Crohn’s disease - all weights
wesleycrouse
2022-04-04
Last updated: 2022-05-24
Checks: 7 0
Knit directory: ctwas_applied/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210726) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d46127d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Untracked files:
Untracked: G_list_ebi-a-GCST004131.RData
Untracked: group_enrichment_results.RData
Untracked: results_summary_inflammatory_bowel_disease.csv
Untracked: z_snp_pos_ebi-a-GCST004131.RData
Untracked: z_snp_pos_ebi-a-GCST004132.RData
Untracked: z_snp_pos_ebi-a-GCST004133.RData
Unstaged changes:
Modified: analysis/ebi-a-GCST004131_allweights.Rmd
Modified: code/automate_Rmd.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ebi-a-GCST004132_allweights.Rmd) and HTML (docs/ebi-a-GCST004132_allweights.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 6f82cb0 | wesleycrouse | 2022-05-13 | matching ons emsembl |
| Rmd | 5c9fc0e | wesleycrouse | 2022-05-13 | additional figures for ulcerative colitis |
| html | 5c9fc0e | wesleycrouse | 2022-05-13 | additional figures for ulcerative colitis |
| Rmd | 3eff970 | wesleycrouse | 2022-05-12 | additional figure |
| Rmd | f65b31c | wesleycrouse | 2022-05-05 | enrichment for known IBD genes and overlap |
| html | f65b31c | wesleycrouse | 2022-05-05 | enrichment for known IBD genes and overlap |
| html | 177114d | wesleycrouse | 2022-05-04 | updating output |
| Rmd | ad70daf | wesleycrouse | 2022-05-04 | fixing file name |
| Rmd | 3354d64 | wesleycrouse | 2022-05-04 | adding known IBD genes from ABC paper |
| html | e6f6eb0 | wesleycrouse | 2022-05-02 | cleanup |
| Rmd | fe4215e | wesleycrouse | 2022-05-02 | cleanup |
| html | ce5831c | wesleycrouse | 2022-05-02 | regenerate reports |
| Rmd | 041c7d2 | wesleycrouse | 2022-05-02 | fixing notebooks after merge |
| Rmd | c048c27 | wesleycrouse | 2022-05-02 | Merge branch ‘master’ of https://github.com/wesleycrouse/ctwas_applied |
| html | c048c27 | wesleycrouse | 2022-05-02 | Merge branch ‘master’ of https://github.com/wesleycrouse/ctwas_applied |
| Rmd | 0fd1bd6 | wesleycrouse | 2022-05-02 | genes nearby and nearest to peak |
| html | 0fd1bd6 | wesleycrouse | 2022-05-02 | genes nearby and nearest to peak |
| html | 46544ec | wesleycrouse | 2022-04-12 | more enrichment |
| html | f40da10 | wesleycrouse | 2022-04-12 | more enrichment |
| Rmd | 107bb6d | wesleycrouse | 2022-04-12 | gene set enrichment for supplied gene sets |
| Rmd | 8c71a43 | wesleycrouse | 2022-04-12 | gene set enrichment for supplied gene sets |
| html | 95e0f8e | wesleycrouse | 2022-04-07 | scroll bar |
| html | 4409757 | wesleycrouse | 2022-04-07 | scroll bar |
| Rmd | a4575d7 | wesleycrouse | 2022-04-07 | formating |
| Rmd | ea23be2 | wesleycrouse | 2022-04-07 | formating |
| Rmd | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| Rmd | cedc812 | wesleycrouse | 2022-04-06 | adding subsections |
| html | d772243 | wesleycrouse | 2022-04-06 | adding subsections |
| html | cedc812 | wesleycrouse | 2022-04-06 | adding subsections |
| html | f7e9822 | wesleycrouse | 2022-04-06 | testing subsections |
| html | 93c8916 | wesleycrouse | 2022-04-06 | testing subsections |
| Rmd | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| Rmd | 981e5bc | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | 60ea899 | wesleycrouse | 2022-04-05 | edge cases for kegg |
| html | 981e5bc | wesleycrouse | 2022-04-05 | edge cases for kegg |
| Rmd | d0f6e53 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| Rmd | 7d883c2 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| html | d0f6e53 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| html | 7d883c2 | wesleycrouse | 2022-04-05 | adding crohn’s disease |
| Rmd | d14af05 | wesleycrouse | 2022-04-04 | kegg results for other traits |
| Rmd | 8222701 | wesleycrouse | 2022-04-04 | kegg results for other traits |
options(width=1000)trait_id <- "ebi-a-GCST004132"
trait_name <- "Crohn's disease"
source("/project2/mstephens/wcrouse/UKB_analysis_allweights/ctwas_config.R")
trait_dir <- paste0("/project2/mstephens/wcrouse/UKB_analysis_allweights/", trait_id)
results_dirs <- list.dirs(trait_dir, recursive=F)Load cTWAS results for all weights
# df <- list()
#
# for (i in 1:length(results_dirs)){
# print(i)
#
# results_dir <- results_dirs[i]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# #load ctwas results
# ctwas_res <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.susieIrss.txt"))
#
# #make unique identifier for regions and effects
# ctwas_res$region_tag <- paste(ctwas_res$region_tag1, ctwas_res$region_tag2, sep="_")
# ctwas_res$region_cs_tag <- paste(ctwas_res$region_tag, ctwas_res$cs_index, sep="_")
#
# #load z scores for SNPs and collect sample size
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# sample_size <- z_snp$ss
# sample_size <- as.numeric(names(which.max(table(sample_size))))
#
# #separate gene and SNP results
# ctwas_gene_res <- ctwas_res[ctwas_res$type == "gene", ]
# ctwas_gene_res <- data.frame(ctwas_gene_res)
# ctwas_snp_res <- ctwas_res[ctwas_res$type == "SNP", ]
# ctwas_snp_res <- data.frame(ctwas_snp_res)
#
# #add gene information to results
# sqlite <- RSQLite::dbDriver("SQLite")
# db = RSQLite::dbConnect(sqlite, paste0("/project2/compbio/predictdb/mashr_models/mashr_", weight, ".db"))
# query <- function(...) RSQLite::dbGetQuery(db, ...)
# gene_info <- query("select gene, genename, gene_type from extra")
# RSQLite::dbDisconnect(db)
#
# ctwas_gene_res <- cbind(ctwas_gene_res, gene_info[sapply(ctwas_gene_res$id, match, gene_info$gene), c("genename", "gene_type")])
#
# #add z scores to results
# load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
# ctwas_gene_res$z <- z_gene[ctwas_gene_res$id,]$z
#
# z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
# ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#
# #merge gene and snp results with added information
# ctwas_snp_res$genename=NA
# ctwas_snp_res$gene_type=NA
#
# ctwas_res <- rbind(ctwas_gene_res,
# ctwas_snp_res[,colnames(ctwas_gene_res)])
#
# #get number of SNPs from s1 results; adjust for thin argument
# ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
# n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
# rm(ctwas_res_s1)
#
# #load estimated parameters
# load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
#
# #estimated group prior
# estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
# names(estimated_group_prior) <- c("gene", "snp")
# estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
#
# #estimated group prior variance
# estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
# names(estimated_group_prior_var) <- c("gene", "snp")
#
# #report group size
# group_size <- c(nrow(ctwas_gene_res), n_snps)
#
# #estimated group PVE
# estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size
# names(estimated_group_pve) <- c("gene", "snp")
#
# #ctwas genes using PIP>0.8
# ctwas_genes_index <- ctwas_gene_res$susie_pip>0.8
# ctwas_genes <- ctwas_gene_res$genename[ctwas_genes_index]
#
# #twas genes using bonferroni threshold
# alpha <- 0.05
# sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#
# twas_genes_index <- abs(ctwas_gene_res$z) > sig_thresh
# twas_genes <- ctwas_gene_res$genename[twas_genes_index]
#
# #gene PIPs and z scores
# gene_pips <- ctwas_gene_res[,c("genename", "region_tag", "susie_pip", "z", "region_cs_tag")]
#
# #total PIPs by region
# regions <- unique(ctwas_gene_res$region_tag)
# region_pips <- data.frame(region=regions, stringsAsFactors=F)
# region_pips$gene_pip <- sapply(regions, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_tag==x])})
# region_pips$snp_pip <- sapply(regions, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_tag==x])})
# region_pips$snp_maxz <- sapply(regions, function(x){max(abs(ctwas_snp_res$z[ctwas_snp_res$region_tag==x]))})
#
# #total PIPs by causal set
# regions_cs <- unique(ctwas_gene_res$region_cs_tag)
# region_cs_pips <- data.frame(region_cs=regions_cs, stringsAsFactors=F)
# region_cs_pips$gene_pip <- sapply(regions_cs, function(x){sum(ctwas_gene_res$susie_pip[ctwas_gene_res$region_cs_tag==x])})
# region_cs_pips$snp_pip <- sapply(regions_cs, function(x){sum(ctwas_snp_res$susie_pip[ctwas_snp_res$region_cs_tag==x])})
#
# df[[weight]] <- list(prior=estimated_group_prior,
# prior_var=estimated_group_prior_var,
# pve=estimated_group_pve,
# ctwas=ctwas_genes,
# twas=twas_genes,
# gene_pips=gene_pips,
# region_pips=region_pips,
# sig_thresh=sig_thresh,
# region_cs_pips=region_cs_pips)
# }
#
# save(df, file=paste(trait_dir, "results_df.RData", sep="/"))
load(paste(trait_dir, "results_df.RData", sep="/"))
output <- data.frame(weight=names(df),
prior_g=unlist(lapply(df, function(x){x$prior["gene"]})),
prior_s=unlist(lapply(df, function(x){x$prior["snp"]})),
prior_var_g=unlist(lapply(df, function(x){x$prior_var["gene"]})),
prior_var_s=unlist(lapply(df, function(x){x$prior_var["snp"]})),
pve_g=unlist(lapply(df, function(x){x$pve["gene"]})),
pve_s=unlist(lapply(df, function(x){x$pve["snp"]})),
n_ctwas=unlist(lapply(df, function(x){length(x$ctwas)})),
n_twas=unlist(lapply(df, function(x){length(x$twas)})),
row.names=NULL,
stringsAsFactors=F)Plot estimated prior parameters and PVE
#plot estimated group prior
output <- output[order(-output$prior_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_g, type="l", ylim=c(0, max(output$prior_g, output$prior_s)*1.1),
xlab="", ylab="Estimated Group Prior", xaxt = "n", col="blue")
lines(output$prior_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)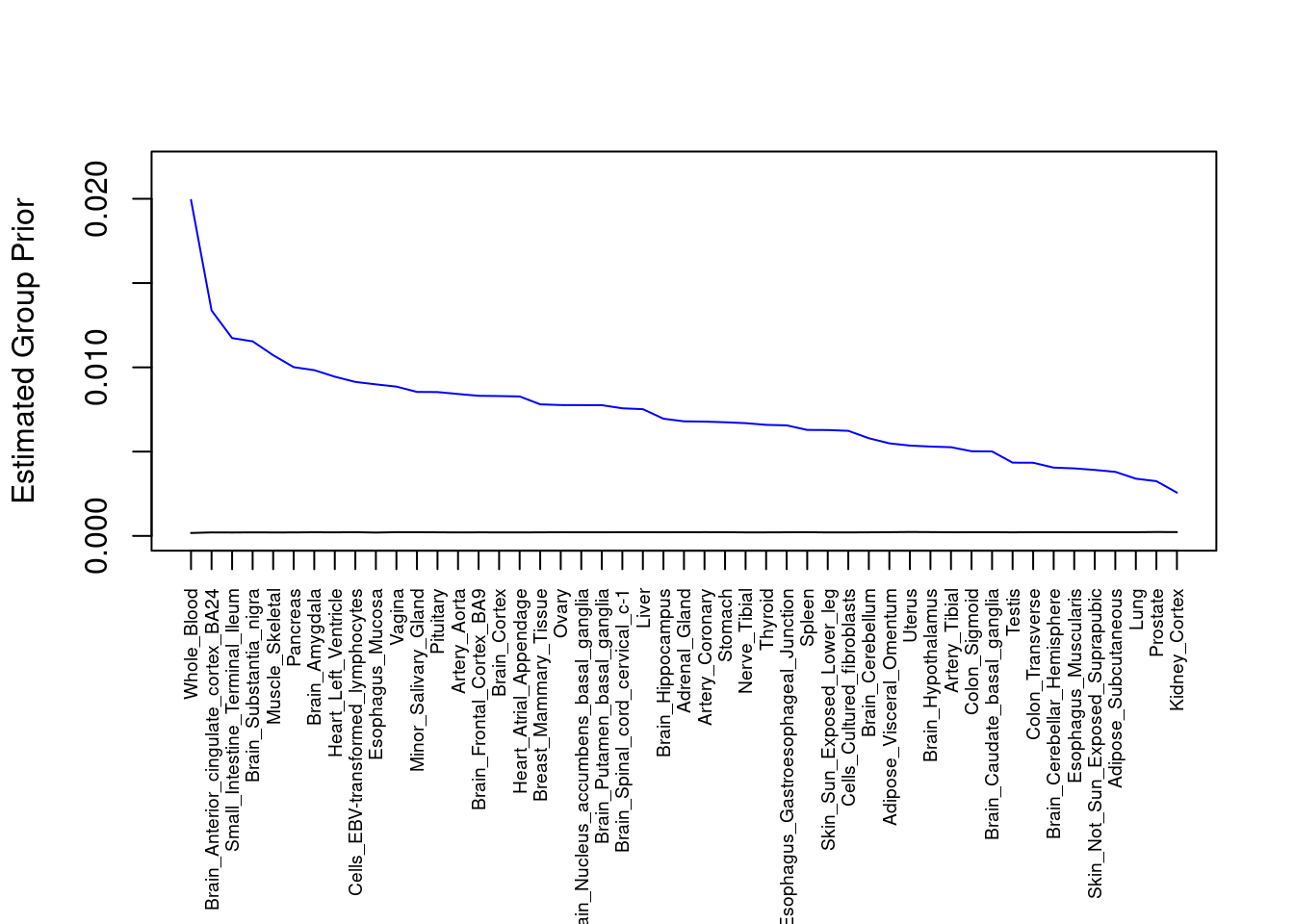
####################
#plot estimated group prior variance
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$prior_var_g, type="l", ylim=c(0, max(output$prior_var_g, output$prior_var_s)*1.1),
xlab="", ylab="Estimated Group Prior Variance", xaxt = "n", col="blue")
lines(output$prior_var_s)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)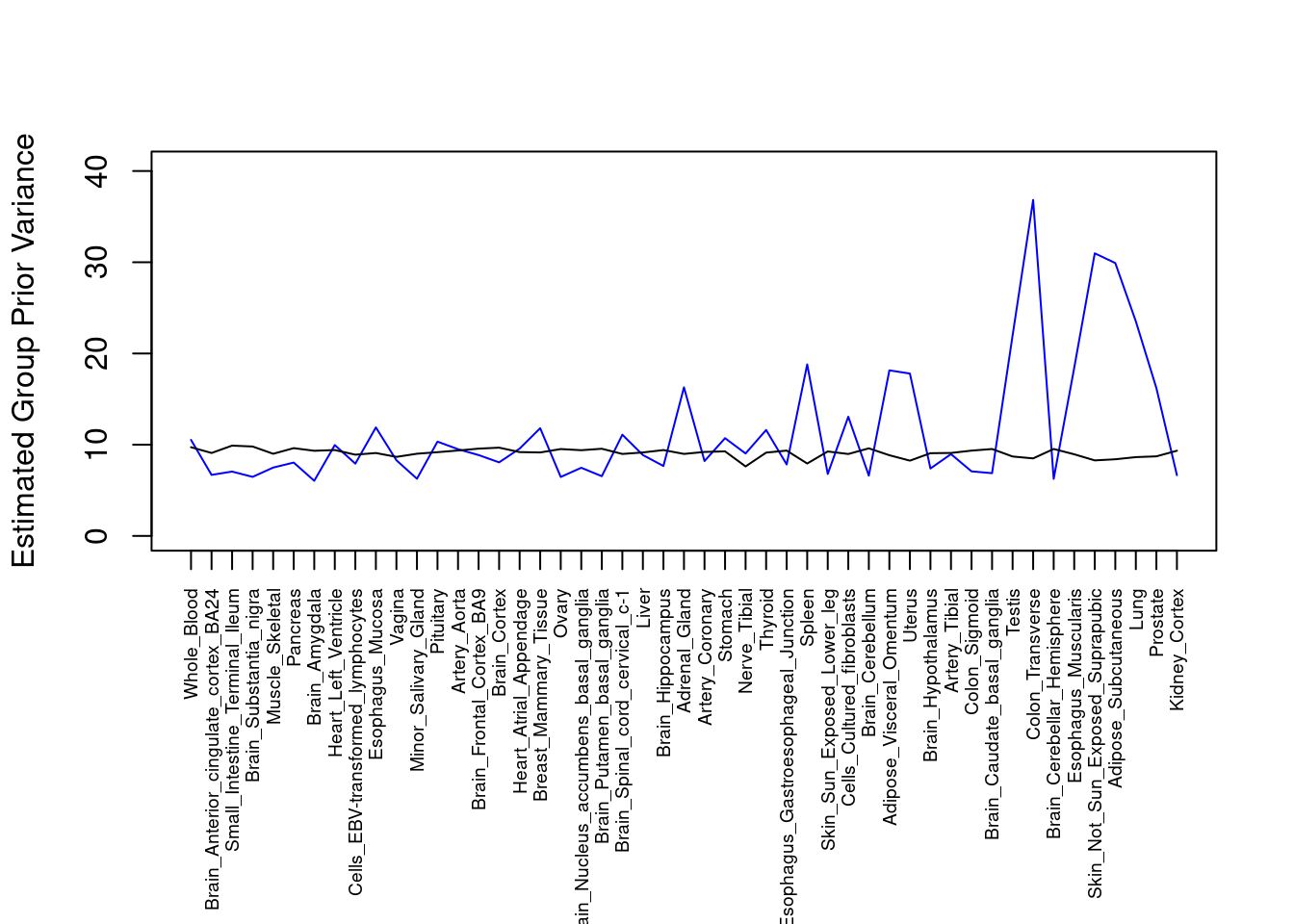
####################
#plot PVE
output <- output[order(-output$pve_g),]
par(mar=c(10.1, 4.1, 4.1, 2.1))
plot(output$pve_g, type="l", ylim=c(0, max(output$pve_g+output$pve_s)*1.1),
xlab="", ylab="Estimated PVE", xaxt = "n", col="blue")
lines(output$pve_s)
lines(output$pve_g+output$pve_s, lty=2)
axis(1, at = 1:nrow(output),
labels = output$weight,
las=2,
cex.axis=0.6)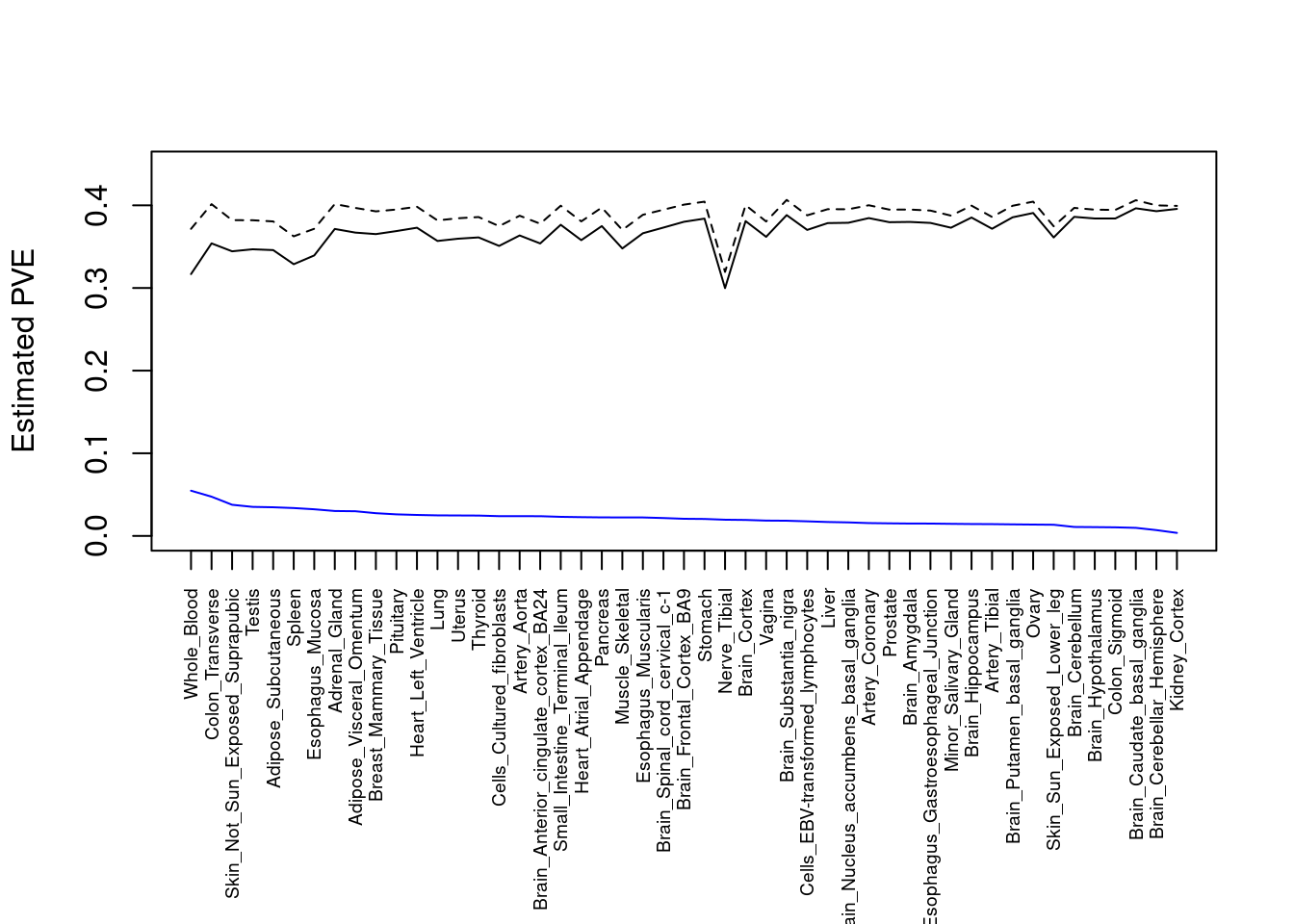
Number of cTWAS and TWAS genes
cTWAS genes are the set of genes with PIP>0.8 in any tissue. TWAS genes are the set of genes with significant z score (Bonferroni within tissue) in any tissue.
#plot number of significant cTWAS and TWAS genes in each tissue
plot(output$n_ctwas, output$n_twas, xlab="Number of cTWAS Genes", ylab="Number of TWAS Genes")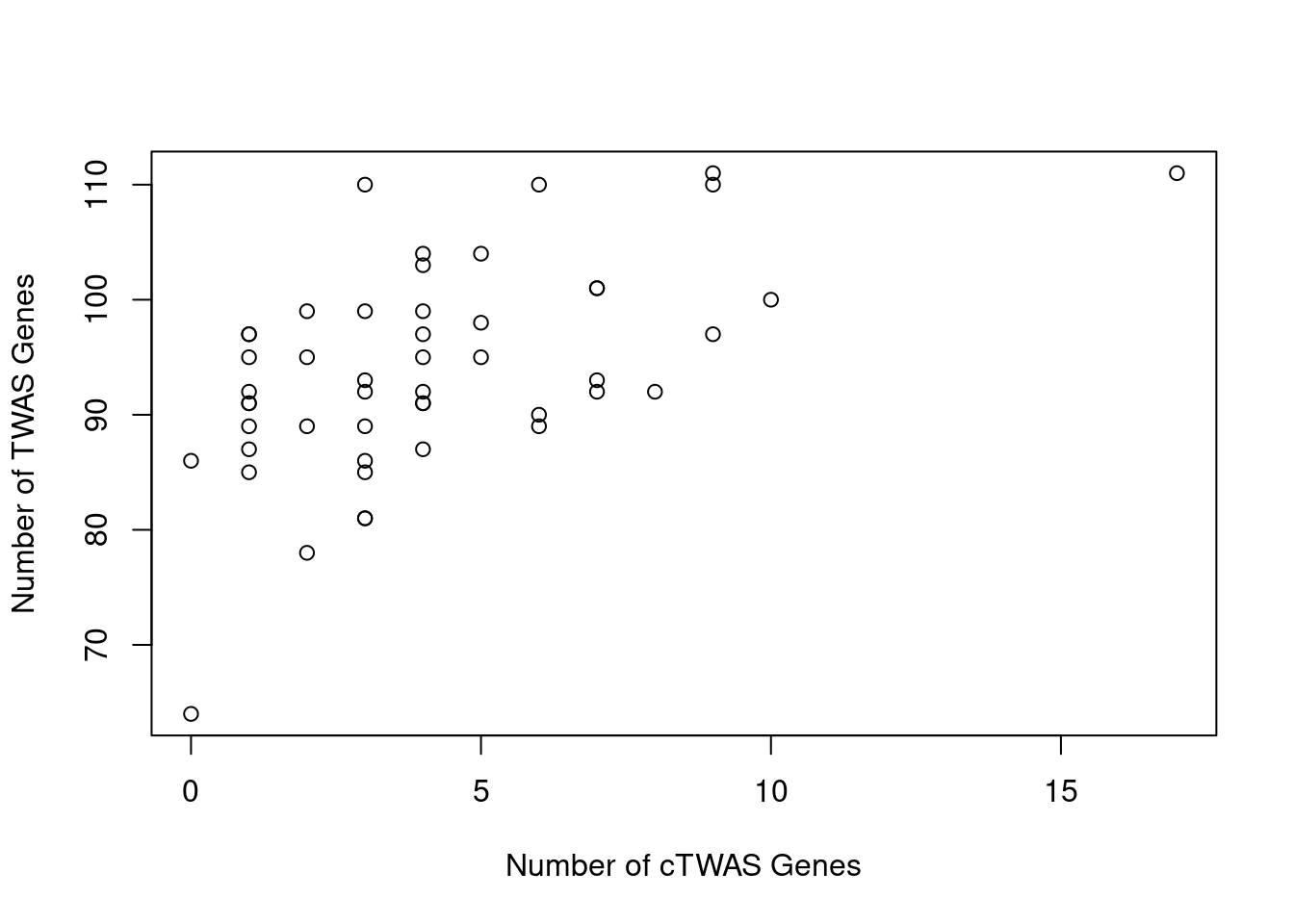
#number of ctwas_genes
ctwas_genes <- unique(unlist(lapply(df, function(x){x$ctwas})))
length(ctwas_genes)[1] 83#number of twas_genes
twas_genes <- unique(unlist(lapply(df, function(x){x$twas})))
length(twas_genes)[1] 465Enrichment analysis for cTWAS genes
GO
#enrichment for cTWAS genes using enrichR
library(enrichR)Welcome to enrichR
Checking connection ... Enrichr ... Connection is Live!
FlyEnrichr ... Connection is available!
WormEnrichr ... Connection is available!
YeastEnrichr ... Connection is available!
