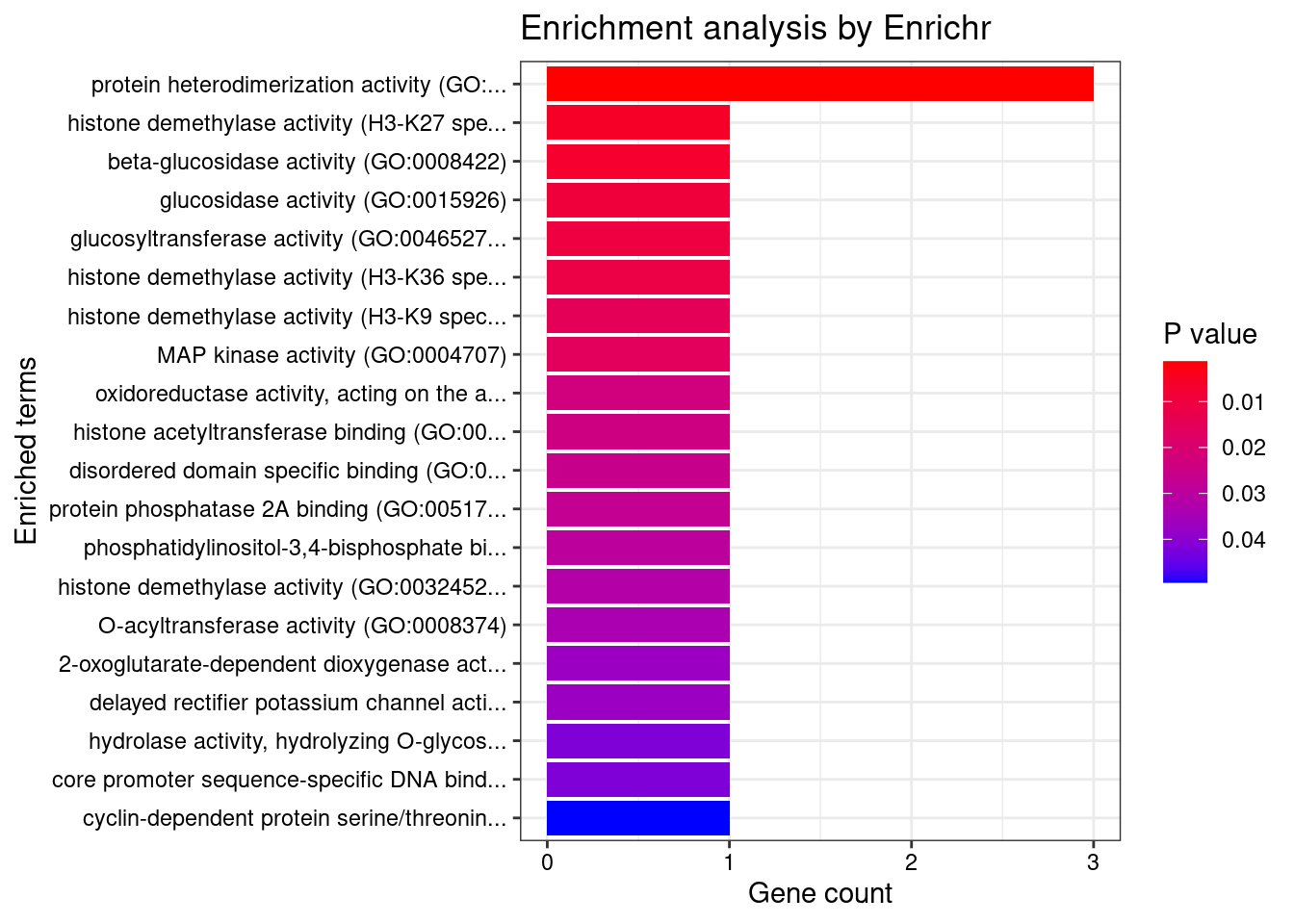
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"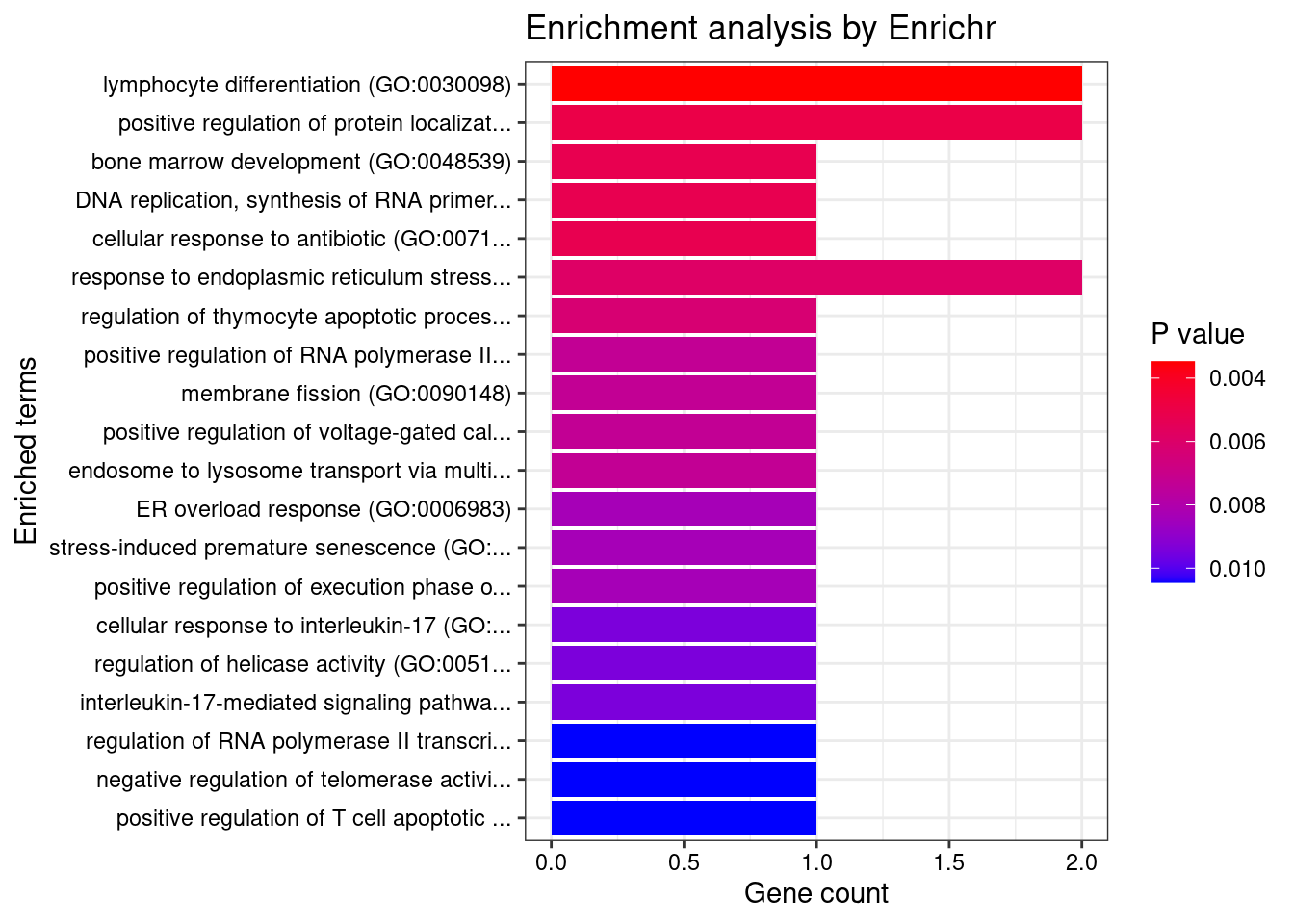
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"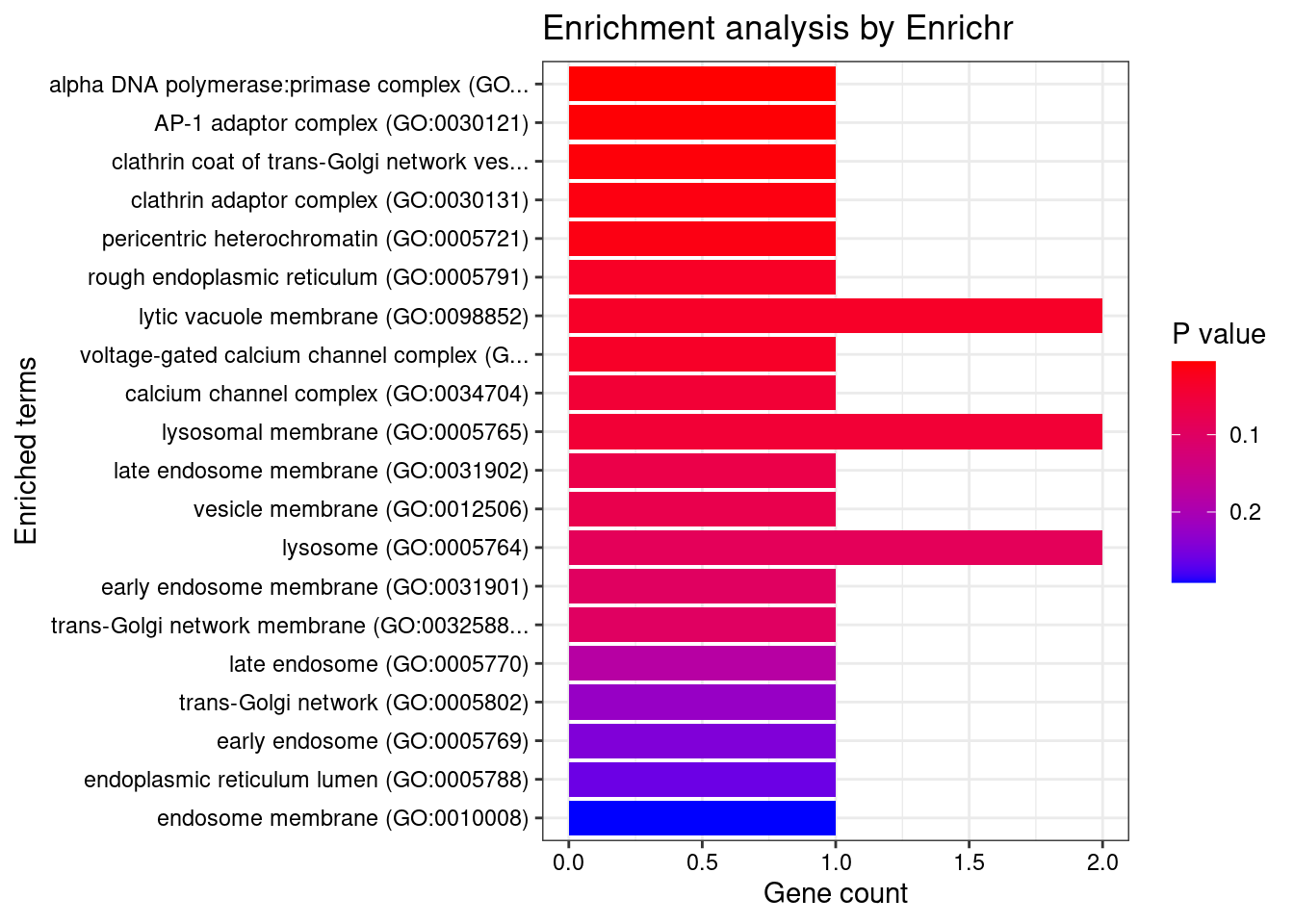
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] enrichR_3.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 compiler_3.6.1 pillar_1.6.1 later_0.8.0
[5] git2r_0.26.1 workflowr_1.6.2 tools_3.6.1 digest_0.6.20
[9] evaluate_0.14 lifecycle_1.0.0 tibble_3.1.2 gtable_0.3.0
[13] pkgconfig_2.0.3 rlang_0.4.11 DBI_1.1.1 curl_3.3
[17] yaml_2.2.0 xfun_0.8 httr_1.4.1 stringr_1.4.0
[21] dplyr_1.0.7 knitr_1.23 generics_0.0.2 fs_1.3.1
[25] vctrs_0.3.8 tidyselect_1.1.0 rprojroot_2.0.2 grid_3.6.1
[29] glue_1.4.2 R6_2.5.0 fansi_0.5.0 rmarkdown_1.13
[33] farver_2.1.0 purrr_0.3.4 ggplot2_3.3.3 magrittr_2.0.1
[37] whisker_0.3-2 scales_1.1.0 promises_1.0.1 htmltools_0.3.6
[41] ellipsis_0.3.2 colorspace_1.4-1 httpuv_1.5.1 labeling_0.3
[45] utf8_1.2.1 stringi_1.4.3 munsell_0.5.0 rjson_0.2.20
[49] crayon_1.4.1