FishEnrichr ... Connection is available!dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(ctwas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 cytokine-mediated signaling pathway (GO:0019221) 12/621 0.007775261 CD40;CIITA;IL18RAP;BCL2L11;SOCS1;TNFSF15;IFNGR2;TNFSF4;LTBR;MMP9;HLA-DQB1;IP6K2
2 regulation of response to interferon-gamma (GO:0060330) 3/14 0.010096697 SOCS1;IFNGR2;PTPN2
3 transmembrane receptor protein tyrosine kinase signaling pathway (GO:0007169) 9/404 0.012482425 GIGYF1;CNKSR1;RGS14;NCF4;TSC2;ITGAV;MMP9;PTPN2;CDK5R1
4 positive regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043123) 6/171 0.016297707 CD40;PPP5C;NDFIP1;CARD9;RBCK1;LTBR
5 regulation of interferon-gamma-mediated signaling pathway (GO:0060334) 3/23 0.019126959 SOCS1;IFNGR2;PTPN2
6 negative regulation of insulin receptor signaling pathway (GO:0046627) 3/27 0.024896799 SOCS1;TSC2;PTPN2
7 negative regulation of cellular response to insulin stimulus (GO:1900077) 3/28 0.024896799 SOCS1;TSC2;PTPN2
8 regulation of cytokine-mediated signaling pathway (GO:0001959) 4/74 0.026454646 SOCS1;IFNGR2;RBCK1;PTPN2
9 regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043122) 6/224 0.031404353 CD40;PPP5C;NDFIP1;CARD9;RBCK1;LTBR
10 response to cytokine (GO:0034097) 5/150 0.033500698 CD40;CIITA;SMAD3;SMPD1;PTPN2
11 positive regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043372) 2/8 0.035455981 SOCS1;TNFSF4
12 positive regulation of production of molecular mediator of immune response (GO:0002702) 3/38 0.036296810 TNFSF4;CD244;VAMP3
13 negative regulation of lipid localization (GO:1905953) 2/9 0.038469088 ITGAV;PTPN2
14 negative regulation of tyrosine phosphorylation of STAT protein (GO:0042532) 2/10 0.041562612 SOCS1;PTPN2
15 regulation of receptor binding (GO:1900120) 2/10 0.041562612 ADAM15;MMP9
16 regulation of insulin receptor signaling pathway (GO:0046626) 3/45 0.044701743 SOCS1;TSC2;PTPN2
17 insulin-like growth factor receptor signaling pathway (GO:0048009) 2/11 0.044701743 GIGYF1;TSC2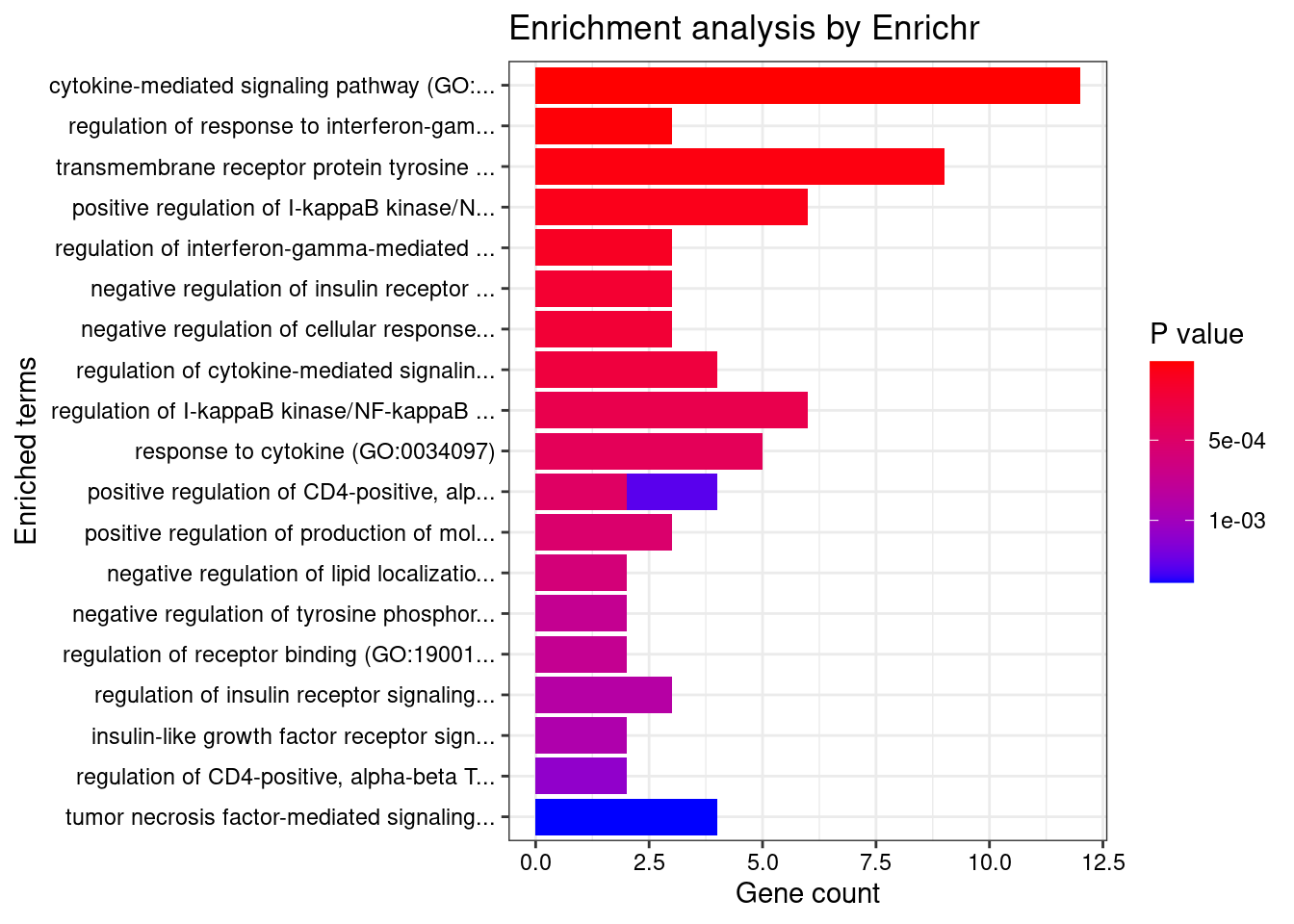
GO_Cellular_Component_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
GO_Molecular_Function_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
KEGG
#enrichment for cTWAS genes using KEGG
library(WebGestaltR)******************************************* ** Welcome to WebGestaltR ! ** *******************************************background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#listGeneSet()
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes, referenceGene=background,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!enrichResult[,c("description", "size", "overlap", "FDR", "userId")]NULLDisGeNET
#enrichment for cTWAS genes using DisGeNET
# devtools::install_bitbucket("ibi_group/disgenet2r")
library(disgenet2r)
disgenet_api_key <- get_disgenet_api_key(
email = "wesleycrouse@gmail.com",
password = "uchicago1" )
Sys.setenv(DISGENET_API_KEY= disgenet_api_key)
res_enrich <- disease_enrichment(entities=ctwas_genes, vocabulary = "HGNC", database = "CURATED")PPP5C gene(s) from the input list not found in DisGeNET CURATEDLINC01623 gene(s) from the input list not found in DisGeNET CURATEDADCY9 gene(s) from the input list not found in DisGeNET CURATEDOSER1 gene(s) from the input list not found in DisGeNET CURATEDLINC02128 gene(s) from the input list not found in DisGeNET CURATEDSDCCAG3 gene(s) from the input list not found in DisGeNET CURATEDC3orf62 gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDSTXBP3 gene(s) from the input list not found in DisGeNET CURATEDSH3D21 gene(s) from the input list not found in DisGeNET CURATEDAGAP5 gene(s) from the input list not found in DisGeNET CURATEDTMEM229B gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDAPBB1 gene(s) from the input list not found in DisGeNET CURATEDFAM171B gene(s) from the input list not found in DisGeNET CURATEDC11orf58 gene(s) from the input list not found in DisGeNET CURATEDADAM15 gene(s) from the input list not found in DisGeNET CURATEDAF064858.11 gene(s) from the input list not found in DisGeNET CURATEDRP11-386E5.1 gene(s) from the input list not found in DisGeNET CURATEDAPEH gene(s) from the input list not found in DisGeNET CURATEDRGS14 gene(s) from the input list not found in DisGeNET CURATEDSPNS2 gene(s) from the input list not found in DisGeNET CURATEDPLEKHH2 gene(s) from the input list not found in DisGeNET CURATEDFGFR1OP gene(s) from the input list not found in DisGeNET CURATEDNDFIP1 gene(s) from the input list not found in DisGeNET CURATEDMUC3A gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDRP11-574K11.29 gene(s) from the input list not found in DisGeNET CURATEDVAMP3 gene(s) from the input list not found in DisGeNET CURATEDGIGYF1 gene(s) from the input list not found in DisGeNET CURATEDAC007383.3 gene(s) from the input list not found in DisGeNET CURATEDSPIRE2 gene(s) from the input list not found in DisGeNET CURATEDC1orf106 gene(s) from the input list not found in DisGeNET CURATEDDEF8 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDif (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
} Description FDR Ratio BgRatio
171 Crohn's disease of large bowel 6.685211e-06 6/48 44/9703
197 Crohn's disease of the ileum 6.685211e-06 6/48 44/9703
267 Regional enteritis 6.685211e-06 6/48 44/9703
314 IIeocolitis 6.685211e-06 6/48 44/9703
41 Crohn Disease 1.177333e-05 6/48 50/9703
35 Ulcerative Colitis 3.996803e-05 6/48 63/9703
73 Inflammatory Bowel Diseases 1.446890e-03 4/48 35/9703
9 Rheumatoid Arthritis 1.030784e-02 6/48 174/9703
19 Brain Diseases 1.103892e-02 3/48 25/9703
159 Encephalopathies 1.254727e-02 3/48 27/9703
80 Fibroid Tumor 1.323532e-02 2/48 6/9703
6 Aortic Aneurysm 1.451288e-02 2/48 7/9703
156 Uterine Fibroids 1.451288e-02 2/48 7/9703
366 Juvenile pauciarticular chronic arthritis 1.451288e-02 2/48 7/9703Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes=ctwas_genes)
#background is union of genes analyzed in all tissue
background <- unique(unlist(lapply(df, function(x){x$gene_pips$genename})))
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background]})
####################
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
hyp_score_padj gene_set nset ngenes percent padj
1 gwascatalog 5971 43 0.007201474 1.241162e-05
2 mgi_essential 2305 14 0.006073753 1.364914e-01
3 clinvar_path_likelypath 2771 12 0.004330567 4.894235e-01
4 fda_approved_drug_targets 352 2 0.005681818 4.894235e-01
5 core_essentials_hart 265 0 0.000000000 1.000000e+00Enrichment analysis for TWAS genes
#enrichment for TWAS genes
dbs <- c("GO_Biological_Process_2021", "GO_Cellular_Component_2021", "GO_Molecular_Function_2021")
GO_enrichment <- enrichr(twas_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 cytokine-mediated signaling pathway (GO:0019221) 56/621 7.135165e-15 CIITA;CD40;TNFRSF6B;IL23R;IL27;RPS6KA4;IL18RAP;PSMD3;IL12B;TNFSF11;MAP3K8;JAK2;IL6R;RIPK2;IFNGR2;IL13;HLA-B;HLA-C;FOS;HLA-A;TYK2;HLA-G;MMP9;PSMA6;IRF1;CSNK2B;LTA;IRF6;HLA-DQB1;CCL13;CCL11;CUL1;NOD2;IL1RL1;MUC1;BCL2L11;SOCS1;CAMK2G;HLA-DQA1;IL12RB2;IP6K2;STAT5A;STAT5B;TNFSF15;STAT3;LIF;PSMB8;PSMB9;IL4;POMC;TNFSF4;IL2RA;TNFSF8;LTBR;TRIM31;IL18R1
2 cellular response to cytokine stimulus (GO:0071345) 37/482 2.807673e-07 CCL13;CD40;CCL11;IL23R;GBA;AIF1;ZFP36L2;ZFP36L1;SBNO2;MUC1;BCL2L11;SOCS1;IL12B;TNFSF11;JAK2;IL6R;IL12RB2;STAT5A;STAT5B;SMAD3;IFNGR2;STAT3;IL13;LIF;FOS;TYK2;MMP9;IRGM;RHOA;IL4;POMC;IL2RA;IRF1;ARHGEF2;SLC26A6;PTPN2;IL18R1
3 cellular response to interferon-gamma (GO:0071346) 17/121 2.565480e-06 CCL13;CIITA;CCL11;IFNGR2;HLA-B;HLA-C;HLA-A;HLA-G;AIF1;IRF1;IRF6;JAK2;TRIM31;CAMK2G;HLA-DQA1;SLC26A6;HLA-DQB1
4 interferon-gamma-mediated signaling pathway (GO:0060333) 13/68 2.956734e-06 CIITA;IFNGR2;HLA-B;HLA-C;HLA-A;HLA-G;IRF1;IRF6;JAK2;TRIM31;CAMK2G;HLA-DQA1;HLA-DQB1
5 positive regulation of cytokine production (GO:0001819) 28/335 2.956734e-06 PTGER4;CD40;IL23R;IL27;PARK7;NOD2;AIF1;AGER;RASGRP1;FLOT1;POLR2E;IL12B;IL6R;IL12RB2;RIPK2;IL13;CARD9;STAT3;HLA-A;HLA-G;CCDC88B;IL4;LACC1;TNFSF4;IRF1;ARHGEF2;IL18R1;CD244
6 antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590) 13/78 1.155899e-05 NCF4;HLA-B;TAP2;HLA-C;TAP1;LNPEP;HLA-A;HLA-G;PSMB8;PSMB9;PSMA6;PSMD3;ITGAV
7 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (GO:0002479) 12/73 3.989181e-05 PSMA6;NCF4;PSMD3;HLA-B;TAP2;HLA-C;TAP1;ITGAV;HLA-A;HLA-G;PSMB8;PSMB9
8 cellular response to tumor necrosis factor (GO:0071356) 19/194 4.625282e-05 CCL13;CD40;TNFRSF6B;CCL11;TNFSF15;GBA;PSMB8;ZFP36L2;PSMB9;ZFP36L1;PSMA6;TNFSF4;PSMD3;LTA;TNFSF11;TNFSF8;ARHGEF2;LTBR;JAK2
9 antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent (GO:0002480) 5/8 9.896016e-05 HLA-B;HLA-C;LNPEP;HLA-A;HLA-G
10 antigen processing and presentation of peptide antigen via MHC class I (GO:0002474) 8/33 1.697050e-04 ERAP2;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G;SEC24C
11 interleukin-23-mediated signaling pathway (GO:0038155) 5/9 1.786914e-04 IL23R;STAT3;IL12B;TYK2;JAK2
12 regulation of inflammatory response (GO:0050727) 18/206 3.504661e-04 PTGER4;IL13;GBA;PARK7;NOD2;MMP9;IL4;CYLD;LACC1;PSMA6;SBNO2;FNDC4;TNFSF4;IL12B;JAK2;GPSM3;PTPN2;BRD4
13 tumor necrosis factor-mediated signaling pathway (GO:0033209) 13/116 5.807390e-04 CD40;TNFRSF6B;TNFSF15;PSMB8;PSMB9;PSMA6;TNFSF4;PSMD3;LTA;TNFSF11;TNFSF8;LTBR;JAK2
14 regulation of lymphocyte proliferation (GO:0050670) 6/19 5.807390e-04 LST1;IL12B;IL27;TNFSF8;ZNF335;IKZF3
15 regulation of tyrosine phosphorylation of STAT protein (GO:0042509) 10/68 6.159570e-04 IL4;CD40;SOCS1;IL23R;STAT3;LIF;IL12B;JAK2;IL6R;PTPN2
16 positive regulation of interferon-gamma production (GO:0032729) 9/57 9.725232e-04 IL23R;TNFSF4;IL12B;IL27;HLA-A;RASGRP1;CD244;IL18R1;IL12RB2
17 positive regulation of T cell activation (GO:0050870) 10/75 1.090759e-03 CCDC88B;IL4;HLA-DMB;IL23R;TNFSF4;IL12B;TNFSF11;HLA-A;NOD2;AIF1
18 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway (GO:0002484) 4/7 1.090759e-03 HLA-B;HLA-C;HLA-A;HLA-G
19 antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent (GO:0002486) 4/7 1.090759e-03 HLA-B;HLA-C;HLA-A;HLA-G
20 nucleotide-binding oligomerization domain containing 2 signaling pathway (GO:0070431) 4/7 1.090759e-03 LACC1;RIPK2;NOD2;IRGM
21 regulation of T cell proliferation (GO:0042129) 10/76 1.090759e-03 CCDC88B;IL4;HLA-DMB;IL23R;TNFSF4;IL12B;IL27;TNFSF8;HLA-G;AIF1
22 positive regulation of interleukin-12 production (GO:0032735) 7/34 1.090759e-03 CD40;RIPK2;IL23R;TNFSF4;IL12B;AGER;HLA-G
23 regulation of interferon-gamma-mediated signaling pathway (GO:0060334) 6/23 1.090759e-03 SOCS1;IFNGR2;CDC37;JAK2;IRGM;PTPN2
24 antigen processing and presentation of endogenous peptide antigen (GO:0002483) 5/14 1.090759e-03 ERAP2;TAP2;TAP1;HLA-A;HLA-G
25 regulation of response to interferon-gamma (GO:0060330) 5/14 1.090759e-03 SOCS1;IFNGR2;CDC37;JAK2;PTPN2
26 immune response-regulating cell surface receptor signaling pathway (GO:0002768) 5/14 1.090759e-03 BAG6;CD40;NCR3;HLA-G;MICB
27 regulation of interleukin-10 production (GO:0032653) 8/48 1.258434e-03 IL4;IL23R;TNFSF4;IL13;STAT3;IL12B;NOD2;AGER
28 regulation of MAP kinase activity (GO:0043405) 11/97 1.420677e-03 CD40;LRRK2;SMPD1;GBA;ERBB2;TNFSF11;MST1R;NOD2;TRIB1;RASGRP1;LIME1
29 cellular response to type I interferon (GO:0071357) 9/65 1.532567e-03 IRF1;HLA-B;HLA-C;HLA-A;TYK2;IRF6;HLA-G;PSMB8;IP6K2
30 type I interferon signaling pathway (GO:0060337) 9/65 1.532567e-03 IRF1;HLA-B;HLA-C;HLA-A;TYK2;IRF6;HLA-G;PSMB8;IP6K2
31 antigen processing and presentation of endogenous peptide antigen via MHC class I (GO:0019885) 4/8 1.532567e-03 ERAP2;TAP2;TAP1;HLA-A
32 regulation of immune response (GO:0050776) 15/179 1.541258e-03 DENND1B;CD40;ITGA4;HLA-B;HLA-C;ICAM5;HLA-A;HLA-G;ADCY7;IL4;NCR3;TNFSF4;IRF1;MICA;MICB
33 T cell receptor signaling pathway (GO:0050852) 14/158 1.541258e-03 DENND1B;MOG;RIPK2;CUL1;HLA-A;BTNL2;PSMB8;PSMB9;PSMA6;PSMD3;ICOSLG;HLA-DQA1;LAT;HLA-DQB1
34 positive regulation of T cell proliferation (GO:0042102) 9/66 1.541258e-03 CCDC88B;IL4;HLA-DMB;IL23R;TNFSF4;IL12B;AGER;AIF1;ICOSLG
35 regulation of interleukin-12 production (GO:0032655) 8/51 1.541258e-03 CD40;RIPK2;IL23R;TNFSF4;HLA-B;IL12B;HLA-G;AGER
36 regulation of defense response (GO:0031347) 10/83 1.573890e-03 PSMA6;CYLD;LACC1;SBNO2;IRF1;TNFSF4;PARK7;NOD2;JAK2;BRD4
37 antigen receptor-mediated signaling pathway (GO:0050851) 15/185 2.016590e-03 DENND1B;MOG;RIPK2;CUL1;HLA-A;BTNL2;LIME1;PSMB8;PSMB9;PSMA6;PSMD3;ICOSLG;HLA-DQA1;LAT;HLA-DQB1
38 regulation of B cell activation (GO:0050864) 6/28 2.487345e-03 IL4;TNFSF4;NOD2;IKZF3;ZFP36L2;ZFP36L1
39 regulation of T cell mediated cytotoxicity (GO:0001914) 6/29 2.996196e-03 IL23R;HLA-B;IL12B;HLA-A;AGER;HLA-G
40 regulation of cytokine production (GO:0001817) 13/150 3.162270e-03 PTGER4;PPP1R11;MOG;CARD9;HLA-B;BTNL2;RPS6KA4;CCDC88B;SYT11;TNFSF4;FLOT1;IL12B;ICOSLG
41 regulation of interleukin-6 production (GO:0032675) 11/110 3.173543e-03 SYT11;TNFSF4;GBA;CARD9;STAT3;HLA-B;ARHGEF2;NOD2;IL6R;AGER;AIF1
42 positive regulation of leukocyte mediated cytotoxicity (GO:0001912) 7/43 3.307537e-03 NCR3;IL23R;HLA-B;IL12B;HLA-A;RASGRP1;HLA-G
43 positive regulation of lymphocyte proliferation (GO:0050671) 9/75 3.491779e-03 CCDC88B;IL4;CD40;HLA-DMB;IL23R;TNFSF4;IL12B;ZNF335;AIF1
44 positive regulation of tyrosine phosphorylation of STAT protein (GO:0042531) 8/59 3.629478e-03 IL4;CD40;IL23R;STAT3;LIF;IL12B;JAK2;IL6R
45 positive regulation of DNA-binding transcription factor activity (GO:0051091) 17/246 3.828986e-03 CD40;SMAD3;RIPK2;CARD9;STAT3;ARID5B;PARK7;NOD2;AGER;RPS6KA4;PSMA6;IL18RAP;FLOT1;TNFSF11;ARHGEF2;TRIM31;IL18R1
46 positive regulation of NF-kappaB transcription factor activity (GO:0051092) 13/155 3.860275e-03 CD40;RIPK2;CARD9;STAT3;NOD2;AGER;RPS6KA4;PSMA6;IL18RAP;FLOT1;TNFSF11;ARHGEF2;IL18R1
47 positive regulation of interleukin-10 production (GO:0032733) 6/32 4.413663e-03 IL4;TNFSF4;IL13;STAT3;IL12B;NOD2
48 inositol phosphate biosynthetic process (GO:0032958) 4/11 4.413663e-03 ITPKC;IPMK;IP6K1;IP6K2
49 MAPK cascade (GO:0000165) 19/303 4.936416e-03 PTGER4;LRRK2;CUL1;RASGRP1;PSMB8;ZFP36L2;PSMB9;ZFP36L1;PSMA6;PPP5C;IL2RA;PSMD3;RASA2;ERBB2;MAP3K8;ITGAV;JAK2;MARK3;LAT
50 positive regulation of T cell cytokine production (GO:0002726) 5/21 5.039053e-03 IL4;DENND1B;TNFSF4;HLA-A;IL18R1
51 inflammatory response (GO:0006954) 16/230 5.082930e-03 PTGER4;CCL13;CD40;CIITA;CCL11;RIPK2;STAT3;FOS;AIF1;IL4;PRDX5;NCR3;TNFSF4;IL2RA;REL;LAT
52 positive regulation of lymphocyte activation (GO:0051251) 6/35 6.823807e-03 CCDC88B;TNFSF4;IL12B;TNFSF11;NOD2;ZNF335
53 regulation of tumor necrosis factor production (GO:0032680) 11/124 7.282952e-03 IL4;POMC;SYT11;NFKBIL1;STAT3;IL12B;ARHGEF2;NOD2;JAK2;AGER;RASGRP1
54 macrophage activation (GO:0042116) 6/36 7.591349e-03 IL4;SBNO2;IL13;JAK2;AGER;AIF1
55 positive regulation of T cell mediated immunity (GO:0002711) 6/36 7.591349e-03 IL23R;TNFSF4;HLA-B;IL12B;HLA-A;HLA-G
56 regulation of interferon-gamma production (GO:0032649) 9/86 7.838982e-03 IL23R;TNFSF4;IL12B;IL27;HLA-A;RASGRP1;CD244;IL18R1;IL12RB2
57 positive regulation of protein serine/threonine kinase activity (GO:0071902) 10/106 8.099408e-03 CD40;CCNY;LRRK2;ERBB2;TNFSF11;MST1R;NOD2;RASGRP1;IRGM;RHOA
58 response to cytokine (GO:0034097) 12/150 9.013725e-03 CD40;CIITA;SMAD3;IL23R;SMPD1;STAT3;REL;JAK2;IL6R;RHOA;IL18R1;PTPN2
59 regulation of response to external stimulus (GO:0032101) 11/130 9.498991e-03 CYLD;LACC1;PSMA6;SBNO2;TNFSF4;IRF1;SAG;PARK7;NOD2;JAK2;BRD4
60 glycolipid transport (GO:0046836) 3/6 9.498991e-03 CLN3;RFT1;PLTP
61 protection from natural killer cell mediated cytotoxicity (GO:0042270) 3/6 9.498991e-03 HLA-B;HLA-A;HLA-G
62 growth hormone receptor signaling pathway via JAK-STAT (GO:0060397) 4/14 9.498991e-03 STAT5A;STAT5B;STAT3;JAK2
63 positive regulation of granulocyte macrophage colony-stimulating factor production (GO:0032725) 4/14 9.498991e-03 IL23R;CARD9;IL12B;RASGRP1
64 positive regulation of type 2 immune response (GO:0002830) 4/14 9.498991e-03 IL4;DENND1B;TNFSF4;NOD2
65 response to peptide (GO:1901652) 6/39 1.014438e-02 STAT5A;STAT5B;STAT3;NOD2;AGER;MMP9
66 cellular response to interleukin-1 (GO:0071347) 12/155 1.073928e-02 RPS6KA4;PSMA6;CCL13;CD40;CCL11;RIPK2;PSMD3;CUL1;MAP3K8;NOD2;PSMB8;PSMB9
67 regulation of regulatory T cell differentiation (GO:0045589) 5/26 1.088277e-02 SOCS1;IL2RA;IRF1;TNFSF4;HLA-G
68 positive regulation of T cell mediated cytotoxicity (GO:0001916) 5/26 1.088277e-02 IL23R;HLA-B;IL12B;HLA-A;HLA-G
69 positive regulation of peptidyl-tyrosine phosphorylation (GO:0050731) 11/134 1.105406e-02 EFNA1;IL4;CD40;RIPK2;IL23R;STAT3;LIF;IL12B;ARHGEF2;JAK2;IL6R
70 interleukin-27-mediated signaling pathway (GO:0070106) 4/15 1.162615e-02 STAT3;IL27;TYK2;JAK2
71 interleukin-1-mediated signaling pathway (GO:0070498) 9/94 1.214697e-02 RPS6KA4;PSMA6;RIPK2;PSMD3;CUL1;MAP3K8;NOD2;PSMB8;PSMB9
72 positive regulation of interleukin-6 production (GO:0032755) 8/76 1.337191e-02 TNFSF4;CARD9;STAT3;ARHGEF2;NOD2;IL6R;AGER;AIF1
73 regulation of T cell tolerance induction (GO:0002664) 3/7 1.348434e-02 IL2RA;HLA-B;HLA-G
74 positive regulation of activation of Janus kinase activity (GO:0010536) 3/7 1.348434e-02 IL23R;IL12B;IL6R
75 nucleotide-binding oligomerization domain containing signaling pathway (GO:0070423) 5/28 1.348434e-02 CYLD;LACC1;RIPK2;NOD2;IRGM
76 cellular response to corticosteroid stimulus (GO:0071384) 4/16 1.348434e-02 BCL2L11;ZFP36L2;UBE2L3;ZFP36L1
77 regulation of granulocyte macrophage colony-stimulating factor production (GO:0032645) 4/16 1.348434e-02 IL23R;CARD9;IL12B;RASGRP1
78 negative regulation of natural killer cell mediated cytotoxicity (GO:0045953) 4/16 1.348434e-02 HLA-B;HLA-A;HLA-G;MICA
79 polyol biosynthetic process (GO:0046173) 4/16 1.348434e-02 ITPKC;IPMK;IP6K1;IP6K2
80 positive regulation of T cell differentiation (GO:0045582) 6/43 1.424041e-02 IL4;SOCS1;IL23R;TNFSF4;IL12B;HLA-G
81 positive regulation of protein phosphorylation (GO:0001934) 20/371 1.458685e-02 CD40;RIPK2;LRRK2;LIF;ITLN1;PARK7;AGER;RASGRP1;MMP9;IRGM;RPS6KA4;EFNA1;RAD50;CSNK2B;ERBB2;FLOT1;ARHGEF2;JAK2;IL6R;LAT
82 negative regulation of multicellular organismal process (GO:0051241) 14/214 1.572435e-02 PTGER4;PPP1R11;SFMBT1;PLCL1;IL13;LNPEP;AGER;RPS6KA4;EFNA1;IL4;APOM;SYT11;IP6K1;IL18R1
83 stress-activated MAPK cascade (GO:0051403) 7/62 1.748055e-02 PTGER4;RIPK2;CUL1;MAP3K8;NOD2;TRIB1;ZFP36L1
84 cellular response to organic substance (GO:0071310) 10/123 1.825195e-02 STAT5B;SMAD3;RPS6KB1;LRRK2;ERBB2;STAT3;PARK7;RHOA;IL18R1;PTPN2
85 regulation of activation of Janus kinase activity (GO:0010533) 3/8 1.845689e-02 IL23R;IL12B;IL6R
86 regulation of apoptotic cell clearance (GO:2000425) 3/8 1.845689e-02 C4B;C4A;C2
87 positive regulation of apoptotic cell clearance (GO:2000427) 3/8 1.845689e-02 C4B;C4A;C2
88 positive regulation of MHC class II biosynthetic process (GO:0045348) 3/8 1.845689e-02 IL4;CIITA;JAK2
89 cellular response to glucocorticoid stimulus (GO:0071385) 4/18 1.918012e-02 BCL2L11;ZFP36L2;UBE2L3;ZFP36L1
90 positive regulation of response to endoplasmic reticulum stress (GO:1905898) 4/18 1.918012e-02 BAG6;BCL2L11;RNFT1;BOK
91 innate immune response (GO:0045087) 17/302 2.106118e-02 CD40;CIITA;PPP1R14B;RIPK2;IL23R;CARD9;NOD2;AGER;IRGM;IL4;CYLD;ADAM15;SMPD1;REL;RNF39;TRIM31;TRIM10
92 cellular response to interleukin-7 (GO:0098761) 4/19 2.308215e-02 STAT5A;STAT5B;SOCS1;STAT3
93 interleukin-7-mediated signaling pathway (GO:0038111) 4/19 2.308215e-02 STAT5A;STAT5B;SOCS1;STAT3
94 regulation of interleukin-17 production (GO:0032660) 5/33 2.394342e-02 IL23R;TNFSF4;CARD9;IL12B;NOD2
95 positive regulation of cytokine production involved in immune response (GO:0002720) 5/33 2.394342e-02 LACC1;TNFSF4;NOD2;HLA-A;HLA-G
96 cellular response to interleukin-9 (GO:0071355) 3/9 2.394342e-02 STAT5A;STAT5B;STAT3
97 cellular response to muramyl dipeptide (GO:0071225) 3/9 2.394342e-02 RIPK2;ARHGEF2;NOD2
98 regulation of T-helper 1 type immune response (GO:0002825) 3/9 2.394342e-02 IL23R;IL12B;IL27
99 interleukin-9-mediated signaling pathway (GO:0038113) 3/9 2.394342e-02 STAT5A;STAT5B;STAT3
100 positive regulation of memory T cell differentiation (GO:0043382) 3/9 2.394342e-02 IL23R;TNFSF4;IL12B
101 regulation of programmed necrotic cell death (GO:0062098) 4/20 2.533059e-02 CYLD;CDC37;FLOT1;NUPR1
102 growth hormone receptor signaling pathway (GO:0060396) 4/20 2.533059e-02 STAT5A;STAT5B;STAT3;JAK2
103 positive regulation of activated T cell proliferation (GO:0042104) 4/20 2.533059e-02 IL23R;IL12B;AGER;ICOSLG
104 positive regulation of alpha-beta T cell activation (GO:0046635) 4/20 2.533059e-02 IL23R;TNFSF4;IL12B;HLA-A
105 response to tumor necrosis factor (GO:0034612) 9/110 2.589131e-02 CCL13;CD40;CCL11;SMPD1;GBA;ARHGEF2;JAK2;ZFP36L2;ZFP36L1
106 positive regulation of MAP kinase activity (GO:0043406) 7/69 2.612842e-02 CD40;LRRK2;ERBB2;TNFSF11;MST1R;NOD2;RASGRP1
107 negative regulation of cytokine production (GO:0001818) 12/182 2.775504e-02 RPS6KA4;PTGER4;CYLD;UBA7;PPP1R11;SYT11;IL23R;TNFSF4;GBA;IL12B;AGER;ADCY7
108 glycolipid metabolic process (GO:0006664) 6/52 2.954595e-02 GALC;PGAP3;SMPD1;GBA;NEU1;FUT2
109 cAMP biosynthetic process (GO:0006171) 3/10 2.994607e-02 ADCY9;ADCY3;ADCY7
110 regulation of epithelial cell apoptotic process (GO:1904035) 3/10 2.994607e-02 NUPR1;BOK;ZFP36L1
111 regulation of memory T cell differentiation (GO:0043380) 3/10 2.994607e-02 IL23R;TNFSF4;IL12B
112 regulation of T-helper 17 type immune response (GO:2000316) 3/10 2.994607e-02 IL23R;CARD9;IL12B
113 regulation of peptidyl-tyrosine phosphorylation (GO:0050730) 8/92 2.994607e-02 EFNA1;RIPK2;IL23R;LIF;IL12B;ARHGEF2;JAK2;IL6R
114 T cell activation (GO:0042110) 8/92 2.994607e-02 IL4;TNFSF4;RASGRP1;ICOSLG;MICA;LAT;PTPN2;MICB
115 negative regulation of mitotic cell cycle phase transition (GO:1901991) 8/92 2.994607e-02 PSMA6;PSMD3;CUL1;BRD7;ZFP36L2;PSMB8;ZFP36L1;PSMB9
116 regulation of osteoclast differentiation (GO:0045670) 5/36 2.995131e-02 IL4;IL23R;IL12B;TNFSF11;TCTA
117 positive regulation of myeloid leukocyte differentiation (GO:0002763) 5/36 2.995131e-02 IL23R;LIF;IL12B;TNFSF11;ZFP36L1
118 extrinsic apoptotic signaling pathway (GO:0097191) 7/72 3.019607e-02 IL4;SMAD3;ITGAV;JAK2;HIPK1;IL6R;BOK
119 regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway (GO:1902235) 4/22 3.194890e-02 BCL2L11;LRRK2;PARK7;BOK
120 microglial cell activation (GO:0001774) 4/22 3.194890e-02 IL4;JAK2;AGER;AIF1
121 positive regulation of protein modification process (GO:0031401) 13/214 3.280620e-02 CD40;LRRK2;GBA;ITLN1;AGER;RASGRP1;MMP9;IRGM;EFNA1;SMPD1;CNEP1R1;ERBB2;FLOT1
122 regulation of cytokine-mediated signaling pathway (GO:0001959) 7/74 3.401353e-02 CYLD;SOCS1;IFNGR2;CDC37;JAK2;HIPK1;PTPN2
123 NIK/NF-kappaB signaling (GO:0038061) 7/74 3.401353e-02 PSMA6;TNFSF15;PSMD3;CUL1;REL;PSMB8;PSMB9
124 regulation of ERAD pathway (GO:1904292) 3/11 3.451964e-02 BAG6;USP19;RNFT1
125 cellular response to interleukin-2 (GO:0071352) 3/11 3.451964e-02 STAT5A;STAT5B;IL2RA
126 interleukin-2-mediated signaling pathway (GO:0038110) 3/11 3.451964e-02 STAT5A;STAT5B;IL2RA
127 interleukin-35-mediated signaling pathway (GO:0070757) 3/11 3.451964e-02 STAT3;JAK2;IL12RB2
128 positive regulation of cellular respiration (GO:1901857) 3/11 3.451964e-02 IL4;NUPR1;PARK7
129 regulation of necroptotic process (GO:0060544) 4/23 3.451964e-02 CYLD;CDC37;FLOT1;ARHGEF2
130 ERK1 and ERK2 cascade (GO:0070371) 4/23 3.451964e-02 PTGER4;ITGAV;ZFP36L2;ZFP36L1
131 positive regulation of histone acetylation (GO:0035066) 4/23 3.451964e-02 RPS6KA4;MUC1;LIF;BRD7
132 positive regulation of interleukin-17 production (GO:0032740) 4/23 3.451964e-02 IL23R;CARD9;IL12B;NOD2
133 organophosphate biosynthetic process (GO:0090407) 5/39 3.800910e-02 ITPKC;IPMK;IP6K1;FADS1;IP6K2
134 cellular response to peptide (GO:1901653) 6/57 3.850812e-02 ITGA4;RIPK2;ARHGEF2;NOD2;AGER;ZFP36L1
135 positive regulation of tumor necrosis factor production (GO:0032760) 7/77 3.902474e-02 STAT3;IL12B;ARHGEF2;NOD2;JAK2;AGER;RASGRP1
136 positive regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043123) 11/171 4.192324e-02 PPP5C;CD40;NDFIP1;RIPK2;CARD9;REL;NOD2;LTBR;LITAF;RHOA;BRD4
137 positive regulation of intracellular signal transduction (GO:1902533) 24/546 4.192324e-02 CD40;NDFIP1;RIPK2;CARD9;LIF;PARK7;NOD2;MST1R;LITAF;AGER;RHOA;PPP5C;BCL2L11;ERBB2;REL;IL12B;TNFSF11;ITGAV;RPL37;NUPR1;LTBR;JAK2;BRD4;BOK
138 regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043370) 3/12 4.192324e-02 SOCS1;TNFSF4;RUNX3
139 regulation of dendritic cell differentiation (GO:2001198) 3/12 4.192324e-02 HLA-B;AGER;HLA-G
140 regulation of MHC class II biosynthetic process (GO:0045346) 3/12 4.192324e-02 IL4;CIITA;JAK2
141 positive regulation of T-helper 1 type immune response (GO:0002827) 3/12 4.192324e-02 IL23R;IL12B;IL18R1
142 positive regulation of T-helper 17 type immune response (GO:2000318) 3/12 4.192324e-02 IL23R;CARD9;IL12B
143 receptor signaling pathway via STAT (GO:0097696) 4/25 4.388167e-02 STAT5A;STAT5B;STAT3;JAK2
144 positive regulation of phosphorylation (GO:0042327) 14/253 4.422908e-02 DDR1;CD40;PRKAA1;LRRK2;ITLN1;MST1R;AGER;RASGRP1;MMP9;IRGM;EFNA1;RAD50;ERBB2;FLOT1
145 response to interferon-gamma (GO:0034341) 7/80 4.523152e-02 CCL13;CD40;CIITA;CCL11;IL23R;AIF1;SLC26A6
146 cellular response to oxidative stress (GO:0034599) 9/125 4.524197e-02 PRDX5;PRKAA1;GPX1;LRRK2;NCF4;PARK7;FOS;MMP9;AIF1
147 positive regulation of tumor necrosis factor superfamily cytokine production (GO:1903557) 7/81 4.735355e-02 STAT3;IL12B;ARHGEF2;NOD2;JAK2;AGER;RASGRP1
148 phosphatidylinositol phosphate biosynthetic process (GO:0046854) 5/42 4.735355e-02 ITPKC;SOCS1;IP6K1;EFR3B;IP6K2
149 antigen processing and presentation of exogenous peptide antigen (GO:0002478) 8/103 4.735355e-02 HLA-DMA;HLA-DMB;HLA-A;HLA-DOA;HLA-DOB;SEC24C;HLA-DQA1;HLA-DQB1
150 cellular response to interleukin-15 (GO:0071350) 3/13 4.971461e-02 STAT5A;STAT5B;STAT3
151 response to muramyl dipeptide (GO:0032495) 3/13 4.971461e-02 RIPK2;NOD2;ARHGEF2
152 interleukin-15-mediated signaling pathway (GO:0035723) 3/13 4.971461e-02 STAT5A;STAT5B;STAT3
153 positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (GO:0002824) 3/13 4.971461e-02 IL23R;CARD9;IL12B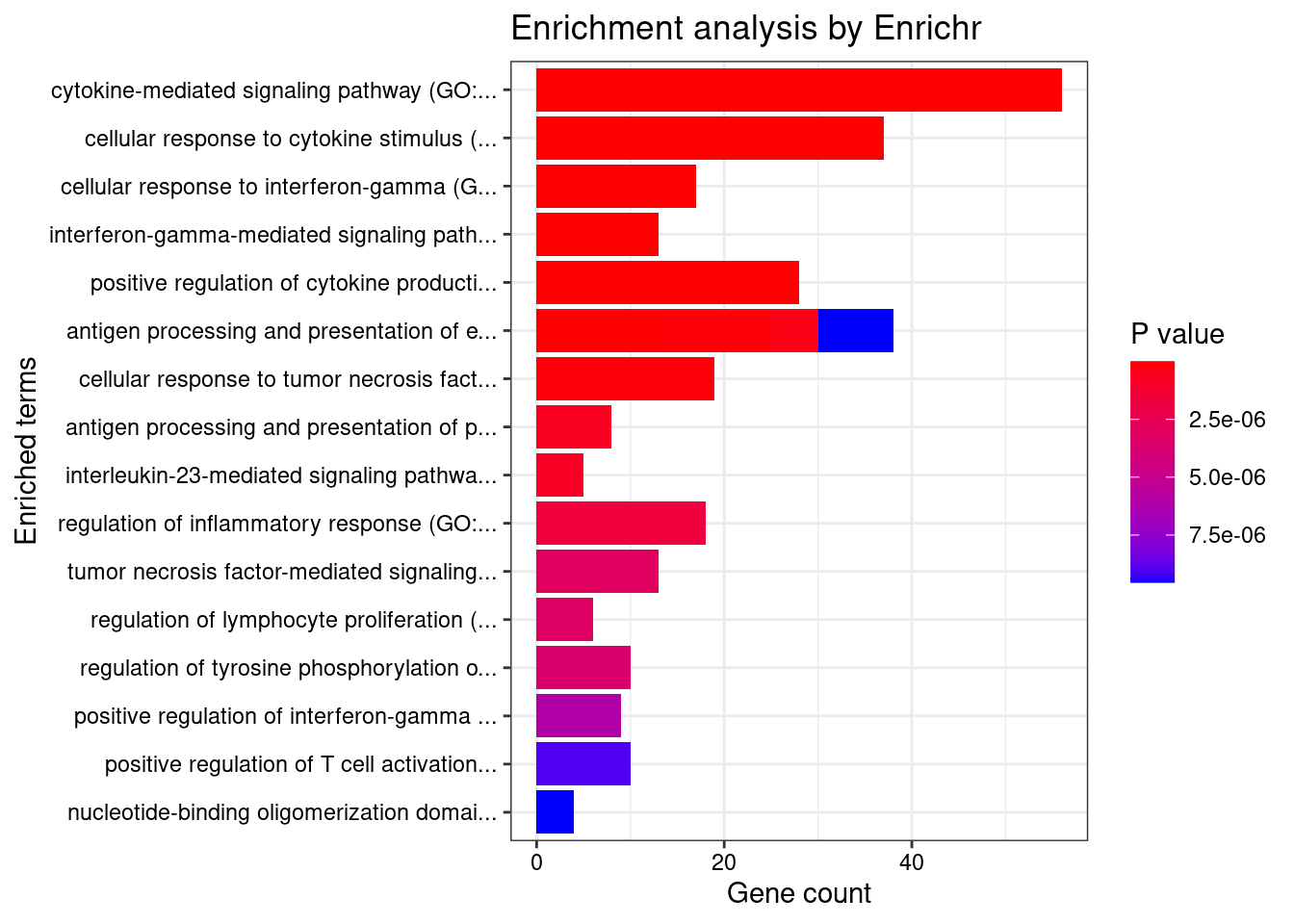
GO_Cellular_Component_2021
Term Overlap Adjusted.P.value Genes
1 MHC protein complex (GO:0042611) 9/20 5.263402e-08 HLA-DMA;HLA-DMB;HLA-B;HLA-C;HLA-A;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB1
2 MHC class II protein complex (GO:0042613) 6/13 2.445134e-05 HLA-DMA;HLA-DMB;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB1
3 ER to Golgi transport vesicle membrane (GO:0012507) 8/54 1.338542e-03 SEC16A;HLA-B;HLA-C;HLA-A;HLA-G;SEC24C;HLA-DQA1;HLA-DQB1
4 integral component of lumenal side of endoplasmic reticulum membrane (GO:0071556) 6/28 1.338542e-03 HLA-B;HLA-C;HLA-A;HLA-G;HLA-DQA1;HLA-DQB1
5 lumenal side of endoplasmic reticulum membrane (GO:0098553) 6/28 1.338542e-03 HLA-B;HLA-C;HLA-A;HLA-G;HLA-DQA1;HLA-DQB1
6 coated vesicle membrane (GO:0030662) 8/55 1.338542e-03 SEC16A;HLA-B;HLA-C;HLA-A;HLA-G;SEC24C;HLA-DQA1;HLA-DQB1
7 transport vesicle membrane (GO:0030658) 8/60 2.178455e-03 SEC16A;HLA-B;HLA-C;HLA-A;HLA-G;SEC24C;HLA-DQA1;HLA-DQB1
8 phagocytic vesicle (GO:0045335) 10/100 2.996865e-03 SYT11;NCF4;HLA-B;TAP2;HLA-C;TAP1;ITGAV;HLA-A;NOD2;HLA-G
9 MHC class I protein complex (GO:0042612) 3/6 5.635935e-03 HLA-B;HLA-C;HLA-A
10 endosome lumen (GO:0031904) 5/26 6.249646e-03 IL12B;LNPEP;JAK2;PDLIM4;AP4B1
11 COPII-coated ER to Golgi transport vesicle (GO:0030134) 8/79 9.626419e-03 SEC16A;HLA-B;HLA-C;HLA-A;HLA-G;SEC24C;HLA-DQA1;HLA-DQB1
12 phagocytic vesicle membrane (GO:0030670) 6/45 1.030053e-02 HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G
13 lysosome (GO:0005764) 23/477 1.400995e-02 STARD3;USP4;LRRK2;RNASET2;GBA;LNPEP;CTSW;LITAF;GALC;CLN3;HLA-DMA;HLA-DMB;NAGLU;SYT11;SMPD1;NEU1;SPNS1;FLOT1;PPT2;HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB1
14 integral component of endoplasmic reticulum membrane (GO:0030176) 10/142 2.571916e-02 CLN3;ATF6B;HLA-B;TAP2;HLA-C;TAP1;HLA-A;HLA-G;HLA-DQA1;HLA-DQB1
15 early endosome membrane (GO:0031901) 8/97 2.571916e-02 CLN3;HLA-B;HLA-C;HLA-A;HLA-G;LITAF;SNX20;BOK
16 lytic vacuole (GO:0000323) 13/219 2.571916e-02 USP4;LRRK2;RNASET2;GBA;CTSW;GALC;CLN3;NAGLU;SYT11;SMPD1;NEU1;PPT2;HLA-DOB
17 recycling endosome membrane (GO:0055038) 6/58 2.804111e-02 HLA-B;HLA-C;HLA-A;HLA-G;SCAMP3;BOK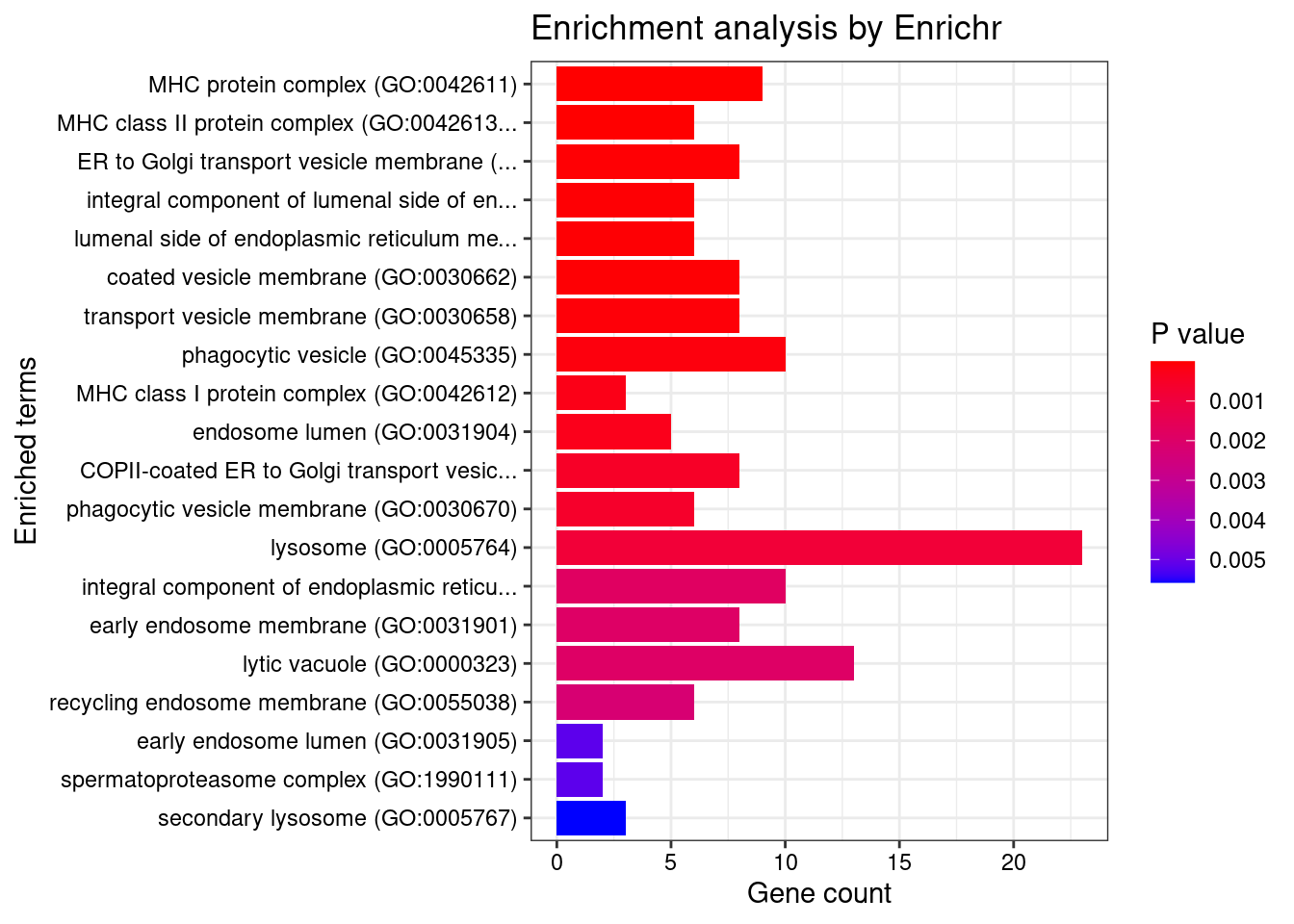
GO_Molecular_Function_2021
Term Overlap Adjusted.P.value Genes
1 cytokine receptor binding (GO:0005126) 11/105 0.00627540 IL4;SMAD3;IL23R;TNFSF4;LIF;IL12B;TNFSF11;IL27;TYK2;JAK2;IL6R
2 cytokine receptor activity (GO:0004896) 10/88 0.00627540 IL1RL1;IL18RAP;IL23R;IFNGR2;IL2RA;IL12B;CCR6;IL6R;IL18R1;IL12RB2
3 MHC class II receptor activity (GO:0032395) 4/10 0.00627540 HLA-DOA;HLA-DOB;HLA-DQA1;HLA-DQB1
4 kinase activity (GO:0016301) 11/112 0.00627540 CERKL;ITPKC;DGKE;DGKD;LRRK2;IPMK;COQ8B;CKB;IP6K1;COASY;IP6K2
5 interleukin-12 receptor binding (GO:0005143) 3/6 0.01962569 IL23R;IL12B;JAK2
6 ribosomal protein S6 kinase activity (GO:0004711) 3/7 0.02482584 RPS6KA4;RPS6KB1;RPS6KA2
7 CARD domain binding (GO:0050700) 4/16 0.02482584 RIPK2;CARD9;NOD2;IRGM
8 MHC class I protein binding (GO:0042288) 4/17 0.02482584 TUBB;TAP2;TAP1;CD244
9 MHC class II protein complex binding (GO:0023026) 4/17 0.02482584 HLA-DMA;HLA-DMB;HLA-DOA;HLA-DOB
10 inositol hexakisphosphate kinase activity (GO:0000828) 3/8 0.02653597 ITPKC;IP6K1;IP6K2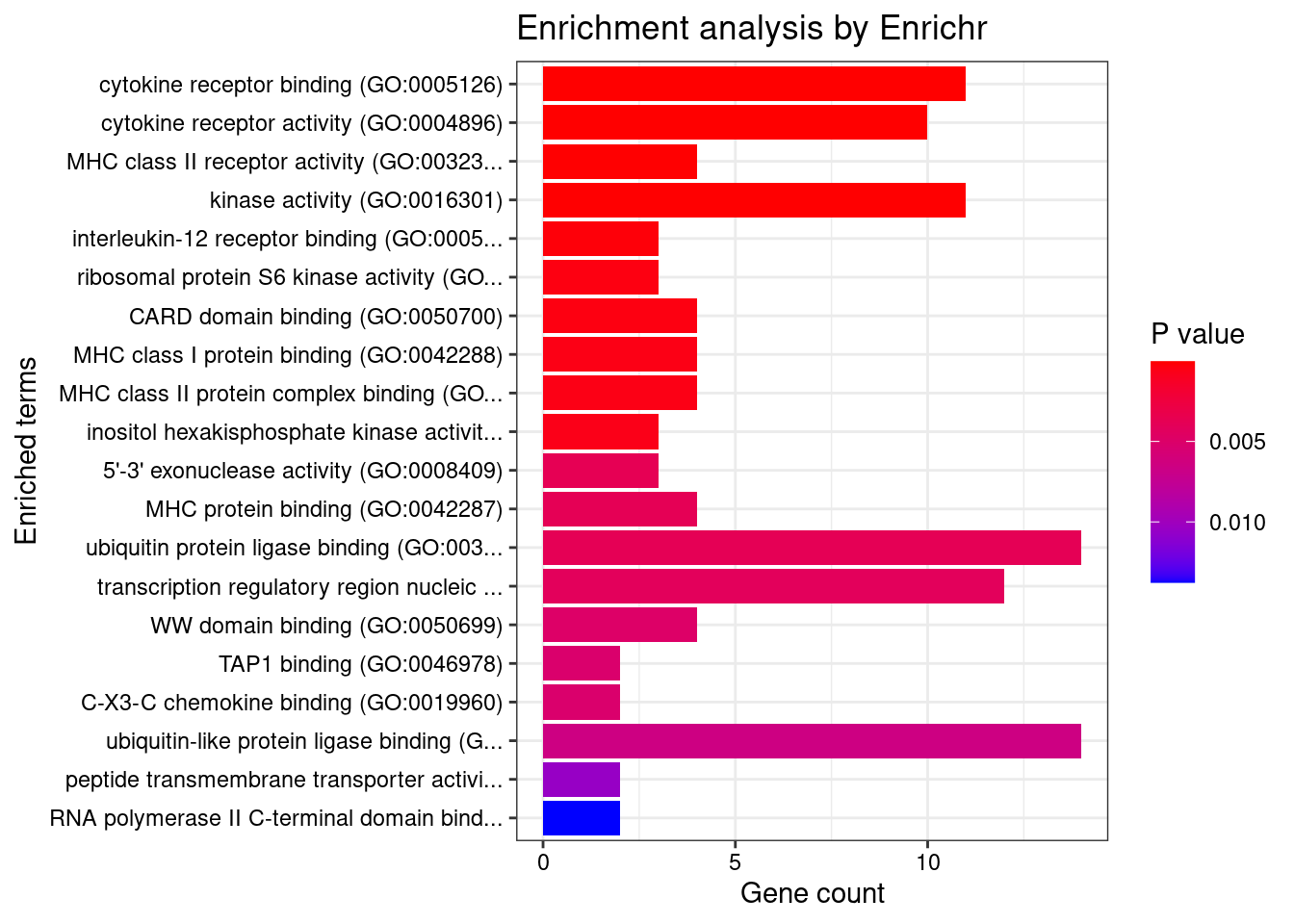
Enrichment analysis for cTWAS genes in top tissues separately
GO
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_tissue, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}
}Whole_Blood
Number of cTWAS Genes in Tissue: 17
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 positive regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043372) 2/8 0.003890511 SOCS1;TNFSF4
2 regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043370) 2/12 0.003890511 SOCS1;TNFSF4
3 positive regulation of CD4-positive, alpha-beta T cell activation (GO:2000516) 2/13 0.003890511 SOCS1;TNFSF4
4 positive regulation of alpha-beta T cell differentiation (GO:0046638) 2/14 0.003890511 SOCS1;TNFSF4
5 regulation of regulatory T cell differentiation (GO:0045589) 2/26 0.011049377 SOCS1;TNFSF4
6 regulation of interleukin-17 production (GO:0032660) 2/33 0.014906963 TNFSF4;CARD9
7 positive regulation of T cell differentiation (GO:0045582) 2/43 0.021743407 SOCS1;TNFSF4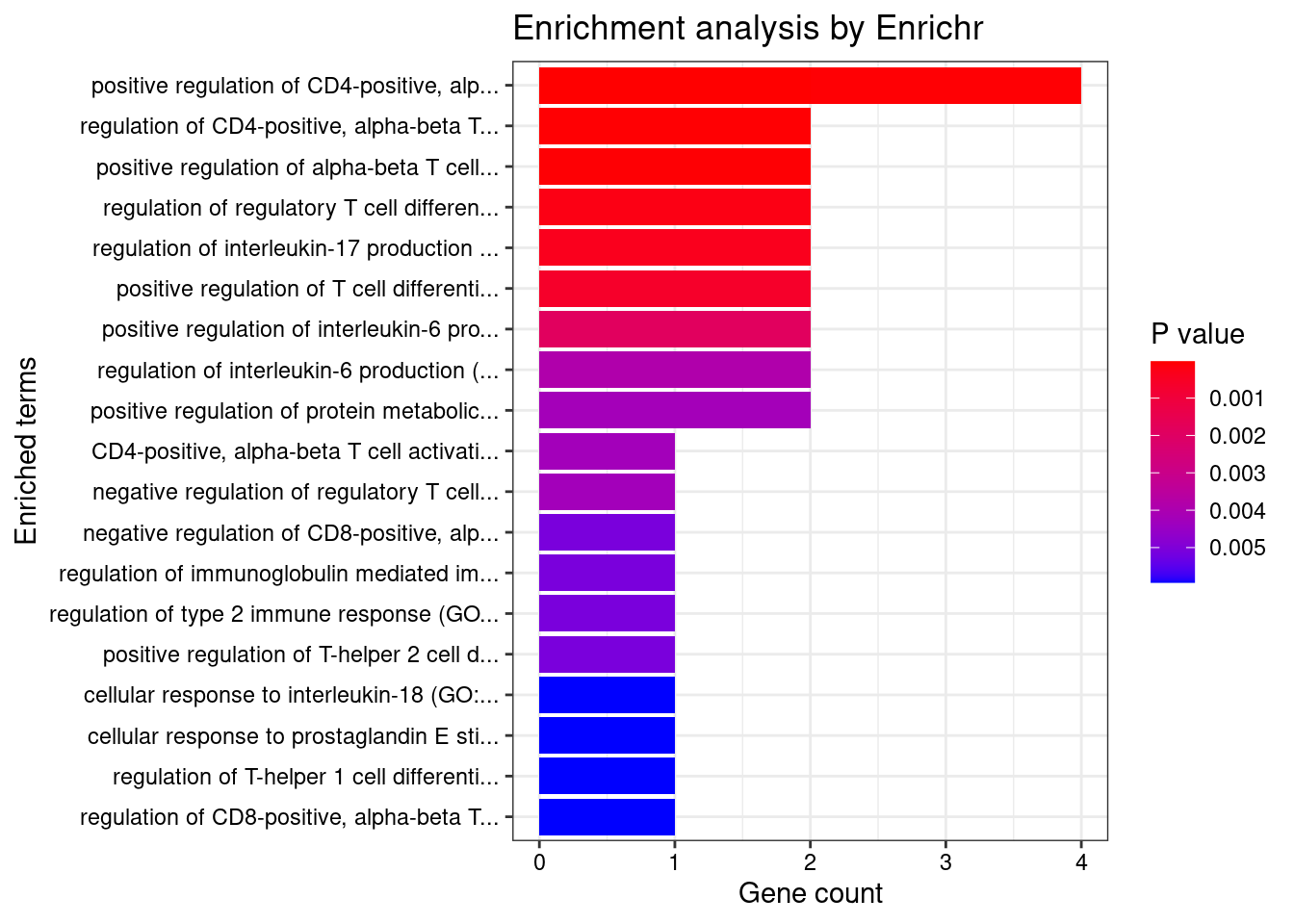
Colon_Transverse
Number of cTWAS Genes in Tissue: 7
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 oocyte development (GO:0048599) 1/5 0.02610937 ZGLP1
2 cleavage furrow formation (GO:0036089) 1/7 0.02610937 SPIRE2
3 parallel actin filament bundle assembly (GO:0030046) 1/7 0.02610937 SPIRE2
4 actin filament network formation (GO:0051639) 1/11 0.03075338 SPIRE2
5 regulation of response to interferon-gamma (GO:0060330) 1/14 0.03129848 IFNGR2
6 actin nucleation (GO:0045010) 1/21 0.03667835 SPIRE2
7 regulation of interferon-gamma-mediated signaling pathway (GO:0060334) 1/23 0.03667835 IFNGR2
8 actin filament polymerization (GO:0030041) 1/29 0.03717093 SPIRE2
9 plasma membrane invagination (GO:0099024) 1/30 0.03717093 SPIRE2
10 positive regulation of double-strand break repair (GO:2000781) 1/40 0.03853371 SPIRE2
11 regulation of double-strand break repair (GO:2000779) 1/42 0.03853371 SPIRE2
12 positive regulation of DNA repair (GO:0045739) 1/48 0.03853371 SPIRE2
13 positive regulation of actin filament polymerization (GO:0030838) 1/49 0.03853371 SPIRE2
14 actin polymerization or depolymerization (GO:0008154) 1/50 0.03853371 SPIRE2
15 autophagosome organization (GO:1905037) 1/56 0.03853371 ATG16L1
16 autophagosome assembly (GO:0000045) 1/58 0.03853371 ATG16L1
17 protein polymerization (GO:0051258) 1/59 0.03853371 SPIRE2
18 interferon-gamma-mediated signaling pathway (GO:0060333) 1/68 0.04098865 IFNGR2
19 cytoskeleton-dependent cytokinesis (GO:0061640) 1/72 0.04098865 SPIRE2
20 regulation of cytokine-mediated signaling pathway (GO:0001959) 1/74 0.04098865 IFNGR2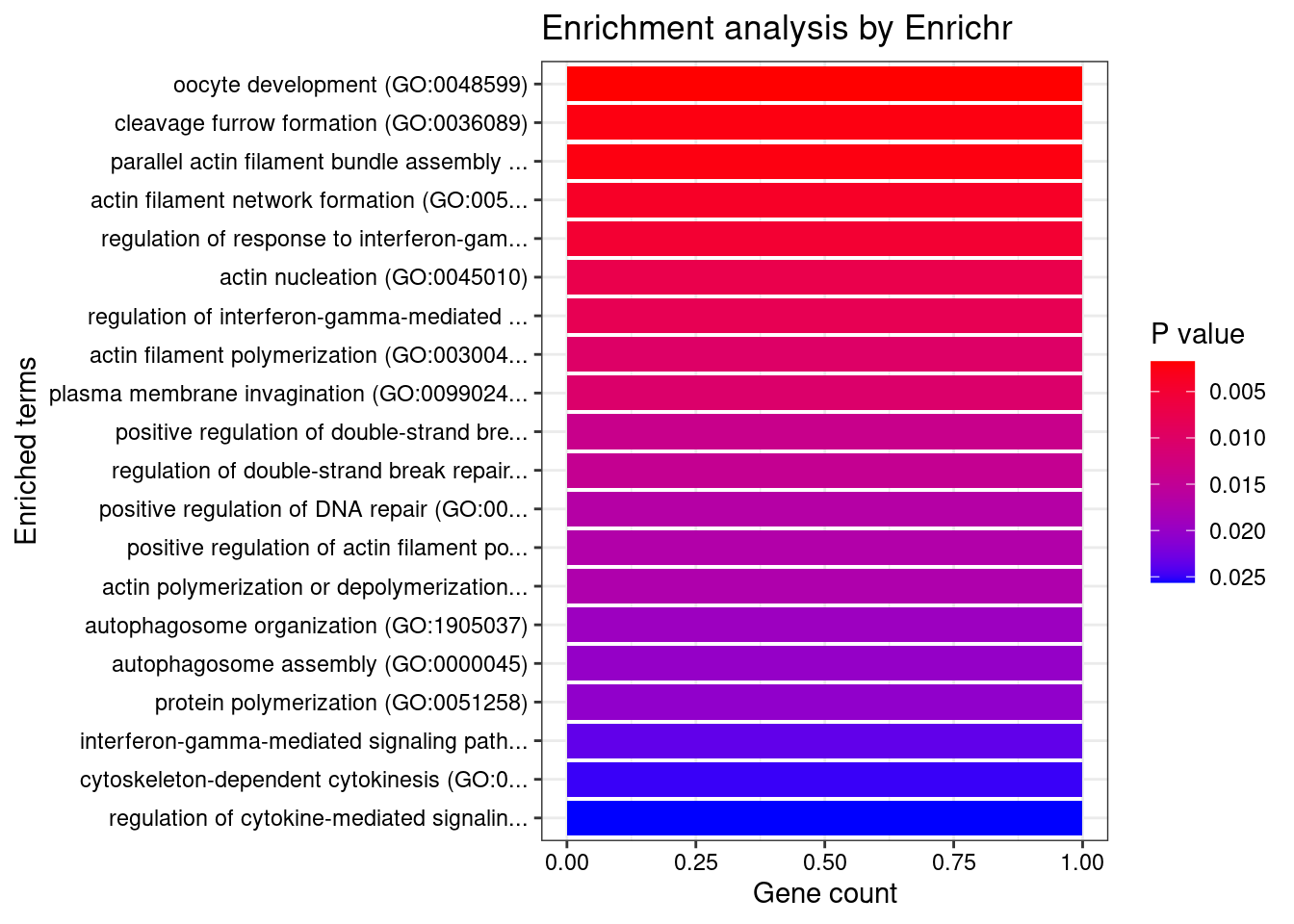
Skin_Not_Sun_Exposed_Suprapubic
Number of cTWAS Genes in Tissue: 9
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)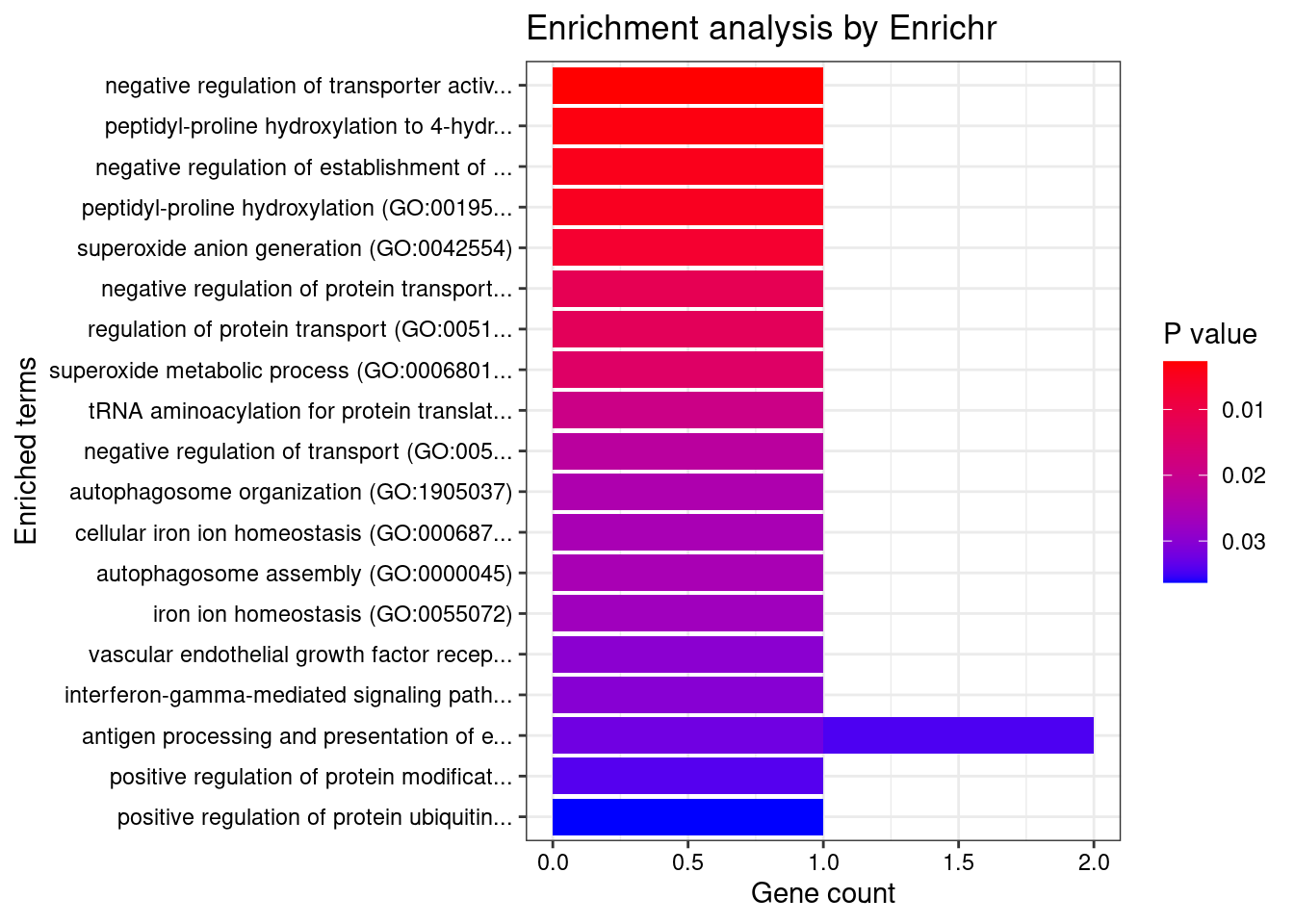
Testis
Number of cTWAS Genes in Tissue: 6
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 quinone biosynthetic process (GO:1901663) 1/14 0.0167683 COQ8B
2 ubiquinone biosynthetic process (GO:0006744) 1/15 0.0167683 COQ8B
3 ubiquinone metabolic process (GO:0006743) 1/15 0.0167683 COQ8B
4 activation of NF-kappaB-inducing kinase activity (GO:0007250) 1/16 0.0167683 TNFSF15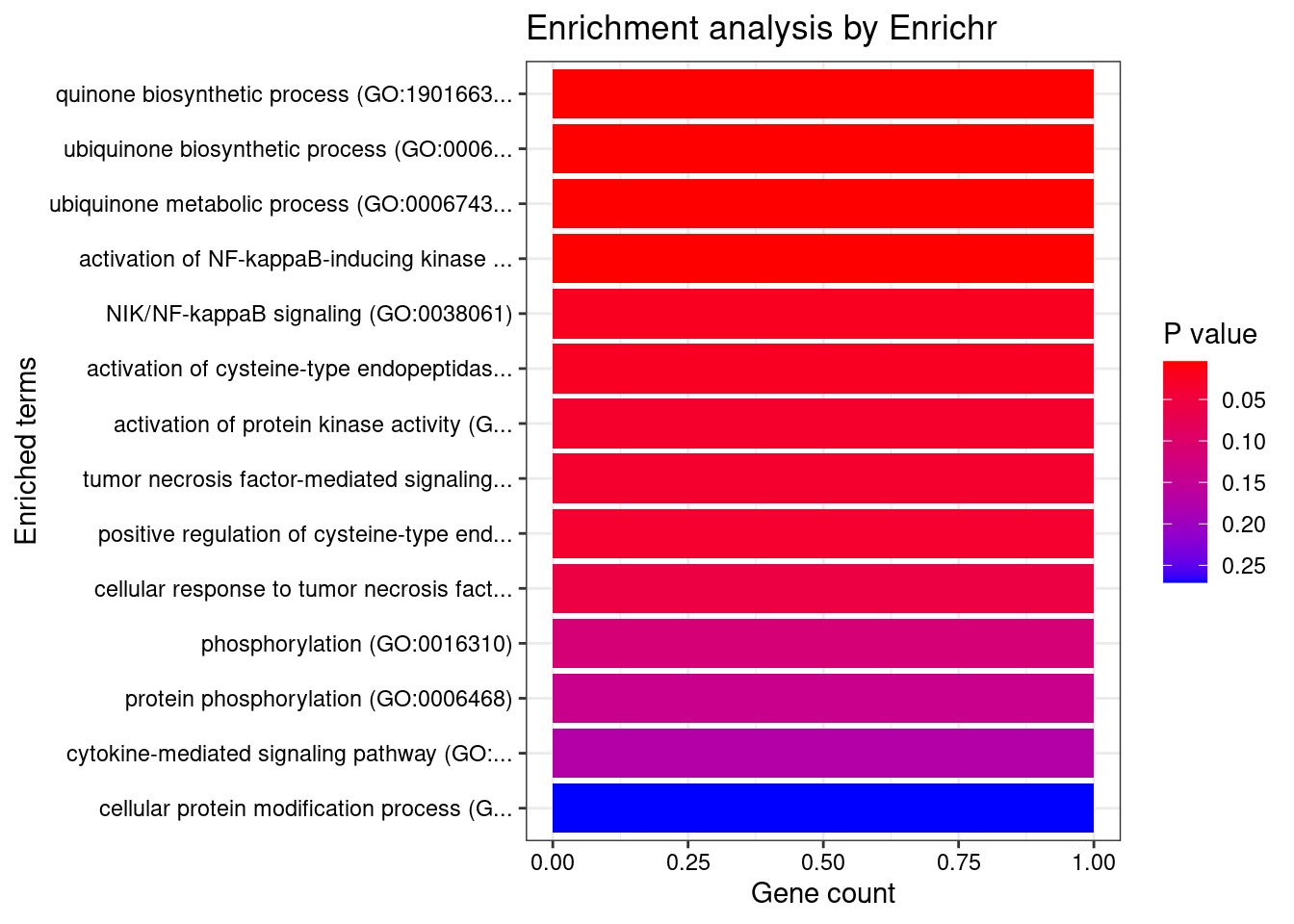
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 6
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 positive regulation of protein metabolic process (GO:0051247) 2/115 0.02287156 CARD9;OAZ3
2 myeloid leukocyte mediated immunity (GO:0002444) 1/8 0.02287156 CARD9
3 regulation of T-helper 17 type immune response (GO:2000316) 1/10 0.02287156 CARD9
4 immunoglobulin mediated immune response (GO:0016064) 1/10 0.02287156 CARD9
5 negative regulation of transmembrane transport (GO:0034763) 1/10 0.02287156 OAZ3
6 B cell mediated immunity (GO:0019724) 1/11 0.02287156 CARD9
7 positive regulation of T-helper 17 type immune response (GO:2000318) 1/12 0.02287156 CARD9
8 positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (GO:0002824) 1/13 0.02287156 CARD9
9 homeostasis of number of cells (GO:0048872) 1/13 0.02287156 CARD9
10 antifungal innate immune response (GO:0061760) 1/13 0.02287156 CARD9
11 positive regulation of granulocyte macrophage colony-stimulating factor production (GO:0032725) 1/14 0.02287156 CARD9
12 regulation of granulocyte macrophage colony-stimulating factor production (GO:0032645) 1/16 0.02309343 CARD9
13 positive regulation of cytokine production involved in inflammatory response (GO:1900017) 1/17 0.02309343 CARD9
14 positive regulation of stress-activated protein kinase signaling cascade (GO:0070304) 1/18 0.02309343 CARD9
15 positive regulation of interleukin-17 production (GO:0032740) 1/23 0.02692217 CARD9
16 defense response to fungus (GO:0050832) 1/24 0.02692217 CARD9
17 modulation by host of symbiont process (GO:0051851) 1/32 0.03286793 CARD9
18 regulation of interleukin-17 production (GO:0032660) 1/33 0.03286793 CARD9
19 regulation of cytokine production involved in inflammatory response (GO:1900015) 1/43 0.04052317 CARD9
20 regulation of stress-activated MAPK cascade (GO:0032872) 1/49 0.04319085 CARD9
21 regulation of cellular amine metabolic process (GO:0033238) 1/51 0.04319085 OAZ3
22 regulation of cellular amino acid metabolic process (GO:0006521) 1/54 0.04319085 OAZ3
23 autophagosome organization (GO:1905037) 1/56 0.04319085 ATG16L1
24 autophagosome assembly (GO:0000045) 1/58 0.04319085 ATG16L1
25 positive regulation of cysteine-type endopeptidase activity (GO:2001056) 1/62 0.04395985 CARD9
26 regulation of cellular ketone metabolic process (GO:0010565) 1/64 0.04395985 OAZ3
27 positive regulation of catabolic process (GO:0009896) 1/67 0.04429941 OAZ3
28 positive regulation of JNK cascade (GO:0046330) 1/73 0.04650785 CARD9
29 positive regulation of interleukin-6 production (GO:0032755) 1/76 0.04673199 CARD9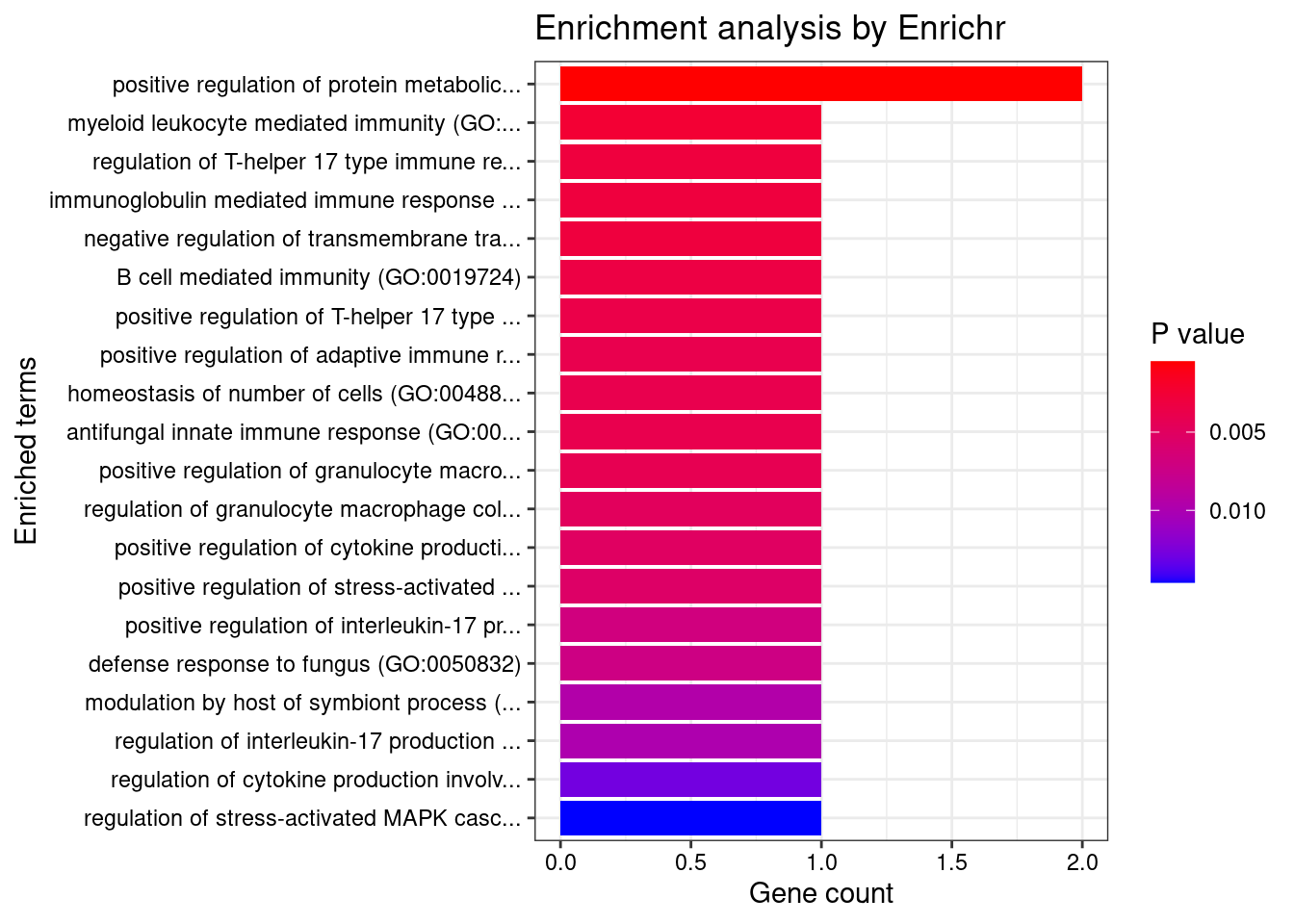
KEGG
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- NULL
try(enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_tissue, referenceGene=background_tissue,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F))
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
} Whole_Blood
Number of cTWAS Genes in Tissue: 17
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Colon_Transverse
Number of cTWAS Genes in Tissue: 7
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Skin_Not_Sun_Exposed_Suprapubic
Number of cTWAS Genes in Tissue: 9
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Testis
Number of cTWAS Genes in Tissue: 6
Loading the functional categories...
Loading the ID list...
ERROR: All IDs in the uploaded list can only annotate to one category, which can not perform the enrichment analysis.Error in .uploadGeneExistingOrganism(organism = organism, dataType = dataType, :
ERROR: All IDs in the uploaded list can only annotate to one category, which can not perform the enrichment analysis.
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 6
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_tissue, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
} Whole_Blood
Number of cTWAS Genes in Tissue: 17TMEM229B gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDFAM171B gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDAF064858.11 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
2 Brain Diseases 0.006953575 2/10 25/9703
30 Encephalopathies 0.006953575 2/10 27/9703
12 Inflammatory Bowel Diseases 0.007823755 2/10 35/9703
38 Deep seated dermatophytosis 0.010822511 1/10 1/9703
4 Ulcerative Colitis 0.012362846 2/10 63/9703
39 Candidiasis, Familial, 2 0.012362846 1/10 2/9703
41 Leukoencephalopathy, Cystic, Without Megalencephaly 0.012362846 1/10 2/9703
8 Graves Disease 0.048520918 1/10 9/9703
Colon_Transverse
Number of cTWAS Genes in Tissue: 7SPIRE2 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDRP11-386E5.1 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
38 Hurthle Cell Tumor 0.007884159 1/3 2/9703
43 Oxyphilic Adenoma 0.007884159 1/3 2/9703
44 Inflammatory Bowel Disease 10 0.007884159 1/3 1/9703
46 IMMUNODEFICIENCY 28 0.007884159 1/3 1/9703
8 Psychosis, Brief Reactive 0.036747247 1/3 14/9703
12 Schizophreniform Disorders 0.036747247 1/3 14/9703
2 Depression, Bipolar 0.038636234 1/3 79/9703
4 Crohn Disease 0.038636234 1/3 50/9703
5 Inflammatory Bowel Diseases 0.038636234 1/3 35/9703
6 Jacksonian Seizure 0.038636234 1/3 101/9703
7 Manic Disorder 0.038636234 1/3 71/9703
9 Psychotic Disorders 0.038636234 1/3 101/9703
10 Schizoaffective Disorder 0.038636234 1/3 29/9703
14 Complex partial seizures 0.038636234 1/3 101/9703
15 Crohn's disease of large bowel 0.038636234 1/3 44/9703
16 Generalized seizures 0.038636234 1/3 101/9703
17 Clonic Seizures 0.038636234 1/3 101/9703
18 Crohn's disease of the ileum 0.038636234 1/3 44/9703
19 Visual seizure 0.038636234 1/3 101/9703
20 Tonic Seizures 0.038636234 1/3 102/9703
21 Epileptic drop attack 0.038636234 1/3 101/9703
23 Manic 0.038636234 1/3 78/9703
24 Seizures, Somatosensory 0.038636234 1/3 101/9703
25 Seizures, Auditory 0.038636234 1/3 101/9703
26 Olfactory seizure 0.038636234 1/3 101/9703
27 Gustatory seizure 0.038636234 1/3 101/9703
28 Vertiginous seizure 0.038636234 1/3 101/9703
29 Tonic - clonic seizures 0.038636234 1/3 104/9703
30 Regional enteritis 0.038636234 1/3 44/9703
31 Non-epileptic convulsion 0.038636234 1/3 101/9703
32 Single Seizure 0.038636234 1/3 101/9703
33 Atonic Absence Seizures 0.038636234 1/3 101/9703
34 Convulsive Seizures 0.038636234 1/3 101/9703
35 Seizures, Focal 0.038636234 1/3 104/9703
36 Seizures, Sensory 0.038636234 1/3 101/9703
37 IIeocolitis 0.038636234 1/3 44/9703
45 Nonepileptic Seizures 0.038636234 1/3 101/9703
47 Convulsions 0.038636234 1/3 102/9703
48 Absence Seizures 0.038636234 1/3 102/9703
49 Epileptic Seizures 0.038636234 1/3 101/9703
50 Myoclonic Seizures 0.038636234 1/3 104/9703
51 Generalized Absence Seizures 0.038636234 1/3 101/9703
3 Renal Cell Carcinoma 0.042388118 1/3 128/9703
39 Chromophobe Renal Cell Carcinoma 0.042388118 1/3 128/9703
40 Sarcomatoid Renal Cell Carcinoma 0.042388118 1/3 128/9703
41 Collecting Duct Carcinoma of the Kidney 0.042388118 1/3 128/9703
42 Papillary Renal Cell Carcinoma 0.042388118 1/3 128/9703
22 Conventional (Clear Cell) Renal Cell Carcinoma 0.047890889 1/3 148/9703
Skin_Not_Sun_Exposed_Suprapubic
Number of cTWAS Genes in Tissue: 9NDFIP1 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
29 Crohn's disease of large bowel 4.665228e-05 3/7 44/9703
33 Crohn's disease of the ileum 4.665228e-05 3/7 44/9703
38 Regional enteritis 4.665228e-05 3/7 44/9703
44 IIeocolitis 4.665228e-05 3/7 44/9703
5 Crohn Disease 5.513042e-05 3/7 50/9703
31 Kleine-Levin Syndrome 4.473304e-03 1/7 1/9703
48 Inflammatory Bowel Disease 10 4.473304e-03 1/7 1/9703
50 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 4.473304e-03 1/7 1/9703
55 COMBINED OXIDATIVE PHOSPHORYLATION DEFICIENCY 20 4.473304e-03 1/7 1/9703
58 MYOPIA 25, AUTOSOMAL DOMINANT 4.473304e-03 1/7 1/9703
17 Megaesophagus 7.453202e-03 1/7 2/9703
62 Limbic encephalitis with leucine-rich glioma-inactivated 1 antibodies 7.453202e-03 1/7 2/9703
23 Bullous pemphigoid 1.031663e-02 1/7 3/9703
8 Esophageal Achalasia 1.051566e-02 1/7 4/9703
21 Oropharyngeal Neoplasms 1.051566e-02 1/7 4/9703
42 Idiopathic achalasia of esophagus 1.051566e-02 1/7 4/9703
49 Oropharyngeal Carcinoma 1.051566e-02 1/7 4/9703
1 Angioedema 1.488797e-02 1/7 6/9703
15 Creutzfeldt-Jakob disease 2.758932e-02 1/7 13/9703
36 New Variant Creutzfeldt-Jakob Disease 2.758932e-02 1/7 13/9703
39 Creutzfeldt-Jakob Disease, Familial 2.758932e-02 1/7 13/9703
18 Moyamoya Disease 3.026835e-02 1/7 17/9703
20 Narcolepsy 3.026835e-02 1/7 17/9703
27 Urticaria 3.026835e-02 1/7 16/9703
41 Narcolepsy-Cataplexy Syndrome 3.026835e-02 1/7 16/9703
26 Systemic Scleroderma 3.250810e-02 1/7 19/9703
6 Drug Eruptions 3.649591e-02 1/7 23/9703
37 Morbilliform Drug Reaction 3.649591e-02 1/7 23/9703
Testis
Number of cTWAS Genes in Tissue: 6RP11-574K11.29 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDRP11-386E5.1 gene(s) from the input list not found in DisGeNET CURATEDLINC01623 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
3 Enteritis 0.0009276438 1/2 1/9703
9 NEPHROTIC SYNDROME, TYPE 9 0.0009276438 1/2 1/9703
5 Inflammatory Bowel Diseases 0.0162053182 1/2 35/9703
8 NEPHROTIC SYNDROME, STEROID-RESISTANT, AUTOSOMAL RECESSIVE 0.0162053182 1/2 30/9703
1 Primary biliary cirrhosis 0.0173983555 1/2 47/9703
2 Ulcerative Colitis 0.0194182686 1/2 63/9703
6 Degenerative polyarthritis 0.0214654486 1/2 93/9703
7 Osteoarthrosis Deformans 0.0214654486 1/2 93/9703
4 Inflammation 0.0260101504 1/2 127/9703
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 6OAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDFGFR1OP gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
5 Inflammatory Bowel Diseases 0.0001804346 2/3 35/9703
9 Crohn's disease of large bowel 0.0001804346 2/3 44/9703
10 Crohn's disease of the ileum 0.0001804346 2/3 44/9703
11 Regional enteritis 0.0001804346 2/3 44/9703
12 IIeocolitis 0.0001804346 2/3 44/9703
3 Crohn Disease 0.0001946273 2/3 50/9703
13 Deep seated dermatophytosis 0.0005797774 1/3 1/9703
15 Inflammatory Bowel Disease 10 0.0005797774 1/3 1/9703
14 Candidiasis, Familial, 2 0.0010306091 1/3 2/9703
8 Ankylosing spondylitis 0.0050967831 1/3 11/9703
4 IGA Glomerulonephritis 0.0142875987 1/3 34/9703
6 Pustulosis of Palms and Soles 0.0202196317 1/3 57/9703
7 Psoriasis 0.0202196317 1/3 57/9703
2 Ulcerative Colitis 0.0207388700 1/3 63/9703Gene sets curated by Macarthur Lab
output <- output[order(-output$pve_g),]
top_tissues <- output$weight[1:5]
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (tissue in top_tissues){
cat(paste0(tissue, "\n\n"))
ctwas_genes_tissue <- df[[tissue]]$ctwas
background_tissue <- df[[tissue]]$gene_pips$genename
cat(paste0("Number of cTWAS Genes in Tissue: ", length(ctwas_genes_tissue), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_tissue=ctwas_genes_tissue)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_tissue]})
##########
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_tissue)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
} Whole_Blood
Number of cTWAS Genes in Tissue: 17
gene_set nset ngenes percent padj
1 gwascatalog 3492 8 0.0022909507 0.8484846
2 mgi_essential 1254 1 0.0007974482 1.0000000
3 core_essentials_hart 153 0 0.0000000000 1.0000000
4 clinvar_path_likelypath 1607 2 0.0012445551 1.0000000
5 fda_approved_drug_targets 177 0 0.0000000000 1.0000000
Colon_Transverse
Number of cTWAS Genes in Tissue: 7
gene_set nset ngenes percent padj
1 gwascatalog 3739 4 0.0010698048 0.71609
2 mgi_essential 1372 1 0.0007288630 1.00000
3 core_essentials_hart 178 0 0.0000000000 1.00000
4 clinvar_path_likelypath 1760 1 0.0005681818 1.00000
5 fda_approved_drug_targets 197 0 0.0000000000 1.00000
Skin_Not_Sun_Exposed_Suprapubic
Number of cTWAS Genes in Tissue: 9
gene_set nset ngenes percent padj
1 gwascatalog 3940 6 0.001522843 0.1602560
2 mgi_essential 1461 3 0.002053388 0.1944222
3 clinvar_path_likelypath 1832 2 0.001091703 0.6465860
4 core_essentials_hart 173 0 0.000000000 1.0000000
5 fda_approved_drug_targets 207 0 0.000000000 1.0000000
Testis
Number of cTWAS Genes in Tissue: 6
gene_set nset ngenes percent padj
1 gwascatalog 4116 1 0.0002429543 1
2 mgi_essential 1516 0 0.0000000000 1
3 core_essentials_hart 164 0 0.0000000000 1
4 clinvar_path_likelypath 1945 0 0.0000000000 1
5 fda_approved_drug_targets 229 0 0.0000000000 1
Adipose_Subcutaneous
Number of cTWAS Genes in Tissue: 6
gene_set nset ngenes percent padj
1 gwascatalog 3851 3 0.0007790184 1
2 mgi_essential 1405 1 0.0007117438 1
3 core_essentials_hart 172 0 0.0000000000 1
4 clinvar_path_likelypath 1789 1 0.0005589715 1
5 fda_approved_drug_targets 206 0 0.0000000000 1Summary of results across tissues
weight_groups <- as.data.frame(matrix(c("Adipose_Subcutaneous", "Adipose",
"Adipose_Visceral_Omentum", "Adipose",
"Adrenal_Gland", "Endocrine",
"Artery_Aorta", "Cardiovascular",
"Artery_Coronary", "Cardiovascular",
"Artery_Tibial", "Cardiovascular",
"Brain_Amygdala", "CNS",
"Brain_Anterior_cingulate_cortex_BA24", "CNS",
"Brain_Caudate_basal_ganglia", "CNS",
"Brain_Cerebellar_Hemisphere", "CNS",
"Brain_Cerebellum", "CNS",
"Brain_Cortex", "CNS",
"Brain_Frontal_Cortex_BA9", "CNS",
"Brain_Hippocampus", "CNS",
"Brain_Hypothalamus", "CNS",
"Brain_Nucleus_accumbens_basal_ganglia", "CNS",
"Brain_Putamen_basal_ganglia", "CNS",
"Brain_Spinal_cord_cervical_c-1", "CNS",
"Brain_Substantia_nigra", "CNS",
"Breast_Mammary_Tissue", "None",
"Cells_Cultured_fibroblasts", "Skin",
"Cells_EBV-transformed_lymphocytes", "Blood or Immune",
"Colon_Sigmoid", "Digestive",
"Colon_Transverse", "Digestive",
"Esophagus_Gastroesophageal_Junction", "Digestive",
"Esophagus_Mucosa", "Digestive",
"Esophagus_Muscularis", "Digestive",
"Heart_Atrial_Appendage", "Cardiovascular",
"Heart_Left_Ventricle", "Cardiovascular",
"Kidney_Cortex", "None",
"Liver", "None",
"Lung", "None",
"Minor_Salivary_Gland", "None",
"Muscle_Skeletal", "None",
"Nerve_Tibial", "None",
"Ovary", "None",
"Pancreas", "None",
"Pituitary", "Endocrine",
"Prostate", "None",
"Skin_Not_Sun_Exposed_Suprapubic", "Skin",
"Skin_Sun_Exposed_Lower_leg", "Skin",
"Small_Intestine_Terminal_Ileum", "Digestive",
"Spleen", "Blood or Immune",
"Stomach", "Digestive",
"Testis", "Endocrine",
"Thyroid", "Endocrine",
"Uterus", "None",
"Vagina", "None",
"Whole_Blood", "Blood or Immune"),
nrow=49, ncol=2, byrow=T), stringsAsFactors=F)
colnames(weight_groups) <- c("weight", "group")
#display tissue groups
print(weight_groups) weight group
1 Adipose_Subcutaneous Adipose
2 Adipose_Visceral_Omentum Adipose
3 Adrenal_Gland Endocrine
4 Artery_Aorta Cardiovascular
5 Artery_Coronary Cardiovascular
6 Artery_Tibial Cardiovascular
7 Brain_Amygdala CNS
8 Brain_Anterior_cingulate_cortex_BA24 CNS
9 Brain_Caudate_basal_ganglia CNS
10 Brain_Cerebellar_Hemisphere CNS
11 Brain_Cerebellum CNS
12 Brain_Cortex CNS
13 Brain_Frontal_Cortex_BA9 CNS
14 Brain_Hippocampus CNS
15 Brain_Hypothalamus CNS
16 Brain_Nucleus_accumbens_basal_ganglia CNS
17 Brain_Putamen_basal_ganglia CNS
18 Brain_Spinal_cord_cervical_c-1 CNS
19 Brain_Substantia_nigra CNS
20 Breast_Mammary_Tissue None
21 Cells_Cultured_fibroblasts Skin
22 Cells_EBV-transformed_lymphocytes Blood or Immune
23 Colon_Sigmoid Digestive
24 Colon_Transverse Digestive
25 Esophagus_Gastroesophageal_Junction Digestive
26 Esophagus_Mucosa Digestive
27 Esophagus_Muscularis Digestive
28 Heart_Atrial_Appendage Cardiovascular
29 Heart_Left_Ventricle Cardiovascular
30 Kidney_Cortex None
31 Liver None
32 Lung None
33 Minor_Salivary_Gland None
34 Muscle_Skeletal None
35 Nerve_Tibial None
36 Ovary None
37 Pancreas None
38 Pituitary Endocrine
39 Prostate None
40 Skin_Not_Sun_Exposed_Suprapubic Skin
41 Skin_Sun_Exposed_Lower_leg Skin
42 Small_Intestine_Terminal_Ileum Digestive
43 Spleen Blood or Immune
44 Stomach Digestive
45 Testis Endocrine
46 Thyroid Endocrine
47 Uterus None
48 Vagina None
49 Whole_Blood Blood or Immunegroups <- unique(weight_groups$group)
df_group <- list()
for (i in 1:length(groups)){
group <- groups[i]
weights <- weight_groups$weight[weight_groups$group==group]
df_group[[group]] <- list(ctwas=unique(unlist(lapply(df[weights], function(x){x$ctwas}))),
background=unique(unlist(lapply(df[weights], function(x){x$gene_pips$genename}))))
}
output <- output[sapply(weight_groups$weight, match, output$weight),,drop=F]
output$group <- weight_groups$group
output$n_ctwas_group <- sapply(output$group, function(x){length(df_group[[x]][["ctwas"]])})
output$n_ctwas_group[output$group=="None"] <- 0
#barplot of number of cTWAS genes in each tissue
output <- output[order(-output$n_ctwas),,drop=F]
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(output$n_ctwas, names.arg=output$weight, las=2, ylab="Number of cTWAS Genes", cex.names=0.6, main="Number of cTWAS Genes by Tissue")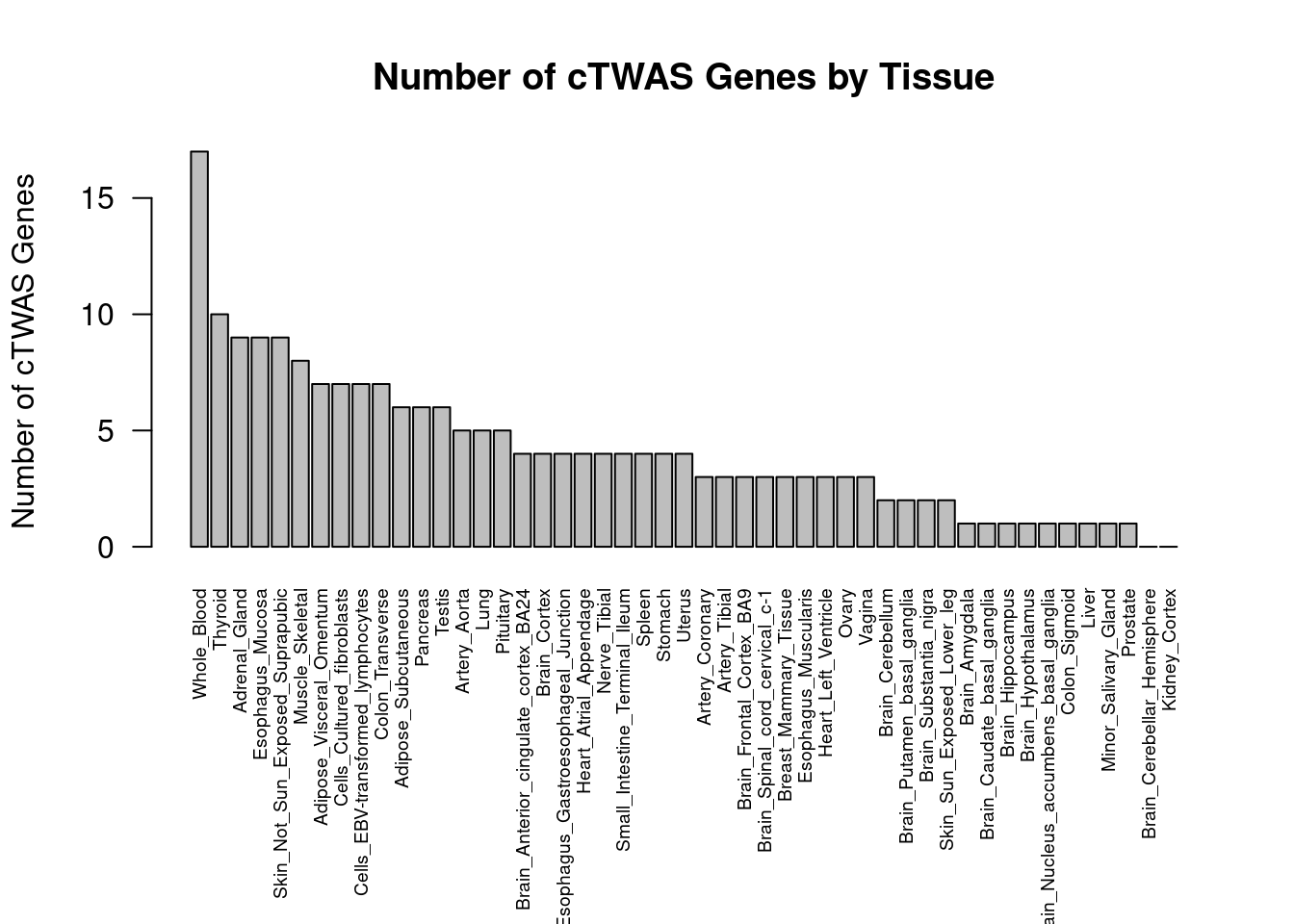
#barplot of number of cTWAS genes in each tissue
df_plot <- -sort(-sapply(groups[groups!="None"], function(x){length(df_group[[x]][["ctwas"]])}))
par(mar=c(10.1, 4.1, 4.1, 2.1))
barplot(df_plot, las=2, ylab="Number of cTWAS Genes", main="Number of cTWAS Genes by Tissue Group")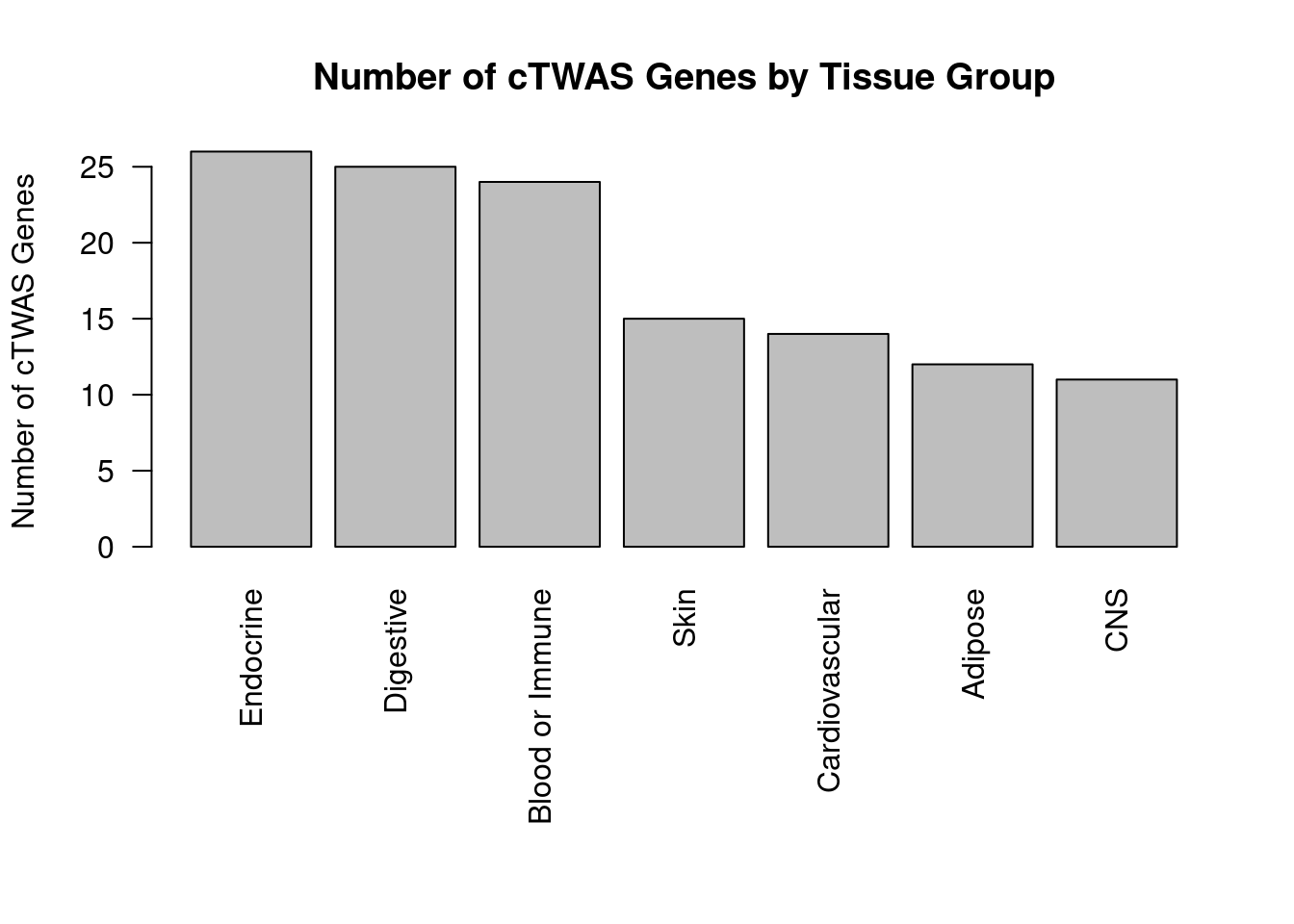
Enrichment analysis for cTWAS genes in each tissue group
GO
suppressWarnings(rm(group_enrichment_results))
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(ctwas_genes_group, dbs)
for (db in dbs){
cat(paste0("\n", db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
if (nrow(enrich_results)>0){
if (!exists("group_enrichment_results")){
group_enrichment_results <- cbind(group, db, enrich_results)
} else {
group_enrichment_results <- rbind(group_enrichment_results, cbind(group, db, enrich_results))
}
}
}
}Adipose
Number of cTWAS Genes in Tissue Group: 12
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)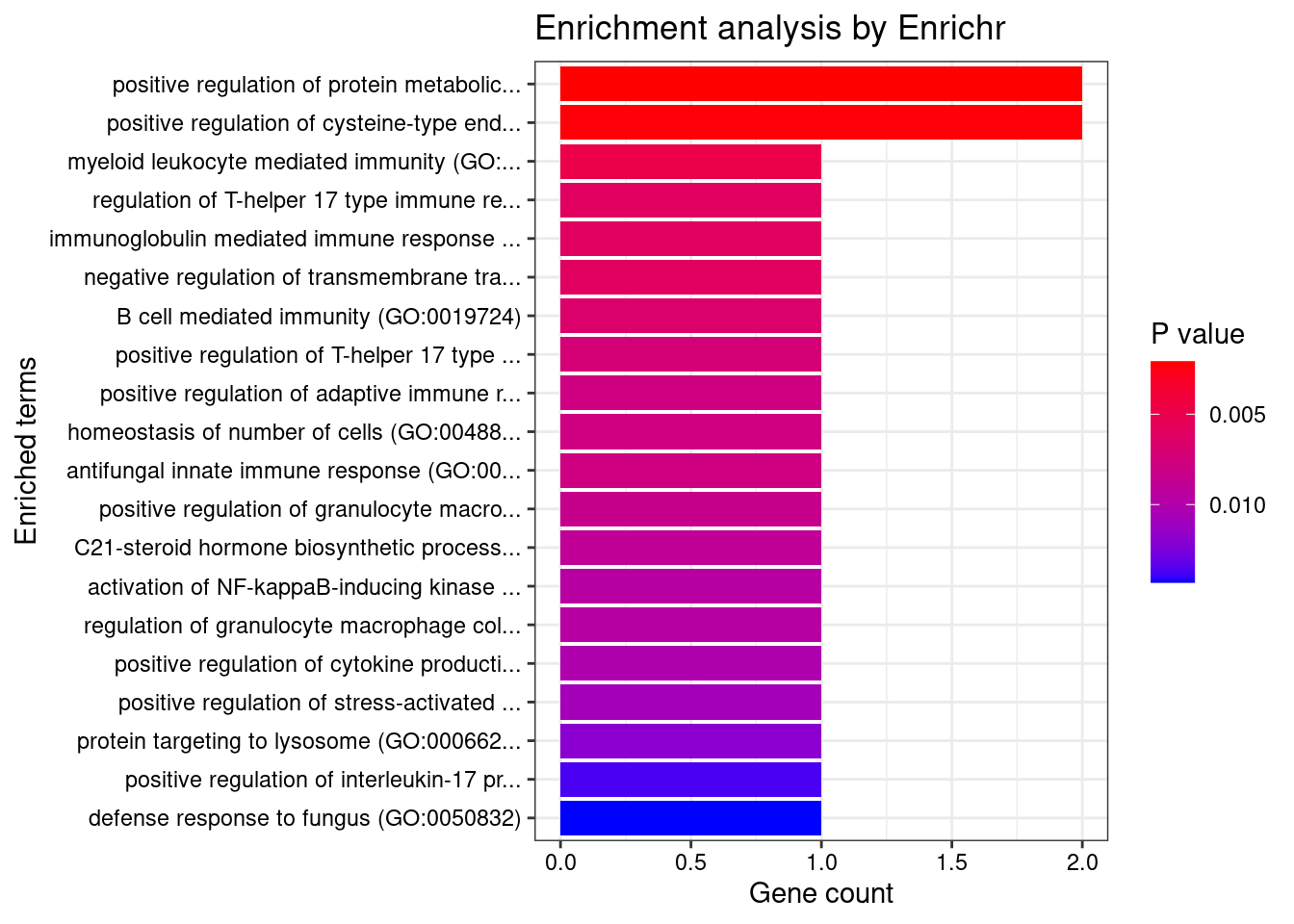
Endocrine
Number of cTWAS Genes in Tissue Group: 26
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)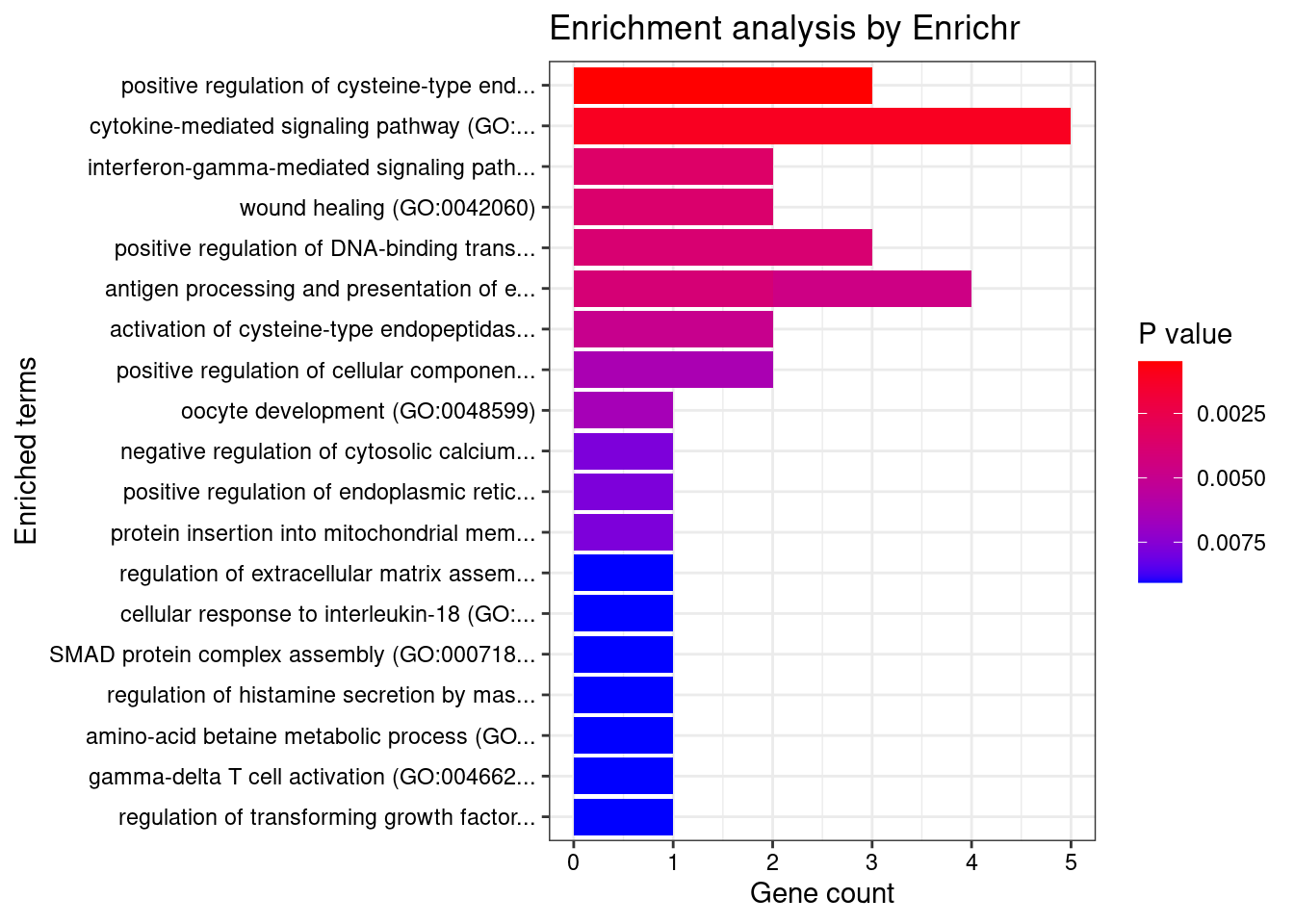
Cardiovascular
Number of cTWAS Genes in Tissue Group: 14
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)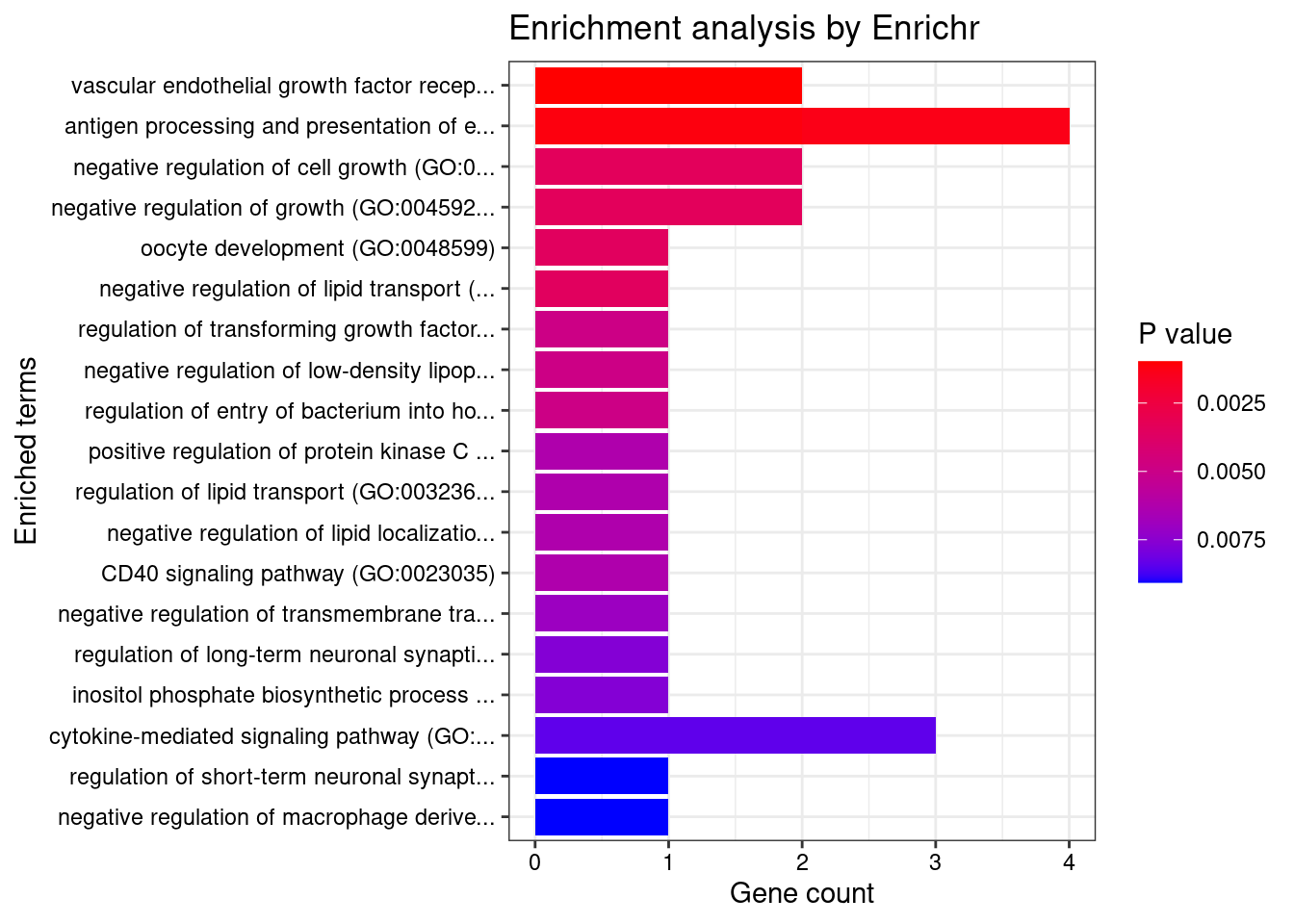
CNS
Number of cTWAS Genes in Tissue Group: 11
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)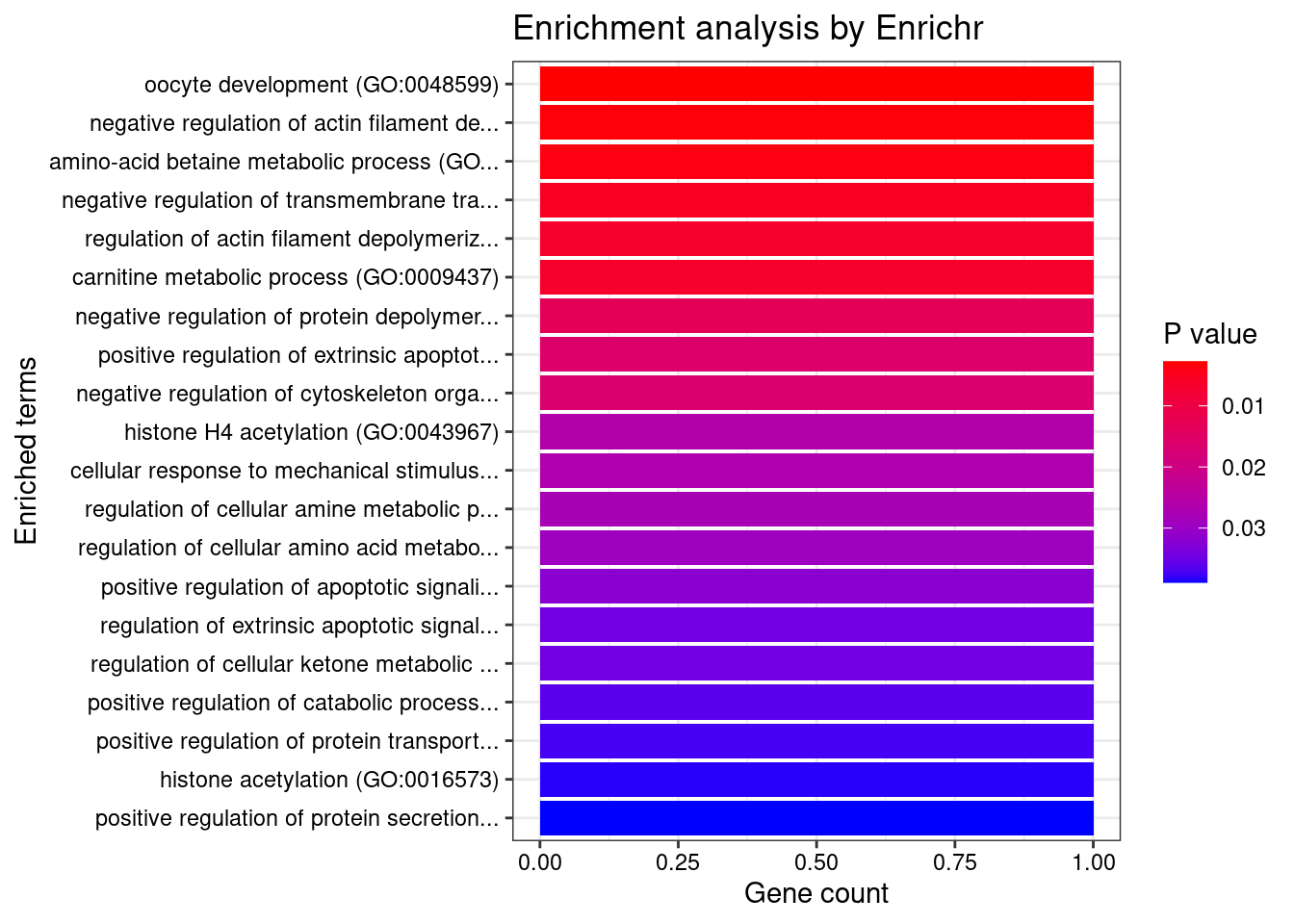
None
Number of cTWAS Genes in Tissue Group: 25
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 insulin-like growth factor receptor signaling pathway (GO:0048009) 2/11 0.009696623 GIGYF1;TSC2
2 interferon-gamma-mediated signaling pathway (GO:0060333) 3/68 0.009696623 CIITA;IFNGR2;HLA-DQB1
3 cytokine-mediated signaling pathway (GO:0019221) 6/621 0.009696623 CIITA;CD40;BCL2L11;TNFSF15;IFNGR2;HLA-DQB1
4 immune response-regulating cell surface receptor signaling pathway (GO:0002768) 2/14 0.010515793 CD40;MICB
5 cellular response to interferon-gamma (GO:0071346) 3/121 0.028036948 CIITA;IFNGR2;HLA-DQB1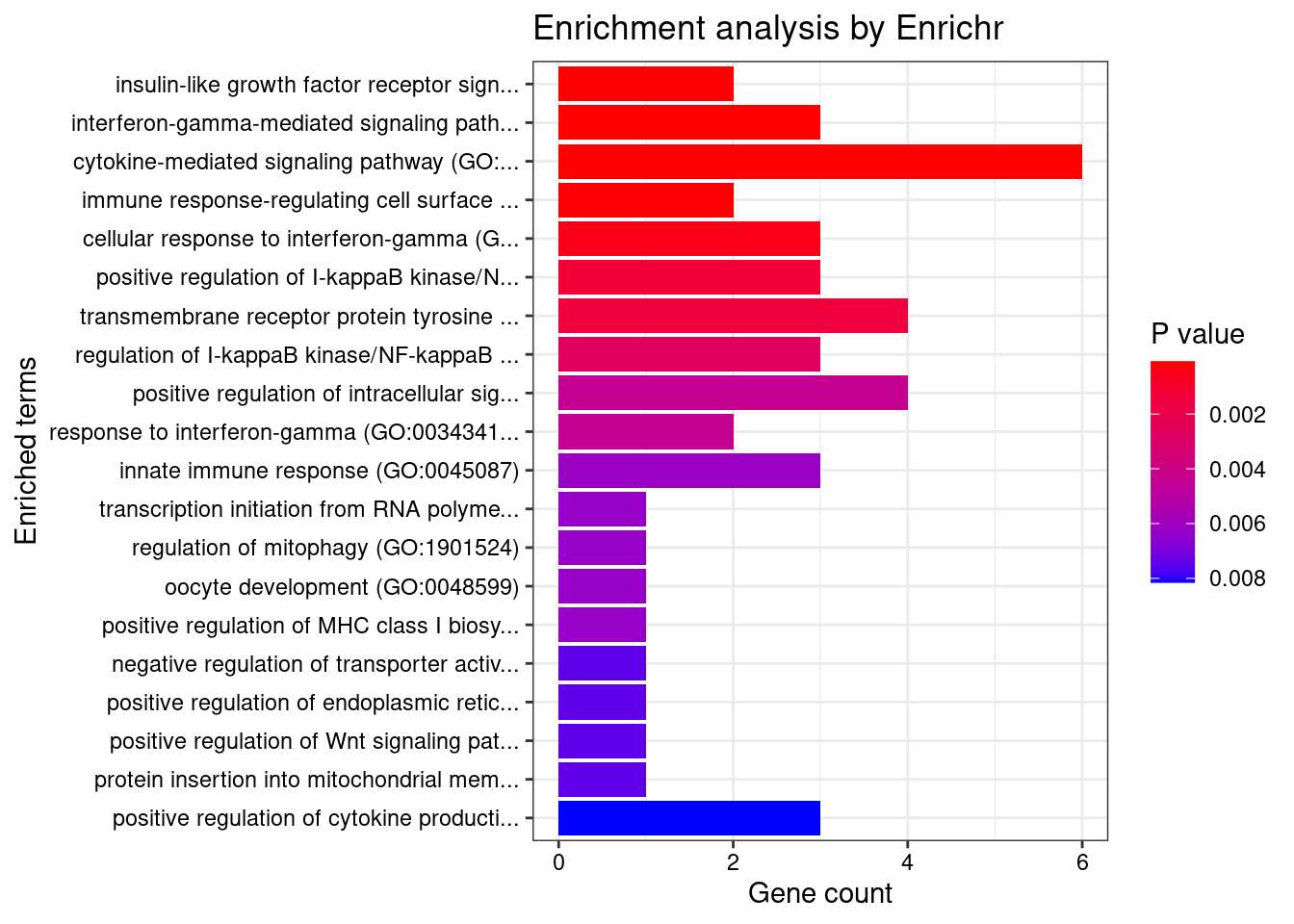
Skin
Number of cTWAS Genes in Tissue Group: 15
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 regulation of receptor binding (GO:1900120) 2/10 0.005085314 ADAM15;MMP9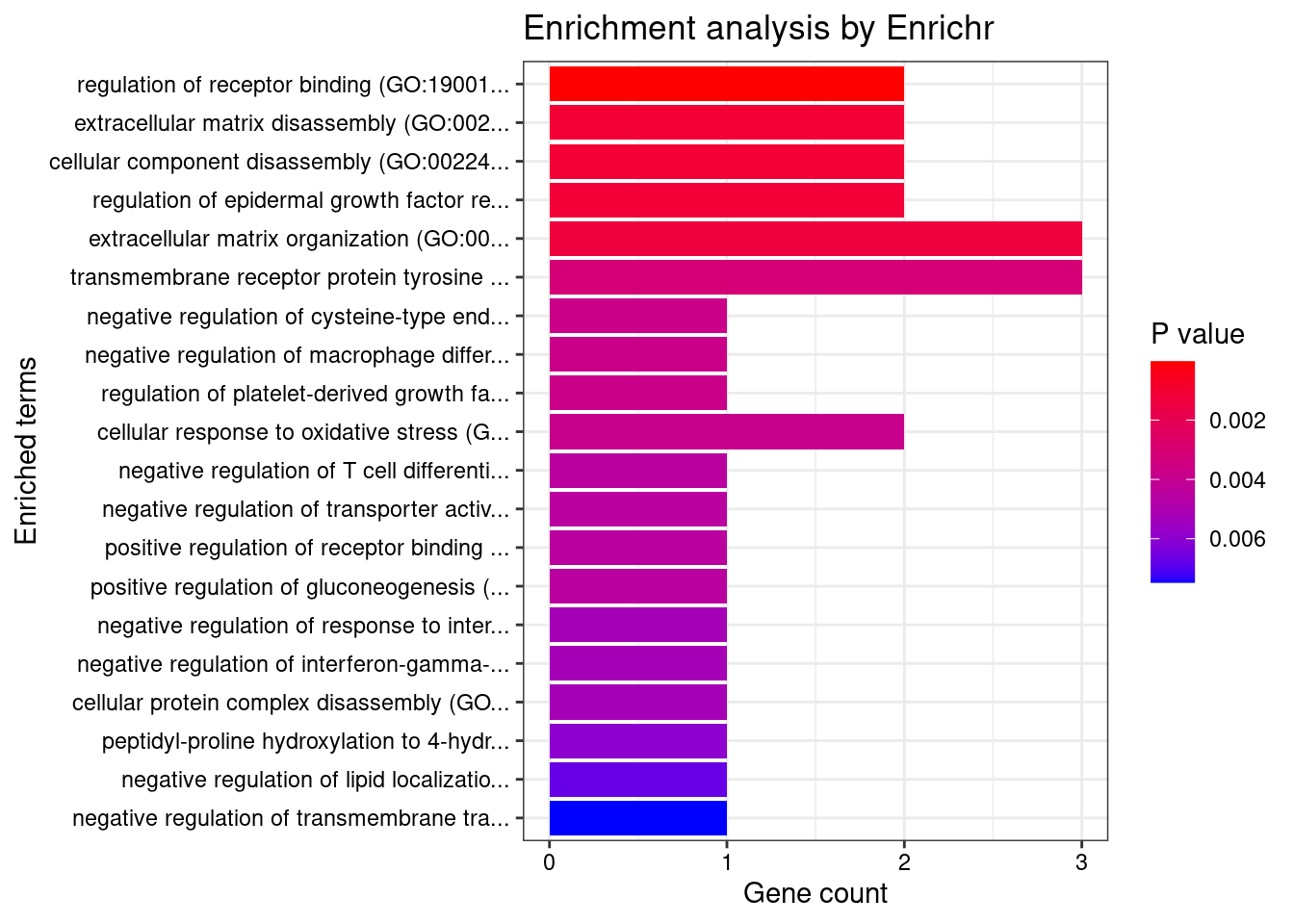
Blood or Immune
Number of cTWAS Genes in Tissue Group: 24
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 positive regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043123) 5/171 0.0004961019 PPP5C;NDFIP1;CARD9;LTBR;RBCK1
2 regulation of I-kappaB kinase/NF-kappaB signaling (GO:0043122) 5/224 0.0009302676 PPP5C;NDFIP1;CARD9;LTBR;RBCK1
3 positive regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043372) 2/8 0.0039624432 SOCS1;TNFSF4
4 regulation of CD4-positive, alpha-beta T cell differentiation (GO:0043370) 2/12 0.0064107688 SOCS1;TNFSF4
5 positive regulation of CD4-positive, alpha-beta T cell activation (GO:2000516) 2/13 0.0064107688 SOCS1;TNFSF4
6 positive regulation of alpha-beta T cell differentiation (GO:0046638) 2/14 0.0064107688 SOCS1;TNFSF4
7 tumor necrosis factor-mediated signaling pathway (GO:0033209) 3/116 0.0151302186 TNFSF15;TNFSF4;LTBR
8 positive regulation of intracellular signal transduction (GO:1902533) 5/546 0.0151302186 PPP5C;NDFIP1;CARD9;LTBR;RBCK1
9 regulation of regulatory T cell differentiation (GO:0045589) 2/26 0.0151302186 SOCS1;TNFSF4
10 regulation of interleukin-17 production (GO:0032660) 2/33 0.0207646020 TNFSF4;CARD9
11 cytokine-mediated signaling pathway (GO:0019221) 5/621 0.0207646020 IL18RAP;SOCS1;TNFSF15;TNFSF4;LTBR
12 positive regulation of NF-kappaB transcription factor activity (GO:0051092) 3/155 0.0211122693 IL18RAP;CARD9;RBCK1
13 positive regulation of T cell differentiation (GO:0045582) 2/43 0.0287438173 SOCS1;TNFSF4
14 cellular response to tumor necrosis factor (GO:0071356) 3/194 0.0345455953 TNFSF15;TNFSF4;LTBR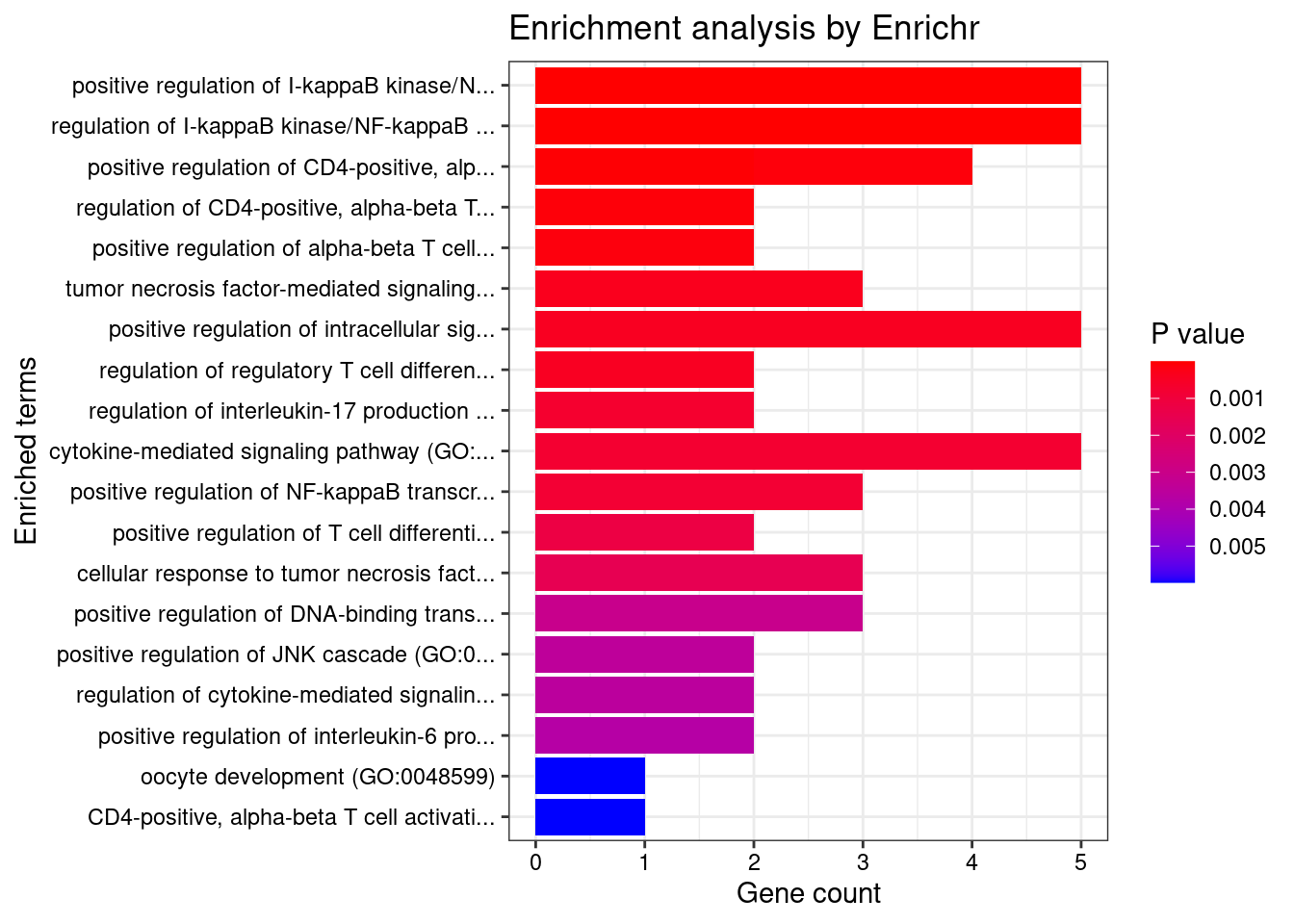
Digestive
Number of cTWAS Genes in Tissue Group: 25
Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.
GO_Biological_Process_2021
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)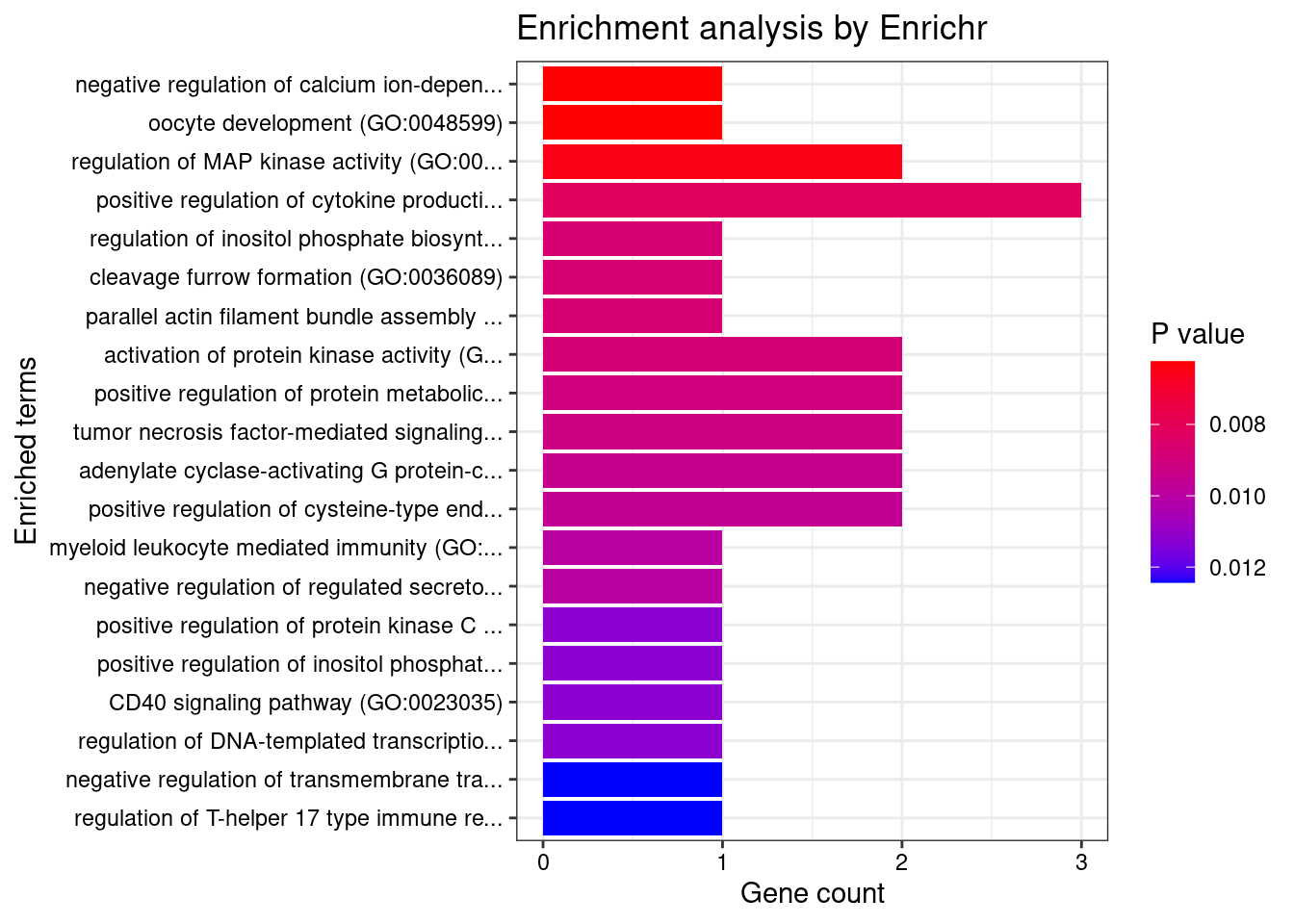
# if (exists("group_enrichment_results")){
# save(group_enrichment_results, file="group_enrichment_results.RData")
# }KEGG
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
databases <- c("pathway_KEGG")
enrichResult <- WebGestaltR(enrichMethod="ORA", organism="hsapiens",
interestGene=ctwas_genes_group, referenceGene=background_group,
enrichDatabase=databases, interestGeneType="genesymbol",
referenceGeneType="genesymbol", isOutput=F)
if (!is.null(enrichResult)){
print(enrichResult[,c("description", "size", "overlap", "FDR", "userId")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 12
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Endocrine
Number of cTWAS Genes in Tissue Group: 26
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...
description size overlap FDR userId
1 Inflammatory bowel disease (IBD) 46 4 0.001152104 IL18RAP;IFNGR2;HLA-DQB1;SMAD3
2 Leishmaniasis 53 3 0.041830029 IFNGR2;HLA-DQB1;NCF4
Cardiovascular
Number of cTWAS Genes in Tissue Group: 14
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
CNS
Number of cTWAS Genes in Tissue Group: 11
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
None
Number of cTWAS Genes in Tissue Group: 25
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...
description size overlap FDR userId
1 Toxoplasmosis 99 4 0.01497774 CIITA;HLA-DQB1;CD40;IFNGR2
2 Tuberculosis 144 4 0.02938877 CARD9;CIITA;HLA-DQB1;IFNGR2
3 Leishmaniasis 61 3 0.02938877 NCF4;HLA-DQB1;IFNGR2
Skin
Number of cTWAS Genes in Tissue Group: 15
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Blood or Immune
Number of cTWAS Genes in Tissue Group: 24
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!
Digestive
Number of cTWAS Genes in Tissue Group: 25
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum = minNum, : No significant gene set is identified based on FDR 0.05!DisGeNET
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
res_enrich <- disease_enrichment(entities=ctwas_genes_group, vocabulary = "HGNC", database = "CURATED")
if (any(res_enrich@qresult$FDR < 0.05)){
print(res_enrich@qresult[res_enrich@qresult$FDR < 0.05, c("Description", "FDR", "Ratio", "BgRatio")])
}
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 12EVA1B gene(s) from the input list not found in DisGeNET CURATEDSDCCAG3 gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDFGFR1OP gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
14 Inflammatory Bowel Diseases 0.0001142692 3/8 35/9703
7 Crohn Disease 0.0043977187 2/8 50/9703
10 Enteritis 0.0043977187 1/8 1/9703
26 Crohn's disease of large bowel 0.0043977187 2/8 44/9703
32 Crohn's disease of the ileum 0.0043977187 2/8 44/9703
35 Regional enteritis 0.0043977187 2/8 44/9703
36 IIeocolitis 0.0043977187 2/8 44/9703
43 Deep seated dermatophytosis 0.0043977187 1/8 1/9703
46 Inflammatory Bowel Disease 10 0.0043977187 1/8 1/9703
6 Ulcerative Colitis 0.0054401014 2/8 63/9703
45 Candidiasis, Familial, 2 0.0065941981 1/8 2/9703
47 Leukoencephalopathy, Cystic, Without Megalencephaly 0.0065941981 1/8 2/9703
12 Graves Disease 0.0221992845 1/8 9/9703
27 Megaconial Myopathies 0.0221992845 1/8 9/9703
28 Pleoconial Myopathies 0.0221992845 1/8 9/9703
37 Luft Disease 0.0221992845 1/8 9/9703
21 Ankylosing spondylitis 0.0241003492 1/8 11/9703
29 Mitochondrial Myopathies 0.0241003492 1/8 11/9703
Endocrine
Number of cTWAS Genes in Tissue Group: 26GIGYF1 gene(s) from the input list not found in DisGeNET CURATEDLINC01623 gene(s) from the input list not found in DisGeNET CURATEDRP11-386E5.1 gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDVAMP3 gene(s) from the input list not found in DisGeNET CURATEDRP11-574K11.29 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
72 Crohn's disease of large bowel 5.843026e-05 4/19 44/9703
81 Crohn's disease of the ileum 5.843026e-05 4/19 44/9703
112 Regional enteritis 5.843026e-05 4/19 44/9703
133 IIeocolitis 5.843026e-05 4/19 44/9703
16 Crohn Disease 7.871305e-05 4/19 50/9703
12 Ulcerative Colitis 1.669433e-04 4/19 63/9703
34 Inflammatory Bowel Diseases 9.849761e-04 3/19 35/9703
58 Systemic Scleroderma 1.309921e-02 2/19 19/9703
21 Enteritis 1.468825e-02 1/19 1/9703
26 IGA Glomerulonephritis 1.468825e-02 2/19 34/9703
49 Niemann-Pick Diseases 1.468825e-02 1/19 1/9703
74 Kleine-Levin Syndrome 1.468825e-02 1/19 1/9703
82 Niemann-Pick Disease, Type A 1.468825e-02 1/19 1/9703
83 Niemann-Pick Disease, Type B 1.468825e-02 1/19 1/9703
84 Niemann-Pick Disease, Type E 1.468825e-02 1/19 1/9703
135 Deep seated dermatophytosis 1.468825e-02 1/19 1/9703
144 LOEYS-DIETZ SYNDROME 3 1.468825e-02 1/19 1/9703
145 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 1.468825e-02 1/19 1/9703
151 NEPHROTIC SYNDROME, TYPE 9 1.468825e-02 1/19 1/9703
153 IMMUNODEFICIENCY 28 1.468825e-02 1/19 1/9703
155 EPILEPSY, IDIOPATHIC GENERALIZED, SUSCEPTIBILITY TO, 14 1.468825e-02 1/19 1/9703
156 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 34 1.468825e-02 1/19 1/9703
157 SPASTIC PARAPLEGIA 73, AUTOSOMAL DOMINANT 1.468825e-02 1/19 1/9703
41 Megaesophagus 2.588656e-02 1/19 2/9703
138 Candidiasis, Familial, 2 2.588656e-02 1/19 2/9703
169 Limbic encephalitis with leucine-rich glioma-inactivated 1 antibodies 2.588656e-02 1/19 2/9703
28 Severe Dengue 3.478067e-02 1/19 3/9703
54 Bullous pemphigoid 3.478067e-02 1/19 3/9703
93 Dengue Shock Syndrome 3.478067e-02 1/19 3/9703
25 Esophageal Achalasia 4.071532e-02 1/19 4/9703
50 Oropharyngeal Neoplasms 4.071532e-02 1/19 4/9703
130 Idiopathic achalasia of esophagus 4.071532e-02 1/19 4/9703
140 Oropharyngeal Carcinoma 4.071532e-02 1/19 4/9703
2 Aneurysm, Dissecting 4.415657e-02 1/19 5/9703
91 Dissection of aorta 4.415657e-02 1/19 5/9703
137 Loeys-Dietz Aortic Aneurysm Syndrome 4.415657e-02 1/19 5/9703
158 Dissection, Blood Vessel 4.415657e-02 1/19 5/9703
165 Loeys-Dietz Syndrome, Type 1a 4.415657e-02 1/19 5/9703
3 Angioedema 4.789699e-02 1/19 6/9703
10 Cholangitis, Sclerosing 4.789699e-02 1/19 6/9703
38 Fibroid Tumor 4.789699e-02 1/19 6/9703
89 Idiopathic generalized epilepsy 4.789699e-02 1/19 6/9703
Cardiovascular
Number of cTWAS Genes in Tissue Group: 14SH3D21 gene(s) from the input list not found in DisGeNET CURATEDAGAP5 gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDAPBB1 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
27 Hyperimmunoglobulin M syndrome 0.02265938 1/8 3/9703
47 Hyper-IgM Immunodeficiency Syndrome, Type 2 0.02265938 1/8 3/9703
48 Hyper-IgM Immunodeficiency Syndrome, Type 3 0.02265938 1/8 3/9703
49 Hyper-IgM Immunodeficiency Syndrome, Type 5 0.02265938 1/8 3/9703
53 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 0.02265938 1/8 1/9703
54 IMMUNODEFICIENCY 28 0.02265938 1/8 1/9703
14 Mucocutaneous Lymph Node Syndrome 0.02588709 1/8 4/9703
CNS
Number of cTWAS Genes in Tissue Group: 11LINC02128 gene(s) from the input list not found in DisGeNET CURATEDAPBB1 gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDPLEKHH2 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDSH3D21 gene(s) from the input list not found in DisGeNET CURATEDC11orf58 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
7 SPASTIC PARAPLEGIA 73, AUTOSOMAL DOMINANT 0.002473717 1/3 1/9703
3 Malaria 0.016459166 1/3 20/9703
5 Systemic Scleroderma 0.016459166 1/3 19/9703
8 Peripheral Nervous System Diseases 0.033213053 1/3 54/9703
1 Alloxan Diabetes 0.037746487 1/3 108/9703
2 Diabetes Mellitus, Experimental 0.037746487 1/3 108/9703
6 Streptozotocin Diabetes 0.037746487 1/3 108/9703
None
Number of cTWAS Genes in Tissue Group: 25GIGYF1 gene(s) from the input list not found in DisGeNET CURATEDMUC3A gene(s) from the input list not found in DisGeNET CURATEDNDFIP1 gene(s) from the input list not found in DisGeNET CURATEDDEF8 gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDC11orf58 gene(s) from the input list not found in DisGeNET CURATEDC3orf62 gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
1 Addison Disease 0.02472174 1/16 3/9703
6 Brain Diseases 0.02472174 2/16 25/9703
12 Ulcerative Colitis 0.02472174 2/16 63/9703
19 Enteritis 0.02472174 1/16 1/9703
24 IGA Glomerulonephritis 0.02472174 2/16 34/9703
27 Severe Dengue 0.02472174 1/16 3/9703
35 Inflammatory Bowel Diseases 0.02472174 2/16 35/9703
43 Megaesophagus 0.02472174 1/16 2/9703
49 Multiple Sclerosis 0.02472174 2/16 45/9703
57 Bullous pemphigoid 0.02472174 1/16 3/9703
65 Tuberous Sclerosis 0.02472174 1/16 3/9703
68 Encephalopathies 0.02472174 2/16 27/9703
75 Kleine-Levin Syndrome 0.02472174 1/16 1/9703
80 Fibrous skin tumor of tuberous sclerosis 0.02472174 1/16 3/9703
82 Petit mal status 0.02472174 2/16 67/9703
87 Hyperimmunoglobulin M syndrome 0.02472174 1/16 3/9703
91 Grand Mal Status Epilepticus 0.02472174 2/16 67/9703
94 Dengue Shock Syndrome 0.02472174 1/16 3/9703
100 Complex Partial Status Epilepticus 0.02472174 2/16 67/9703
128 Multiple Sclerosis, Acute Fulminating 0.02472174 2/16 45/9703
133 Status Epilepticus, Subclinical 0.02472174 2/16 67/9703
134 Non-Convulsive Status Epilepticus 0.02472174 2/16 67/9703
135 Simple Partial Status Epilepticus 0.02472174 2/16 67/9703
149 Deep seated dermatophytosis 0.02472174 1/16 1/9703
154 Hyper-IgM Immunodeficiency Syndrome, Type 2 0.02472174 1/16 3/9703
155 Hyper-IgM Immunodeficiency Syndrome, Type 3 0.02472174 1/16 3/9703
156 Hyper-IgM Immunodeficiency Syndrome, Type 5 0.02472174 1/16 3/9703
157 Polycystic kidneys, severe infantile with tuberous sclerosis 0.02472174 1/16 2/9703
158 FOCAL CORTICAL DYSPLASIA OF TAYLOR 0.02472174 1/16 3/9703
159 Focal Cortical Dysplasia of Taylor, Type IIa 0.02472174 1/16 3/9703
160 Focal Cortical Dysplasia of Taylor, Type IIb 0.02472174 1/16 3/9703
161 TUBEROUS SCLEROSIS 1 (disorder) 0.02472174 1/16 2/9703
163 Candidiasis, Familial, 2 0.02472174 1/16 2/9703
164 Bare Lymphocyte Syndrome, Type II, Complementation Group A 0.02472174 1/16 1/9703
165 TUBEROUS SCLEROSIS 2 (disorder) 0.02472174 1/16 2/9703
169 Leukoencephalopathy, Cystic, Without Megalencephaly 0.02472174 1/16 2/9703
174 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 0.02472174 1/16 1/9703
180 IMMUNODEFICIENCY 28 0.02472174 1/16 1/9703
182 EPILEPSY, IDIOPATHIC GENERALIZED, SUSCEPTIBILITY TO, 14 0.02472174 1/16 1/9703
183 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 34 0.02472174 1/16 1/9703
184 SPASTIC PARAPLEGIA 73, AUTOSOMAL DOMINANT 0.02472174 1/16 1/9703
194 Limbic encephalitis with leucine-rich glioma-inactivated 1 antibodies 0.02472174 1/16 2/9703
64 Status Epilepticus 0.02485467 2/16 68/9703
23 Esophageal Achalasia 0.02579867 1/16 4/9703
48 Mucocutaneous Lymph Node Syndrome 0.02579867 1/16 4/9703
54 Oropharyngeal Neoplasms 0.02579867 1/16 4/9703
137 Idiopathic achalasia of esophagus 0.02579867 1/16 4/9703
168 Oropharyngeal Carcinoma 0.02579867 1/16 4/9703
171 Bare lymphocyte syndrome 2 0.02579867 1/16 4/9703
179 Other License Status 0.02579867 1/16 4/9703
2 Angioedema 0.02658084 1/16 6/9703
11 Cholangitis, Sclerosing 0.02658084 1/16 6/9703
37 Jacksonian Seizure 0.02658084 2/16 101/9703
40 Fibroid Tumor 0.02658084 1/16 6/9703
67 Uterine Fibroids 0.02658084 1/16 7/9703
69 Epilepsy, Cryptogenic 0.02658084 2/16 82/9703
71 Complex partial seizures 0.02658084 2/16 101/9703
76 Generalized seizures 0.02658084 2/16 101/9703
77 Clonic Seizures 0.02658084 2/16 101/9703
78 Aura 0.02658084 2/16 82/9703
83 Visual seizure 0.02658084 2/16 101/9703
84 Tonic Seizures 0.02658084 2/16 102/9703
85 Epileptic drop attack 0.02658084 2/16 101/9703
86 Idiopathic generalized epilepsy 0.02658084 1/16 6/9703
98 Cryptogenic Infantile Spasms 0.02658084 1/16 6/9703
99 Symptomatic Infantile Spasms 0.02658084 1/16 6/9703
102 Seizures, Somatosensory 0.02658084 2/16 101/9703
103 Seizures, Auditory 0.02658084 2/16 101/9703
104 Olfactory seizure 0.02658084 2/16 101/9703
105 Gustatory seizure 0.02658084 2/16 101/9703
106 Vertiginous seizure 0.02658084 2/16 101/9703
110 Nodding spasm 0.02658084 1/16 6/9703
112 Jackknife Seizures 0.02658084 1/16 6/9703
116 Hypsarrhythmia 0.02658084 1/16 6/9703
119 Non-epileptic convulsion 0.02658084 2/16 101/9703
120 Single Seizure 0.02658084 2/16 101/9703
121 Awakening Epilepsy 0.02658084 2/16 82/9703
122 Atonic Absence Seizures 0.02658084 2/16 101/9703
130 Convulsive Seizures 0.02658084 2/16 101/9703
132 Seizures, Sensory 0.02658084 2/16 101/9703
136 Lymphangioleiomyomatosis 0.02658084 1/16 5/9703
152 spasmus nutans 0.02658084 1/16 6/9703
153 Salaam Seizures 0.02658084 1/16 6/9703
175 Nonepileptic Seizures 0.02658084 2/16 101/9703
181 Convulsions 0.02658084 2/16 102/9703
188 Absence Seizures 0.02658084 2/16 102/9703
189 Epileptic Seizures 0.02658084 2/16 101/9703
191 Generalized Absence Seizures 0.02658084 2/16 101/9703
108 Tonic - clonic seizures 0.02667637 2/16 104/9703
131 Seizures, Focal 0.02667637 2/16 104/9703
190 Myoclonic Seizures 0.02667637 2/16 104/9703
113 Primary sclerosing cholangitis 0.02736069 1/16 8/9703
117 Pure Gonadal Dysgenesis, 46, XX 0.02736069 1/16 8/9703
142 Gonadal Dysgenesis, 46,XX 0.02736069 1/16 8/9703
20 Epilepsy 0.02794754 2/16 109/9703
14 Febrile Convulsions 0.02950162 1/16 9/9703
25 Graves Disease 0.02950162 1/16 9/9703
88 Adult Hepatocellular Carcinoma 0.02950162 1/16 9/9703
34 Inflammation 0.03547360 2/16 127/9703
63 Ankylosing spondylitis 0.03547360 1/16 11/9703
38 Creutzfeldt-Jakob disease 0.04041983 1/16 13/9703
95 New Variant Creutzfeldt-Jakob Disease 0.04041983 1/16 13/9703
127 Creutzfeldt-Jakob Disease, Familial 0.04041983 1/16 13/9703
170 Gastro-enteropancreatic neuroendocrine tumor 0.04611859 1/16 15/9703
66 Urticaria 0.04822778 1/16 16/9703
129 Narcolepsy-Cataplexy Syndrome 0.04822778 1/16 16/9703
Skin
Number of cTWAS Genes in Tissue Group: 15NDFIP1 gene(s) from the input list not found in DisGeNET CURATEDAC007383.3 gene(s) from the input list not found in DisGeNET CURATEDAPEH gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATEDADAM15 gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
89 Crohn's disease of large bowel 2.191535e-06 4/9 44/9703
101 Crohn's disease of the ileum 2.191535e-06 4/9 44/9703
133 Regional enteritis 2.191535e-06 4/9 44/9703
157 IIeocolitis 2.191535e-06 4/9 44/9703
27 Crohn Disease 2.966955e-06 4/9 50/9703
94 Kleine-Levin Syndrome 1.619160e-02 1/9 1/9703
168 Inflammatory Bowel Disease 10 1.619160e-02 1/9 1/9703
172 Metaphyseal Anadysplasia 2 1.619160e-02 1/9 1/9703
177 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 1.619160e-02 1/9 1/9703
182 COMBINED OXIDATIVE PHOSPHORYLATION DEFICIENCY 20 1.619160e-02 1/9 1/9703
185 MYOPIA 25, AUTOSOMAL DOMINANT 1.619160e-02 1/9 1/9703
25 Ulcerative Colitis 2.225427e-02 2/9 63/9703
52 Megaesophagus 2.225427e-02 1/9 2/9703
126 Metaphyseal anadysplasia 2.225427e-02 1/9 2/9703
127 Mandibuloacral dysostosis 2.225427e-02 1/9 2/9703
190 Limbic encephalitis with leucine-rich glioma-inactivated 1 antibodies 2.225427e-02 1/9 2/9703
8 Asthma 2.426738e-02 2/9 80/9703
33 Glioblastoma 2.426738e-02 2/9 79/9703
66 Bullous pemphigoid 2.426738e-02 1/9 3/9703
67 Pericementitis 2.426738e-02 1/9 3/9703
116 Giant Cell Glioblastoma 2.426738e-02 2/9 84/9703
118 Stable angina 2.426738e-02 1/9 3/9703
32 Esophageal Achalasia 2.541249e-02 1/9 4/9703
49 Lupus Nephritis 2.541249e-02 1/9 4/9703
63 Oropharyngeal Neoplasms 2.541249e-02 1/9 4/9703
68 Periodontitis 2.541249e-02 1/9 4/9703
152 Idiopathic achalasia of esophagus 2.541249e-02 1/9 4/9703
170 Oropharyngeal Carcinoma 2.541249e-02 1/9 4/9703
3 Angioedema 2.732220e-02 1/9 6/9703
4 Aortic Aneurysm 2.732220e-02 1/9 7/9703
5 Aortic Diseases 2.732220e-02 1/9 6/9703
6 Aortic Rupture 2.732220e-02 1/9 5/9703
14 Bone neoplasms 2.732220e-02 1/9 8/9703
15 Cerebral Edema 2.732220e-02 1/9 8/9703
43 Lead Poisoning 2.732220e-02 1/9 7/9703
44 Leukemia, T-Cell 2.732220e-02 1/9 5/9703
53 Morphine Dependence 2.732220e-02 1/9 7/9703
55 Multiple Organ Failure 2.732220e-02 1/9 8/9703
91 Aortic Aneurysm, Thoracic 2.732220e-02 1/9 7/9703
108 Malignant Bone Neoplasm 2.732220e-02 1/9 8/9703
119 Aortic Aneurysm, Thoracoabdominal 2.732220e-02 1/9 7/9703
128 Vasogenic Cerebral Edema 2.732220e-02 1/9 8/9703
129 Cytotoxic Cerebral Edema 2.732220e-02 1/9 8/9703
132 Morphine Abuse 2.732220e-02 1/9 7/9703
138 Aortic Aneurysm, Ruptured 2.732220e-02 1/9 5/9703
141 Vasogenic Brain Edema 2.732220e-02 1/9 8/9703
142 Cytotoxic Brain Edema 2.732220e-02 1/9 8/9703
163 Brain Edema 2.732220e-02 1/9 8/9703
164 Glioblastoma Multiforme 2.732220e-02 2/9 111/9703
173 Juvenile pauciarticular chronic arthritis 2.732220e-02 1/9 7/9703
174 Neointima 2.732220e-02 1/9 6/9703
175 Neointima Formation 2.732220e-02 1/9 6/9703
192 Marfan Syndrome, Type I 3.014509e-02 1/9 9/9703
90 Aortic Aneurysm, Abdominal 3.286073e-02 1/9 10/9703
7 Rheumatoid Arthritis 3.444693e-02 2/9 174/9703
31 Diabetic Neuropathies 3.444693e-02 1/9 14/9703
42 Creutzfeldt-Jakob disease 3.444693e-02 1/9 13/9703
51 Marfan Syndrome 3.444693e-02 1/9 11/9703
88 Premature Birth 3.444693e-02 1/9 11/9703
95 Centriacinar Emphysema 3.444693e-02 1/9 12/9703
100 Panacinar Emphysema 3.444693e-02 1/9 12/9703
102 Symmetric Diabetic Proximal Motor Neuropathy 3.444693e-02 1/9 14/9703
103 Asymmetric Diabetic Proximal Motor Neuropathy 3.444693e-02 1/9 14/9703
104 Diabetic Mononeuropathy 3.444693e-02 1/9 14/9703
105 Diabetic Polyneuropathies 3.444693e-02 1/9 14/9703
106 Diabetic Amyotrophy 3.444693e-02 1/9 14/9703
107 Diabetic Autonomic Neuropathy 3.444693e-02 1/9 14/9703
122 New Variant Creutzfeldt-Jakob Disease 3.444693e-02 1/9 13/9703
124 Diabetic Asymmetric Polyneuropathy 3.444693e-02 1/9 14/9703
143 Diabetic Neuralgia 3.444693e-02 1/9 14/9703
144 Creutzfeldt-Jakob Disease, Familial 3.444693e-02 1/9 13/9703
171 Focal Emphysema 3.444693e-02 1/9 12/9703
83 Urticaria 3.827237e-02 1/9 16/9703
146 Narcolepsy-Cataplexy Syndrome 3.827237e-02 1/9 16/9703
54 Moyamoya Disease 3.906397e-02 1/9 17/9703
58 Narcolepsy 3.906397e-02 1/9 17/9703
71 Pulmonary Emphysema 3.906397e-02 1/9 17/9703
62 Oral Submucous Fibrosis 4.029811e-02 1/9 18/9703
80 Gastric ulcer 4.029811e-02 1/9 18/9703
77 Systemic Scleroderma 4.198789e-02 1/9 19/9703
9 Astrocytoma 4.592573e-02 1/9 25/9703
28 Drug Eruptions 4.592573e-02 1/9 23/9703
73 Pyemia 4.592573e-02 1/9 24/9703
78 Septicemia 4.592573e-02 1/9 24/9703
93 Subependymal Giant Cell Astrocytoma 4.592573e-02 1/9 25/9703
98 Sepsis 4.592573e-02 1/9 24/9703
109 Juvenile Pilocytic Astrocytoma 4.592573e-02 1/9 25/9703
110 Diffuse Astrocytoma 4.592573e-02 1/9 25/9703
115 Pilocytic Astrocytoma 4.592573e-02 1/9 25/9703
117 Childhood Cerebral Astrocytoma 4.592573e-02 1/9 25/9703
125 Morbilliform Drug Reaction 4.592573e-02 1/9 23/9703
131 Mixed oligoastrocytoma 4.592573e-02 1/9 25/9703
139 Cerebral Astrocytoma 4.592573e-02 1/9 25/9703
140 Intracranial Astrocytoma 4.592573e-02 1/9 25/9703
165 Grade I Astrocytoma 4.592573e-02 1/9 25/9703
166 Severe Sepsis 4.592573e-02 1/9 24/9703
112 Protoplasmic astrocytoma 4.629633e-02 1/9 26/9703
113 Gemistocytic astrocytoma 4.629633e-02 1/9 26/9703
114 Fibrillary Astrocytoma 4.629633e-02 1/9 26/9703
75 Retinal Diseases 4.710555e-02 1/9 27/9703
111 Anaplastic astrocytoma 4.710555e-02 1/9 27/9703
37 Hyperplasia 4.788193e-02 1/9 28/9703
176 Cerebral Hemorrhage 4.788193e-02 1/9 28/9703
Blood or Immune
Number of cTWAS Genes in Tissue Group: 24NDFIP1 gene(s) from the input list not found in DisGeNET CURATEDFAM171B gene(s) from the input list not found in DisGeNET CURATEDPPP5C gene(s) from the input list not found in DisGeNET CURATEDAF064858.11 gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDGIGYF1 gene(s) from the input list not found in DisGeNET CURATEDOR2H2 gene(s) from the input list not found in DisGeNET CURATEDTMEM229B gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
22 Inflammatory Bowel Diseases 0.001616047 3/14 35/9703
5 Ulcerative Colitis 0.004786997 3/14 63/9703
2 Brain Diseases 0.016995350 2/14 25/9703
10 Enteritis 0.016995350 1/14 1/9703
45 Encephalopathies 0.016995350 2/14 27/9703
91 Deep seated dermatophytosis 0.016995350 1/14 1/9703
100 EPILEPSY, IDIOPATHIC GENERALIZED, SUSCEPTIBILITY TO, 14 0.016995350 1/14 1/9703
101 EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 34 0.016995350 1/14 1/9703
106 Polyglucosan body myopathy type 1 0.016995350 1/14 1/9703
92 Candidiasis, Familial, 2 0.025475944 1/14 2/9703
94 Leukoencephalopathy, Cystic, Without Megalencephaly 0.025475944 1/14 2/9703
98 POLYGLUCOSAN BODY MYOPATHY 1 WITH OR WITHOUT IMMUNODEFICIENCY 0.025475944 1/14 2/9703
Digestive
Number of cTWAS Genes in Tissue Group: 25RP11-386E5.1 gene(s) from the input list not found in DisGeNET CURATEDADCY9 gene(s) from the input list not found in DisGeNET CURATEDZGLP1 gene(s) from the input list not found in DisGeNET CURATEDRGS14 gene(s) from the input list not found in DisGeNET CURATEDGIGYF1 gene(s) from the input list not found in DisGeNET CURATEDSTXBP3 gene(s) from the input list not found in DisGeNET CURATEDSPNS2 gene(s) from the input list not found in DisGeNET CURATEDSPIRE2 gene(s) from the input list not found in DisGeNET CURATEDOSER1 gene(s) from the input list not found in DisGeNET CURATEDC1orf106 gene(s) from the input list not found in DisGeNET CURATEDOAZ3 gene(s) from the input list not found in DisGeNET CURATEDEVA1B gene(s) from the input list not found in DisGeNET CURATED Description FDR Ratio BgRatio
20 Inflammatory Bowel Diseases 0.001404992 3/13 35/9703
2 Rheumatoid Arthritis 0.016159544 3/13 174/9703
12 Enteritis 0.016159544 1/13 1/9703
43 Crohn's disease of large bowel 0.016159544 2/13 44/9703
51 Crohn's disease of the ileum 0.016159544 2/13 44/9703
68 Regional enteritis 0.016159544 2/13 44/9703
81 IIeocolitis 0.016159544 2/13 44/9703
92 Deep seated dermatophytosis 0.016159544 1/13 1/9703
101 Inflammatory Bowel Disease 10 0.016159544 1/13 1/9703
104 GRANULOMATOUS DISEASE, CHRONIC, AUTOSOMAL RECESSIVE, CYTOCHROME b-POSITIVE, TYPE III 0.016159544 1/13 1/9703
109 IMMUNODEFICIENCY 28 0.016159544 1/13 1/9703
10 Crohn Disease 0.019094845 2/13 50/9703
83 Hurthle Cell Tumor 0.019584355 1/13 2/9703
94 Oxyphilic Adenoma 0.019584355 1/13 2/9703
100 Candidiasis, Familial, 2 0.019584355 1/13 2/9703
102 Leukoencephalopathy, Cystic, Without Megalencephaly 0.019584355 1/13 2/9703
9 Ulcerative Colitis 0.021278864 2/13 63/9703
56 Hyperimmunoglobulin M syndrome 0.022368278 1/13 3/9703
96 Hyper-IgM Immunodeficiency Syndrome, Type 2 0.022368278 1/13 3/9703
97 Hyper-IgM Immunodeficiency Syndrome, Type 3 0.022368278 1/13 3/9703
98 Hyper-IgM Immunodeficiency Syndrome, Type 5 0.022368278 1/13 3/9703
25 Mucocutaneous Lymph Node Syndrome 0.028451115 1/13 4/9703
103 Juvenile pauciarticular chronic arthritis 0.047536429 1/13 7/9703Gene sets curated by Macarthur Lab
gene_set_dir <- "/project2/mstephens/wcrouse/gene_sets/"
gene_set_files <- c("gwascatalog.tsv",
"mgi_essential.tsv",
"core_essentials_hart.tsv",
"clinvar_path_likelypath.tsv",
"fda_approved_drug_targets.tsv")
for (group in names(df_group)){
cat(paste0(group, "\n\n"))
ctwas_genes_group <- df_group[[group]]$ctwas
background_group <- df_group[[group]]$background
cat(paste0("Number of cTWAS Genes in Tissue Group: ", length(ctwas_genes_group), "\n\n"))
gene_sets <- lapply(gene_set_files, function(x){as.character(read.table(paste0(gene_set_dir, x))[,1])})
names(gene_sets) <- sapply(gene_set_files, function(x){unlist(strsplit(x, "[.]"))[1]})
gene_lists <- list(ctwas_genes_group=ctwas_genes_group)
#genes in gene_sets filtered to ensure inclusion in background
gene_sets <- lapply(gene_sets, function(x){x[x %in% background_group]})
#hypergeometric test
hyp_score <- data.frame()
size <- c()
ngenes <- c()
for (i in 1:length(gene_sets)) {
for (j in 1:length(gene_lists)){
group1 <- length(gene_sets[[i]])
group2 <- length(as.vector(gene_lists[[j]]))
size <- c(size, group1)
Overlap <- length(intersect(gene_sets[[i]],as.vector(gene_lists[[j]])))
ngenes <- c(ngenes, Overlap)
Total <- length(background_group)
hyp_score[i,j] <- phyper(Overlap-1, group2, Total-group2, group1,lower.tail=F)
}
}
rownames(hyp_score) <- names(gene_sets)
colnames(hyp_score) <- names(gene_lists)
#multiple testing correction
hyp_score_padj <- apply(hyp_score,2, p.adjust, method="BH", n=(nrow(hyp_score)*ncol(hyp_score)))
hyp_score_padj <- as.data.frame(hyp_score_padj)
hyp_score_padj$gene_set <- rownames(hyp_score_padj)
hyp_score_padj$nset <- size
hyp_score_padj$ngenes <- ngenes
hyp_score_padj$percent <- ngenes/size
hyp_score_padj <- hyp_score_padj[order(hyp_score_padj$ctwas_genes),]
colnames(hyp_score_padj)[1] <- "padj"
hyp_score_padj <- hyp_score_padj[,c(2:5,1)]
rownames(hyp_score_padj)<- NULL
print(hyp_score_padj)
cat("\n")
}Adipose
Number of cTWAS Genes in Tissue Group: 12
gene_set nset ngenes percent padj
1 gwascatalog 4578 5 0.0010921800 0.6802722
2 mgi_essential 1717 2 0.0011648224 0.6802722
3 clinvar_path_likelypath 2137 2 0.0009358914 0.6802722
4 fda_approved_drug_targets 258 1 0.0038759690 0.6802722
5 core_essentials_hart 207 0 0.0000000000 1.0000000
Endocrine
Number of cTWAS Genes in Tissue Group: 26
gene_set nset ngenes percent padj
1 gwascatalog 5394 13 0.002410085 0.08449444
2 mgi_essential 2022 4 0.001978239 0.50248159
3 clinvar_path_likelypath 2486 5 0.002011263 0.50248159
4 core_essentials_hart 236 0 0.000000000 1.00000000
5 fda_approved_drug_targets 305 0 0.000000000 1.00000000
Cardiovascular
Number of cTWAS Genes in Tissue Group: 14
gene_set nset ngenes percent padj
1 gwascatalog 5194 7 0.001347709 0.5666681
2 mgi_essential 1970 3 0.001522843 0.5666681
3 clinvar_path_likelypath 2404 3 0.001247920 0.5666681
4 core_essentials_hart 242 0 0.000000000 1.0000000
5 fda_approved_drug_targets 287 0 0.000000000 1.0000000
CNS
Number of cTWAS Genes in Tissue Group: 11
gene_set nset ngenes percent padj
1 gwascatalog 5426 4 0.0007371913 1
2 mgi_essential 2090 0 0.0000000000 1
3 core_essentials_hart 244 0 0.0000000000 1
4 clinvar_path_likelypath 2529 0 0.0000000000 1
5 fda_approved_drug_targets 316 0 0.0000000000 1
None
Number of cTWAS Genes in Tissue Group: 25
gene_set nset ngenes percent padj
1 gwascatalog 5635 14 0.002484472 0.03297449
2 clinvar_path_likelypath 2609 7 0.002683020 0.12827451
3 mgi_essential 2146 5 0.002329916 0.25705333
4 core_essentials_hart 255 0 0.000000000 1.00000000
5 fda_approved_drug_targets 323 0 0.000000000 1.00000000
Skin
Number of cTWAS Genes in Tissue Group: 15
gene_set nset ngenes percent padj
1 gwascatalog 5107 9 0.001762287 0.1054853
2 mgi_essential 1921 3 0.001561687 0.4312792
3 fda_approved_drug_targets 276 1 0.003623188 0.4312792
4 clinvar_path_likelypath 2341 3 0.001281504 0.4636974
5 core_essentials_hart 227 0 0.000000000 1.0000000
Blood or Immune
Number of cTWAS Genes in Tissue Group: 24
gene_set nset ngenes percent padj
1 gwascatalog 4763 13 0.002729372 0.09379103
2 mgi_essential 1773 2 0.001128032 1.00000000
3 core_essentials_hart 216 0 0.000000000 1.00000000
4 clinvar_path_likelypath 2189 3 0.001370489 1.00000000
5 fda_approved_drug_targets 255 0 0.000000000 1.00000000
Digestive
Number of cTWAS Genes in Tissue Group: 25
gene_set nset ngenes percent padj
1 gwascatalog 5399 15 0.002778292 0.01300787
2 mgi_essential 2053 5 0.002435460 0.41344197
3 clinvar_path_likelypath 2491 5 0.002007226 0.47246213
4 core_essentials_hart 243 0 0.000000000 1.00000000
5 fda_approved_drug_targets 308 0 0.000000000 1.00000000Analysis of TWAS False Positives by Region
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_tag, function(x){unlist(region_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from region total to get combined pip for other genes in region
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip < 0.5)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < 0.5 & gene_pips$snp_pip > 0.5)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > 0.5 & gene_pips$snp_pip < 0.5)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
Analysis of TWAS False Positives by Credible Set
cTWAS is using susie settings that mask credible sets consisting of variables with minimum pairwise correlations below a specified threshold. The default threshold is 0.5. I think this is intended to mask credible sets with “diffuse” support. As a consequence, many of the genes considered here (TWAS false positives; significant z score but low PIP) are not assigned to a credible set (have cs_index=0). For this reason, the first figure is not really appropriate for answering the question “are TWAS false positives due to SNPs or genes”.
The second figure includes only TWAS genes that are assigned to a reported causal set (i.e. they are in a “pure” causal set with high pairwise correlations). I think that this figure is closer to the intended analysis. However, it may be biased in some way because we have excluded many TWAS false positive genes that are in “impure” credible sets.
Some alternatives to these figures include the region-based analysis in the previous section; or re-analysis with lower/no minimum pairwise correlation threshold (“min_abs_corr” option in susie_get_cs) for reporting credible sets.
library(ggplot2)
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
####################
#using only genes assigned to a credible set
pip_threshold <- 0.5
df_plot <- data.frame(Outcome=c("SNPs", "Genes", "Both", "Neither"), Frequency=rep(0,4))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$twas,,drop=F]
gene_pips <- gene_pips[gene_pips$susie_pip < pip_threshold,,drop=F]
#exclude genes that are not assigned to a credible set, cs_index==0
gene_pips <- gene_pips[as.numeric(sapply(gene_pips$region_cs_tag, function(x){rev(unlist(strsplit(x, "_")))[1]}))!=0,]
region_cs_pips <- df[[i]]$region_cs_pips
rownames(region_cs_pips) <- region_cs_pips$region_cs
gene_pips <- cbind(gene_pips, t(sapply(gene_pips$region_cs_tag, function(x){unlist(region_cs_pips[x,c("gene_pip", "snp_pip")])})))
gene_pips$gene_pip <- gene_pips$gene_pip - gene_pips$susie_pip #subtract gene pip from causal set total to get combined pip for other genes in causal set
plot_cutoff <- 0.5
df_plot$Frequency[df_plot$Outcome=="Neither"] <- df_plot$Frequency[df_plot$Outcome=="Neither"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip < plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Both"] <- df_plot$Frequency[df_plot$Outcome=="Both"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="SNPs"] <- df_plot$Frequency[df_plot$Outcome=="SNPs"] + sum(gene_pips$gene_pip < plot_cutoff & gene_pips$snp_pip > plot_cutoff)
df_plot$Frequency[df_plot$Outcome=="Genes"] <- df_plot$Frequency[df_plot$Outcome=="Genes"] + sum(gene_pips$gene_pip > plot_cutoff & gene_pips$snp_pip < plot_cutoff)
}
pie <- ggplot(df_plot, aes(x="", y=Frequency, fill=Outcome)) + geom_bar(width = 1, stat = "identity")
pie <- pie + coord_polar("y", start=0) + theme_minimal() + theme(axis.title.y=element_blank())
pie
cTWAS genes without genome-wide significant SNP nearby
novel_genes <- data.frame(genename=as.character(), weight=as.character(), susie_pip=as.numeric(), snp_maxz=as.numeric())
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips[df[[i]]$gene_pips$genename %in% df[[i]]$ctwas,,drop=F]
region_pips <- df[[i]]$region_pips
rownames(region_pips) <- region_pips$region
gene_pips <- cbind(gene_pips, sapply(gene_pips$region_tag, function(x){region_pips[x,"snp_maxz"]}))
names(gene_pips)[ncol(gene_pips)] <- "snp_maxz"
if (nrow(gene_pips)>0){
gene_pips$weight <- names(df)[i]
gene_pips <- gene_pips[gene_pips$snp_maxz < qnorm(1-(5E-8/2), lower=T),c("genename", "weight", "susie_pip", "snp_maxz")]
novel_genes <- rbind(novel_genes, gene_pips)
}
}
novel_genes_summary <- data.frame(genename=unique(novel_genes$genename))
novel_genes_summary$nweights <- sapply(novel_genes_summary$genename, function(x){length(novel_genes$weight[novel_genes$genename==x])})
novel_genes_summary$weights <- sapply(novel_genes_summary$genename, function(x){paste(novel_genes$weight[novel_genes$genename==x],collapse=", ")})
novel_genes_summary <- novel_genes_summary[order(-novel_genes_summary$nweights),]
novel_genes_summary[,c("genename","nweights")] genename nweights
1 EVA1B 31
2 OAZ3 11
6 CPT1C 4
13 GIGYF1 4
5 BCL2L11 2
8 APBB1 2
10 SH3D21 2
11 C11orf58 2
22 CNKSR1 2
3 M6PR 1
4 VAMP3 1
7 IP6K2 1
9 FBXO45 1
12 AC007383.3 1
14 RBCK1 1
15 SPIRE2 1
16 ADCY9 1
17 ITGAV 1
18 MUC3A 1
19 TSC2 1
20 POLR3H 1
21 DEF8 1
23 ANKRD55 1
24 RGS14 1
25 STXBP3 1
26 SPNS2 1
27 LINC01623 1
28 COQ8B 1
29 SMPD1 1
30 TNFSF4 1
31 FAM171B 1
32 TIMD4 1
33 FAM53B 1
34 CAMKK1 1
35 CDK5R1 1
36 PPP5C 1
37 AF064858.11 1Tissue-specificity for cTWAS genes
gene_pips_by_weight <- data.frame(genename=as.character(ctwas_genes))
for (i in 1:length(df)){
gene_pips <- df[[i]]$gene_pips
gene_pips <- gene_pips[match(ctwas_genes, gene_pips$genename),,drop=F]
gene_pips_by_weight <- cbind(gene_pips_by_weight, gene_pips$susie_pip)
names(gene_pips_by_weight)[ncol(gene_pips_by_weight)] <- names(df)[i]
}
gene_pips_by_weight <- as.matrix(gene_pips_by_weight[,-1])
rownames(gene_pips_by_weight) <- ctwas_genes
#handling missing values
gene_pips_by_weight_bkup <- gene_pips_by_weight
gene_pips_by_weight[is.na(gene_pips_by_weight)] <- 0
#number of tissues with PIP>0.5 for cTWAS genes
ctwas_frequency <- rowSums(gene_pips_by_weight>0.5)
hist(ctwas_frequency, col="grey", breaks=0:max(ctwas_frequency), xlim=c(0,ncol(gene_pips_by_weight)),
xlab="Number of Tissues with PIP>0.5",
ylab="Number of cTWAS Genes",
main="Tissue Specificity for cTWAS Genes")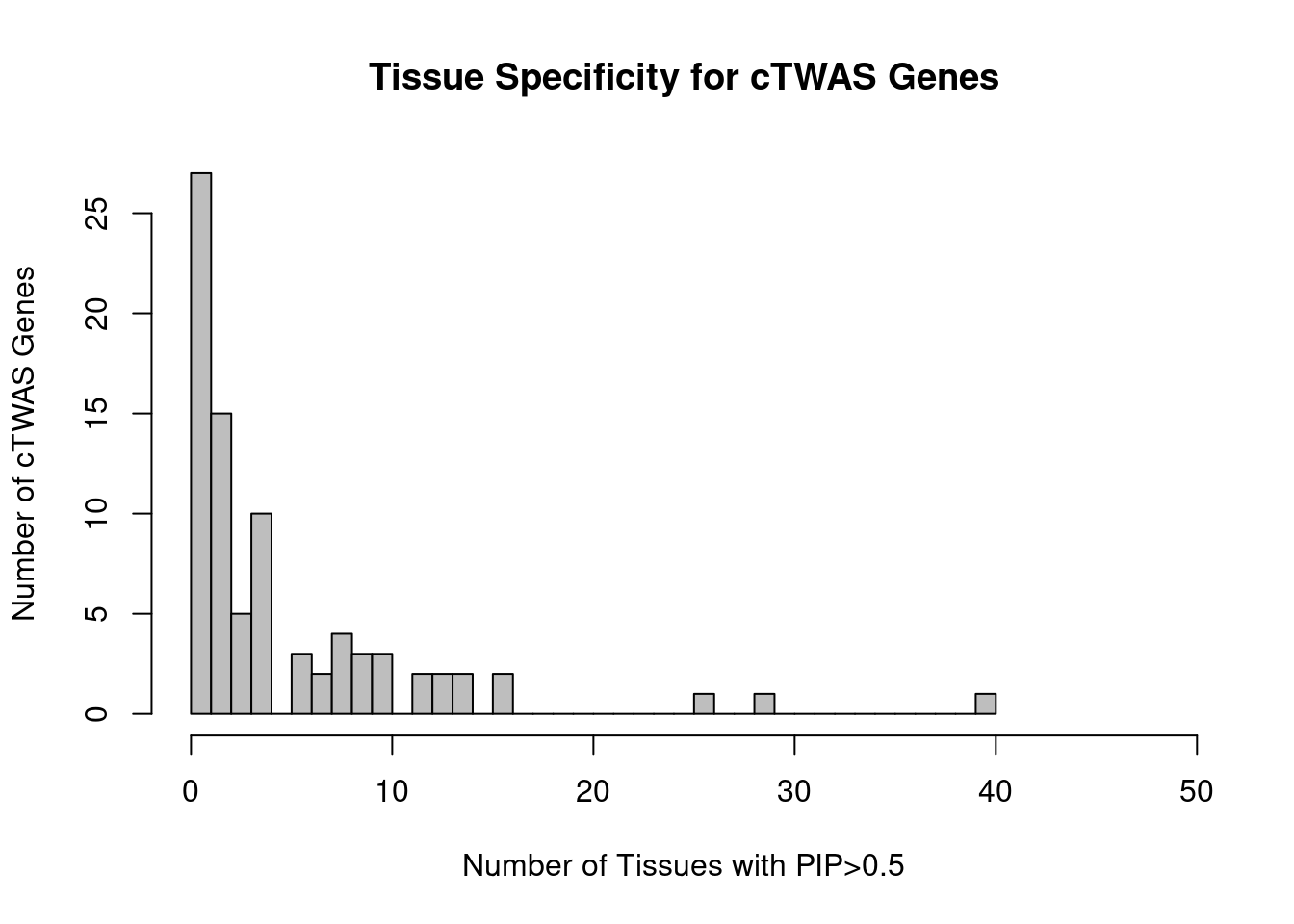
#heatmap of gene PIPs
cluster_ctwas_genes <- hclust(dist(gene_pips_by_weight))
cluster_ctwas_weights <- hclust(dist(t(gene_pips_by_weight)))
plot(cluster_ctwas_weights, cex=0.6)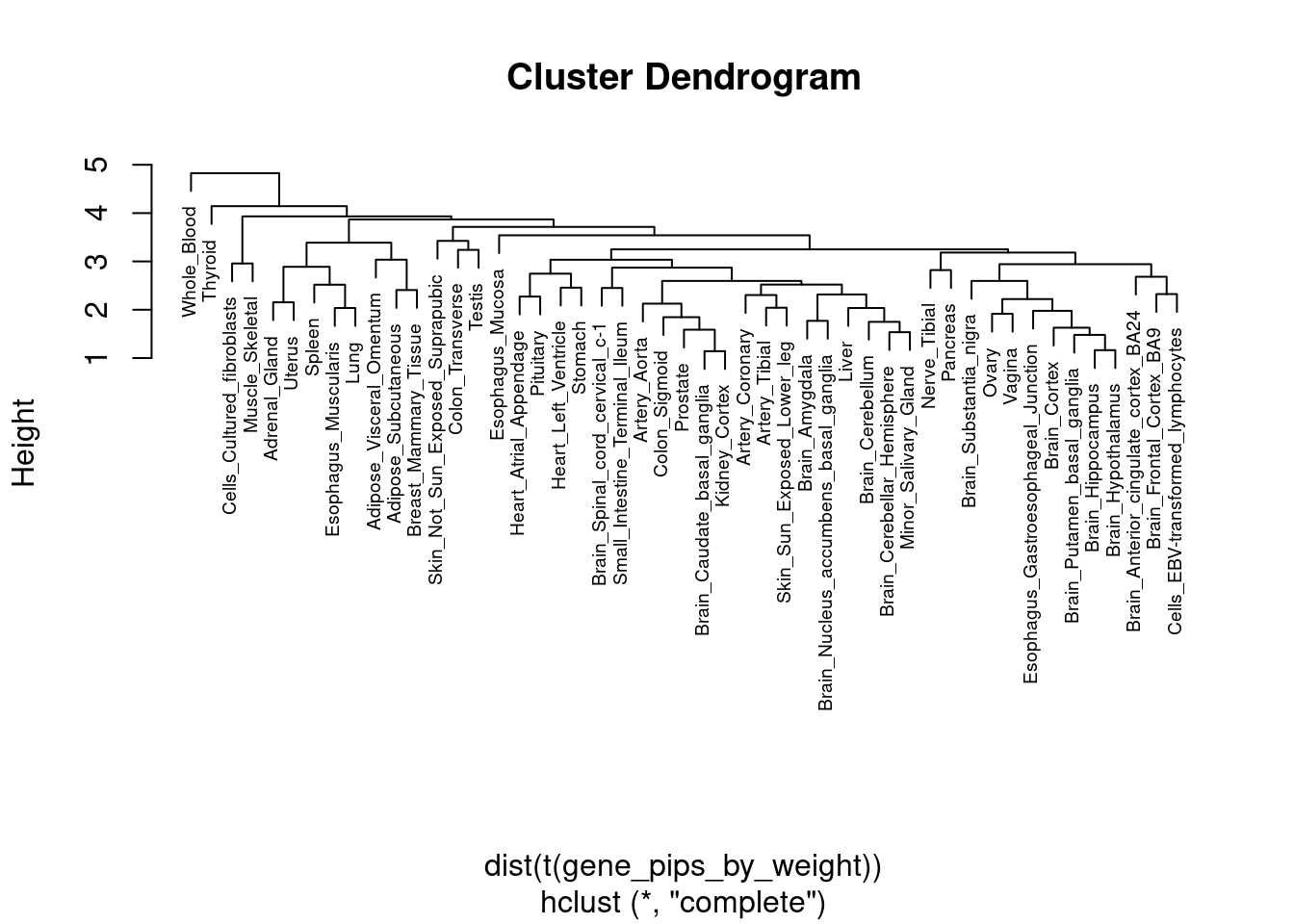
plot(cluster_ctwas_genes, cex=0.6, labels=F)
par(mar=c(14.1, 4.1, 4.1, 2.1))
image(t(gene_pips_by_weight[rev(cluster_ctwas_genes$order),rev(cluster_ctwas_weights$order)]),
axes=F)
mtext(text=colnames(gene_pips_by_weight)[cluster_ctwas_weights$order], side=1, line=0.3, at=seq(0,1,1/(ncol(gene_pips_by_weight)-1)), las=2, cex=0.8)
mtext(text=rownames(gene_pips_by_weight)[cluster_ctwas_genes$order], side=2, line=0.3, at=seq(0,1,1/(nrow(gene_pips_by_weight)-1)), las=1, cex=0.4)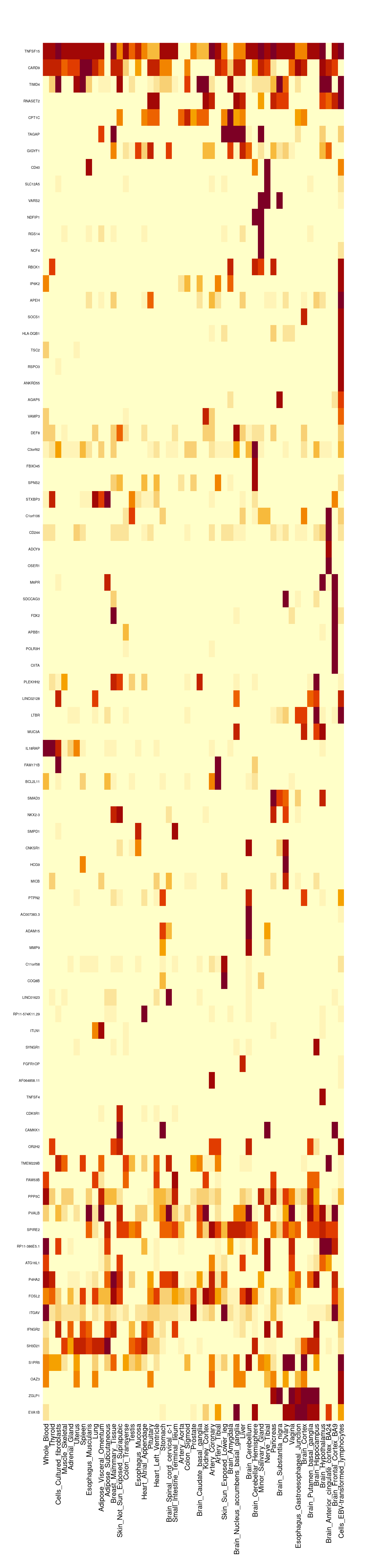
cTWAS genes with highest proportion of total PIP on a single tissue
#genes with highest proportion of PIP on a single tissue
gene_pips_proportion <- gene_pips_by_weight/rowSums(gene_pips_by_weight)
proportion_table <- data.frame(genename=as.character(rownames(gene_pips_proportion)))
proportion_table$max_pip_prop <- apply(gene_pips_proportion,1,max)
proportion_table$max_weight <- colnames(gene_pips_proportion)[apply(gene_pips_proportion,1,which.max)]
proportion_table[order(-proportion_table$max_pip_prop),] genename max_pip_prop max_weight
67 LINC01623 0.97247577 Testis
53 RSPO3 0.92858899 Muscle_Skeletal
83 AF064858.11 0.85841503 Whole_Blood
36 PTPN2 0.77397389 Cells_Cultured_fibroblasts
43 PVALB 0.77172391 Colon_Transverse
73 TNFSF4 0.75390158 Whole_Blood
70 NKX2-3 0.61826068 Thyroid
72 SMAD3 0.58642209 Thyroid
47 ADCY9 0.56715208 Esophagus_Mucosa
33 AC007383.3 0.51397677 Cells_Cultured_fibroblasts
10 SDCCAG3 0.50619366 Adipose_Visceral_Omentum
63 ANKRD55 0.50450742 Small_Intestine_Terminal_Ileum
40 RP11-386E5.1 0.50000049 Colon_Transverse
50 AGAP5 0.47700286 Heart_Atrial_Appendage
25 LINC02128 0.43863512 Brain_Spinal_cord_cervical_c-1
46 C1orf106 0.43026507 Esophagus_Mucosa
66 SPNS2 0.42900153 Stomach
61 P4HA2 0.41925232 Skin_Not_Sun_Exposed_Suprapubic
13 VAMP3 0.41546645 Adrenal_Gland
76 OR2H2 0.41078591 Whole_Blood
24 FBXO45 0.40205869 Artery_Tibial
56 TSC2 0.39473028 Nerve_Tibial
81 CDK5R1 0.38995891 Whole_Blood
5 FGFR1OP 0.38258791 Adipose_Subcutaneous
41 SPIRE2 0.36239573 Colon_Transverse
71 SMPD1 0.35875362 Thyroid
7 HCG9 0.35599971 Thyroid
45 CD244 0.35267263 Esophagus_Mucosa
80 CAMKK1 0.34824234 Whole_Blood
21 SYNGR1 0.33840377 Artery_Aorta
23 APBB1 0.31198087 Artery_Coronary
44 FOSL2 0.29562681 Skin_Not_Sun_Exposed_Suprapubic
58 DEF8 0.29105836 Ovary
3 ATG16L1 0.28938869 Colon_Transverse
65 STXBP3 0.28468148 Stomach
12 FDX2 0.28058909 Adipose_Visceral_Omentum
31 CIITA 0.26981844 Breast_Mammary_Tissue
54 MUC3A 0.26861349 Muscle_Skeletal
59 CNKSR1 0.26736316 Thyroid
52 MICB 0.26528071 Muscle_Skeletal
51 C3orf62 0.25799298 Liver
55 HLA-DQB1 0.25619626 Skin_Not_Sun_Exposed_Suprapubic
11 M6PR 0.22482737 Adipose_Visceral_Omentum
77 FAM53B 0.21421075 Whole_Blood
29 PLEKHH2 0.21308842 Brain_Spinal_cord_cervical_c-1
37 MMP9 0.21133465 Cells_Cultured_fibroblasts
69 COQ8B 0.20715342 Testis
57 POLR3H 0.19297600 Nerve_Tibial
78 TMEM229B 0.19028923 Whole_Blood
48 OSER1 0.17836773 Esophagus_Mucosa
74 FAM171B 0.17559872 Whole_Blood
82 PPP5C 0.17394604 Whole_Blood
14 IL18RAP 0.17179183 Adrenal_Gland
32 ADAM15 0.16447339 Cells_Cultured_fibroblasts
60 ITLN1 0.16121199 Pituitary
15 BCL2L11 0.16007577 Adrenal_Gland
28 LTBR 0.15697959 Cells_EBV-transformed_lymphocytes
30 C11orf58 0.14339831 Brain_Substantia_nigra
79 SOCS1 0.13937682 Whole_Blood
42 IFNGR2 0.12875596 Colon_Transverse
22 IP6K2 0.12496787 Artery_Coronary
62 VARS2 0.12432507 Skin_Not_Sun_Exposed_Suprapubic
6 CARD9 0.12001833 Lung
68 RP11-574K11.29 0.11656891 Testis
27 SH3D21 0.10743837 Heart_Left_Ventricle
75 TIMD4 0.10711075 Whole_Blood
34 APEH 0.10459054 Cells_Cultured_fibroblasts
9 TNFSF15 0.10243607 Esophagus_Muscularis
35 NDFIP1 0.10161253 Cells_Cultured_fibroblasts
49 ITGAV 0.09212845 Heart_Atrial_Appendage
18 S1PR5 0.08665937 Whole_Blood
38 GIGYF1 0.08373594 Thyroid
4 TAGAP 0.07861164 Adrenal_Gland
16 CPT1C 0.07280377 Brain_Substantia_nigra
8 RNASET2 0.06949360 Adipose_Visceral_Omentum
17 SLC12A5 0.06666738 Pancreas
2 OAZ3 0.06228623 Muscle_Skeletal
39 RBCK1 0.06214499 Cells_EBV-transformed_lymphocytes
20 NCF4 0.05517163 Esophagus_Mucosa
19 CD40 0.04981559 Pancreas
64 RGS14 0.04514276 Small_Intestine_Terminal_Ileum
26 ZGLP1 0.03872411 Brain_Cortex
1 EVA1B 0.02839951 Whole_BloodGenes nearby and nearest to GWAS peaks
#####load positions for all genes on autosomes in ENSEMBL, subset to only protein coding and lncRNA with non-missing HGNC symbol
# library(biomaRt)
#
# ensembl <- useEnsembl(biomart="ENSEMBL_MART_ENSEMBL", dataset="hsapiens_gene_ensembl")
# G_list <- getBM(filters= "chromosome_name", attributes= c("hgnc_symbol","chromosome_name","start_position","end_position","gene_biotype", "ensembl_gene_id"), values=1:22, mart=ensembl)
#
# save(G_list, file=paste0("G_list_", trait_id, ".RData"))
load(paste0("G_list_", trait_id, ".RData"))
G_list <- G_list[G_list$gene_biotype %in% c("protein_coding","lncRNA"),]
#####load z scores from the analysis and add positions from the LD reference
# results_dir <- results_dirs[1]
# weight <- rev(unlist(strsplit(results_dir, "/")))[1]
# analysis_id <- paste(trait_id, weight, sep="_")
#
# load(paste0(results_dir, "/", analysis_id, "_expr_z_snp.Rd"))
#
# LDR_dir <- "/project2/mstephens/wcrouse/UKB_LDR_0.1/"
# LDR_files <- list.files(LDR_dir)
# LDR_files <- LDR_files[grep(".Rvar" ,LDR_files)]
#
# z_snp$chrom <- as.integer(NA)
# z_snp$pos <- as.integer(NA)
#
# ptm <- proc.time()
#
# for (i in 1:length(LDR_files)){
# print(i)
#
# LDR_info <- read.table(paste0(LDR_dir, LDR_files[i]), header=T)
# z_snp_index <- which(z_snp$id %in% LDR_info$id)
# z_snp[z_snp_index,c("chrom", "pos")] <- t(sapply(z_snp_index, function(x){unlist(LDR_info[match(z_snp$id[x], LDR_info$id),c("chrom", "pos")])}))
# }
#
#
# proc.time() - ptm
#
# z_snp <- z_snp[,c("id", "z", "chrom","pos")]
# save(z_snp, file=paste0("z_snp_pos_", trait_id, ".RData"))
load(paste0("z_snp_pos_", trait_id, ".RData"))
####################
#identify genes within 500kb of genome-wide significant variant ("nearby")
G_list$nearby <- NA
window_size <- 500000
for (chr in 1:22){
#index genes on chromosome
G_list_index <- which(G_list$chromosome_name==chr)
#subset z_snp to chromosome, then subset to significant genome-wide significant variants
z_snp_chr <- z_snp[z_snp$chrom==chr,,drop=F]
z_snp_chr <- z_snp_chr[abs(z_snp_chr$z)>qnorm(1-(5E-8/2), lower=T),,drop=F]
#iterate over genes on chromsome and check if a genome-wide significant SNP is within the window
for (i in G_list_index){
window_start <- G_list$start_position[i] - window_size
window_end <- G_list$end_position[i] + window_size
G_list$nearby[i] <- any(z_snp_chr$pos>=window_start & z_snp_chr$pos<=window_end)
}
}
####################
#identify genes that are nearest to lead genome-wide significant variant ("nearest")
G_list$nearest <- F
window_size <- 500000
for (chr in 1:22){
#index genes on chromosome
G_list_index <- which(G_list$chromosome_name==chr)
#subset z_snp to chromosome, then subset to significant genome-wide significant variants
z_snp_chr <- z_snp[z_snp$chrom==chr,,drop=F]
z_snp_chr <- z_snp_chr[abs(z_snp_chr$z)>qnorm(1-(5E-8/2), lower=T),,drop=F]
count <- 0
while (nrow(z_snp_chr)>0){
lead_index <- which.max(abs(z_snp_chr$z))
lead_position <- z_snp_chr$pos[lead_index]
distances <- sapply(G_list_index, function(i){
if (lead_position >= G_list$start_position[i] & lead_position <= G_list$end_position[i]){
distance <- 0
} else {
distance <- min(abs(G_list$start_position[i] - lead_position), abs(G_list$end_position[i] - lead_position))
}
distance
})
min_distance <- min(distances)
G_list$nearest[G_list_index[distances==min_distance]] <- 1
window_start <- lead_position - window_size
window_end <- lead_position + window_size
z_snp_chr <- z_snp_chr[!(z_snp_chr$pos>=window_start & z_snp_chr$pos<=window_end),,drop=F]
}
}Enrichment analysis using known IBD genes from ABC paper
known_genes <- data.table::fread("nasser_2021_ABC_IBD_genes.txt")
known_genes <- unique(known_genes$KnownGene)
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(known_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.for (db in dbs){
cat(paste0(db, "\n\n"))
enrich_results <- GO_enrichment[[db]]
enrich_results <- enrich_results[enrich_results$Adjusted.P.value<0.05,c("Term", "Overlap", "Adjusted.P.value", "Genes")]
print(enrich_results)
print(plotEnrich(GO_enrichment[[db]]))
}GO_Biological_Process_2021
Term Overlap Adjusted.P.value Genes
1 cytokine-mediated signaling pathway (GO:0019221) 12/621 3.159212e-09 IL10;CD40;NOS2;IFNG;TNFSF15;IL23R;IL2RA;RORC;STAT4;IL12B;JAK2;IL6ST
2 cellular response to cytokine stimulus (GO:0071345) 11/482 3.159212e-09 IL10;CD40;SMAD3;NOS2;IL23R;IL2RA;RORC;STAT4;IL12B;JAK2;IL6ST
3 positive regulation of tyrosine phosphorylation of STAT protein (GO:0042531) 6/59 2.963998e-08 CD40;IFNG;IL23R;IL12B;JAK2;IL6ST
4 regulation of tyrosine phosphorylation of STAT protein (GO:0042509) 6/68 4.484450e-08 CD40;IFNG;IL23R;IL12B;JAK2;IL6ST
5 interleukin-23-mediated signaling pathway (GO:0038155) 4/9 4.484450e-08 IL23R;STAT4;IL12B;JAK2
6 interleukin-12-mediated signaling pathway (GO:0035722) 5/46 4.334439e-07 IL10;IFNG;STAT4;IL12B;JAK2
7 cellular response to interleukin-12 (GO:0071349) 5/48 4.632848e-07 IL10;IFNG;STAT4;IL12B;JAK2
8 regulation of interleukin-12 production (GO:0032655) 5/51 5.546642e-07 IL10;CD40;IFNG;IL23R;IL12B
9 positive regulation of cytokine production (GO:0001819) 8/335 6.063177e-07 PTGER4;IL10;CD40;NOS2;IFNG;IL23R;IL12B;IL6ST
10 positive regulation of peptidyl-tyrosine phosphorylation (GO:0050731) 6/134 1.328228e-06 CD40;IFNG;IL23R;IL12B;JAK2;IL6ST
11 response to molecule of bacterial origin (GO:0002237) 5/73 2.530185e-06 IL10;NOS2;IL23R;TNFAIP3;JAK2
12 positive regulation of immune response (GO:0050778) 5/75 2.660413e-06 IL10;IFNG;IL23R;IL12B;IL6ST
13 positive regulation of immune effector process (GO:0002699) 4/32 4.824287e-06 IL10;IFNG;IL23R;IL12B
14 negative regulation of smooth muscle cell proliferation (GO:0048662) 4/33 5.093107e-06 IL10;IFNG;TNFAIP3;IL12B
15 positive regulation of interleukin-12 production (GO:0032735) 4/34 5.154051e-06 CD40;IFNG;IL23R;IL12B
16 negative regulation of cytokine production (GO:0001818) 6/182 5.154051e-06 PTGER4;IL10;IFNG;IL23R;TNFAIP3;IL12B
17 regulation of inflammatory response (GO:0050727) 6/206 1.008944e-05 PTGER4;IL10;IFNG;TNFAIP3;IL12B;JAK2
18 interleukin-35-mediated signaling pathway (GO:0070757) 3/11 1.414897e-05 STAT4;IL6ST;JAK2
19 regulation of MHC class II biosynthetic process (GO:0045346) 3/12 1.785704e-05 IL10;IFNG;JAK2
20 regulation of tumor necrosis factor production (GO:0032680) 5/124 1.994800e-05 IL10;IFNG;TNFAIP3;IL12B;JAK2
21 T-helper cell differentiation (GO:0042093) 3/14 2.668554e-05 PTGER4;RORC;IL12B
22 positive regulation of osteoclast differentiation (GO:0045672) 3/15 3.181334e-05 IFNG;IL23R;IL12B
23 regulation of cytokine production (GO:0001817) 5/150 4.269432e-05 PTGER4;IL10;NOS2;IFNG;IL12B
24 response to cytokine (GO:0034097) 5/150 4.269432e-05 CD40;SMAD3;IL23R;JAK2;IL6ST
25 response to lipopolysaccharide (GO:0032496) 5/159 5.462762e-05 IL10;NOS2;IL23R;TNFAIP3;JAK2
26 positive regulation of lymphocyte proliferation (GO:0050671) 4/75 7.850430e-05 CD40;IL23R;IL12B;IL6ST
27 regulation of T cell proliferation (GO:0042129) 4/76 7.972634e-05 IL10;IL23R;IL12B;IL6ST
28 positive regulation of multicellular organismal process (GO:0051240) 6/345 1.209518e-04 PTGER4;IL10;SMAD3;IFNG;IL12B;JAK2
29 receptor signaling pathway via STAT (GO:0097696) 3/25 1.209518e-04 IFNG;STAT4;JAK2
30 positive regulation of inflammatory response (GO:0050729) 4/89 1.350005e-04 PTGER4;IFNG;IL12B;IL6ST
31 regulation of transcription by RNA polymerase II (GO:0006357) 12/2206 1.768790e-04 CD40;SMAD3;IFNG;HNF4A;NRIP1;RORC;STAT4;IKZF1;PRDM1;BACH2;NKX2-3;JAZF1
32 negative regulation of cell population proliferation (GO:0008285) 6/379 1.851566e-04 IL10;IFNG;HNF4A;TNFAIP3;IL12B;JAK2
33 receptor signaling pathway via JAK-STAT (GO:0007259) 3/31 2.066586e-04 IFNG;STAT4;JAK2
34 regulation of interleukin-17 production (GO:0032660) 3/33 2.368256e-04 IFNG;IL23R;IL12B
35 inflammatory response (GO:0006954) 5/230 2.368256e-04 PTGER4;CD40;NOS2;IFNG;IL2RA
36 regulation of interleukin-6 production (GO:0032675) 4/110 2.611320e-04 IL10;NOS2;IFNG;TNFAIP3
37 regulation of defense response to virus by host (GO:0050691) 3/36 2.765671e-04 IL23R;TNFAIP3;IL12B
38 regulation of osteoclast differentiation (GO:0045670) 3/36 2.765671e-04 IFNG;IL23R;IL12B
39 positive regulation of myeloid leukocyte differentiation (GO:0002763) 3/36 2.765671e-04 IFNG;IL23R;IL12B
40 positive regulation of signaling receptor activity (GO:2000273) 3/37 2.931931e-04 IL10;IFNG;JAK2
41 positive regulation of signal transduction (GO:0009967) 5/252 3.004671e-04 LRRK2;TNFAIP3;RSPO3;JAK2;IL6ST
42 positive regulation of cell fate commitment (GO:0010455) 2/5 3.004671e-04 IL23R;IL12B
43 positive regulation of natural killer cell proliferation (GO:0032819) 2/5 3.004671e-04 IL23R;IL12B
44 regulation of T cell differentiation (GO:0045580) 3/39 3.129611e-04 IFNG;IL2RA;PRDM1
45 negative regulation of transcription, DNA-templated (GO:0045892) 8/948 3.189470e-04 SMAD3;IFNG;HNF4A;NRIP1;RORC;IKZF1;PRDM1;JAZF1
46 regulation of nitric oxide biosynthetic process (GO:0045428) 3/40 3.233473e-04 SMAD3;IFNG;JAK2
47 regulation of protein secretion (GO:0050708) 4/125 3.313583e-04 NOS2;IFNG;HNF4A;IL12B
48 positive regulation of leukocyte mediated cytotoxicity (GO:0001912) 3/43 3.797001e-04 NOS2;IL23R;IL12B
49 regulation of NK T cell activation (GO:0051133) 2/6 3.797001e-04 IL23R;IL12B
50 regulation of T-helper 17 cell lineage commitment (GO:2000328) 2/6 3.797001e-04 IL23R;IL12B
51 positive regulation of killing of cells of other organism (GO:0051712) 2/6 3.797001e-04 IFNG;NOS2
52 regulation of endothelial cell apoptotic process (GO:2000351) 3/44 3.821106e-04 IL10;CD40;TNFAIP3
53 regulation of natural killer cell proliferation (GO:0032817) 2/7 4.828489e-04 IL23R;IL12B
54 positive regulation of activation of Janus kinase activity (GO:0010536) 2/7 4.828489e-04 IL23R;IL12B
55 positive regulation of NK T cell activation (GO:0051135) 2/7 4.828489e-04 IL23R;IL12B
56 positive regulation of T-helper 17 cell differentiation (GO:2000321) 2/7 4.828489e-04 IL23R;IL12B
57 regulation of smooth muscle cell proliferation (GO:0048660) 3/49 4.828489e-04 IFNG;TNFAIP3;IL12B
58 regulation of activation of Janus kinase activity (GO:0010533) 2/8 6.116927e-04 IL23R;IL12B
59 positive regulation of MHC class II biosynthetic process (GO:0045348) 2/8 6.116927e-04 IFNG;JAK2
60 regulation of smooth muscle cell apoptotic process (GO:0034391) 2/9 7.359406e-04 IFNG;IL12B
61 regulation of T-helper 1 type immune response (GO:0002825) 2/9 7.359406e-04 IL23R;IL12B
62 positive regulation of memory T cell differentiation (GO:0043382) 2/9 7.359406e-04 IL23R;IL12B
63 positive regulation of smooth muscle cell apoptotic process (GO:0034393) 2/9 7.359406e-04 IFNG;IL12B
64 regulation of memory T cell differentiation (GO:0043380) 2/10 8.909091e-04 IL23R;IL12B
65 regulation of T-helper 17 type immune response (GO:2000316) 2/10 8.909091e-04 IL23R;IL12B
66 positive regulation of T cell proliferation (GO:0042102) 3/66 1.020663e-03 IL23R;IL12B;IL6ST
67 negative regulation of interleukin-17 production (GO:0032700) 2/11 1.040019e-03 IFNG;IL12B
68 positive regulation of nitric-oxide synthase biosynthetic process (GO:0051770) 2/11 1.040019e-03 IFNG;JAK2
69 regulation of natural killer cell activation (GO:0032814) 2/12 1.194336e-03 IL12B;PRDM1
70 positive regulation of T-helper 1 type immune response (GO:0002827) 2/12 1.194336e-03 IL23R;IL12B
71 positive regulation of T-helper 17 type immune response (GO:2000318) 2/12 1.194336e-03 IL23R;IL12B
72 extrinsic apoptotic signaling pathway (GO:0097191) 3/72 1.213111e-03 SMAD3;IFNG;JAK2
73 defense response to Gram-negative bacterium (GO:0050829) 3/73 1.246696e-03 NOS2;IL23R;IL12B
74 regulation of cytokine-mediated signaling pathway (GO:0001959) 3/74 1.263635e-03 IFNG;TNFAIP3;JAK2
75 regulation of peptide hormone secretion (GO:0090276) 3/74 1.263635e-03 NOS2;IFNG;HNF4A
76 positive regulation of T cell activation (GO:0050870) 3/75 1.267540e-03 IL23R;IL12B;IL6ST
77 negative regulation of hydrogen peroxide-induced cell death (GO:1903206) 2/13 1.267540e-03 IL10;LRRK2
78 positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (GO:0002824) 2/13 1.267540e-03 IL23R;IL12B
79 protein localization to mitochondrion (GO:0070585) 2/13 1.267540e-03 LRRK2;RNF186
80 positive regulation of tumor necrosis factor production (GO:0032760) 3/77 1.333364e-03 IFNG;IL12B;JAK2
81 regulation of nitric-oxide synthase biosynthetic process (GO:0051769) 2/14 1.406405e-03 IFNG;JAK2
82 regulation of response to interferon-gamma (GO:0060330) 2/14 1.406405e-03 IFNG;JAK2
83 positive regulation of granulocyte macrophage colony-stimulating factor production (GO:0032725) 2/14 1.406405e-03 IL23R;IL12B
84 activation of cysteine-type endopeptidase activity involved in apoptotic process (GO:0006919) 3/81 1.458679e-03 SMAD3;TNFSF15;JAK2
85 positive regulation of tumor necrosis factor superfamily cytokine production (GO:1903557) 3/81 1.458679e-03 IFNG;IL12B;JAK2
86 regulation of interleukin-1 beta production (GO:0032651) 3/83 1.512167e-03 IFNG;TNFAIP3;JAK2
87 positive regulation of cell killing (GO:0031343) 2/15 1.512167e-03 NOS2;IFNG
88 interleukin-27-mediated signaling pathway (GO:0070106) 2/15 1.512167e-03 IL6ST;JAK2
89 positive regulation of natural killer cell activation (GO:0032816) 2/15 1.512167e-03 IL23R;IL12B
90 cellular response to lipid (GO:0071396) 4/219 1.532524e-03 IL10;NRIP1;RORC;TNFAIP3
91 negative regulation of defense response (GO:0031348) 3/85 1.571864e-03 PTGER4;IL10;TNFAIP3
92 regulation of granulocyte macrophage colony-stimulating factor production (GO:0032645) 2/16 1.670502e-03 IL23R;IL12B
93 negative regulation of transcription by RNA polymerase II (GO:0000122) 6/684 1.719239e-03 SMAD3;IFNG;NRIP1;RORC;PRDM1;JAZF1
94 negative regulation of interleukin-10 production (GO:0032693) 2/17 1.831985e-03 IL23R;IL12B
95 regulation of cellular respiration (GO:0043457) 2/17 1.831985e-03 NOS2;IFNG
96 regulation of peptidyl-tyrosine phosphorylation (GO:0050730) 3/92 1.883050e-03 IL23R;IL12B;JAK2
97 interleukin-6-mediated signaling pathway (GO:0070102) 2/18 2.016877e-03 IL6ST;JAK2
98 positive regulation of defense response (GO:0031349) 3/98 2.222594e-03 PTGER4;IFNG;IL12B
99 regulation of transcription, DNA-templated (GO:0006355) 10/2244 2.354954e-03 IL10;SMAD3;IFNG;HNF4A;RORC;STAT4;IKZF1;PRDM1;BACH2;NKX2-3
100 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (GO:0002460) 2/20 2.354954e-03 IL12B;BACH2
101 positive regulation of activated T cell proliferation (GO:0042104) 2/20 2.354954e-03 IL23R;IL12B
102 positive regulation of alpha-beta T cell activation (GO:0046635) 2/20 2.354954e-03 IL23R;IL12B
103 mesoderm development (GO:0007498) 2/20 2.354954e-03 IKZF1;JAK2
104 regulation of insulin secretion (GO:0050796) 3/104 2.494654e-03 NOS2;IFNG;HNF4A
105 positive regulation of protein serine/threonine kinase activity (GO:0071902) 3/106 2.613149e-03 CD40;IFNG;LRRK2
106 regulation of cell population proliferation (GO:0042127) 6/764 2.676650e-03 IL10;IFNG;HNF4A;STAT4;JAK2;IL6ST
107 negative regulation of signal transduction (GO:0009968) 4/267 2.676650e-03 IL10;LRRK2;TNFAIP3;IL12B
108 negative regulation of B cell activation (GO:0050869) 2/22 2.676650e-03 IL10;TNFAIP3
109 regulation of membrane protein ectodomain proteolysis (GO:0051043) 2/22 2.676650e-03 IL10;IFNG
110 microglial cell activation (GO:0001774) 2/22 2.676650e-03 IFNG;JAK2
111 regulation of Wnt signaling pathway (GO:0030111) 3/111 2.829920e-03 LRRK2;TNFAIP3;RSPO3
112 regulation of interferon-gamma-mediated signaling pathway (GO:0060334) 2/23 2.851461e-03 IFNG;JAK2
113 positive regulation of interleukin-17 production (GO:0032740) 2/23 2.851461e-03 IL23R;IL12B
114 response to lipid (GO:0033993) 3/114 2.979406e-03 NOS2;IL23R;JAK2
115 positive regulation of protein metabolic process (GO:0051247) 3/115 3.029999e-03 IL23R;TNFAIP3;IL12B
116 tumor necrosis factor-mediated signaling pathway (GO:0033209) 3/116 3.080974e-03 CD40;TNFSF15;JAK2
117 negative regulation of response to external stimulus (GO:0032102) 3/118 3.211275e-03 PTGER4;IL10;TNFAIP3
118 positive regulation of cysteine-type endopeptidase activity involved in apoptotic process (GO:0043280) 3/119 3.263596e-03 SMAD3;TNFSF15;JAK2
119 regulation of regulatory T cell differentiation (GO:0045589) 2/26 3.441017e-03 IFNG;IL2RA
120 positive regulation of T cell mediated cytotoxicity (GO:0001916) 2/26 3.441017e-03 IL23R;IL12B
121 negative regulation of endothelial cell apoptotic process (GO:2000352) 2/27 3.652458e-03 IL10;TNFAIP3
122 positive regulation of nitric oxide biosynthetic process (GO:0045429) 2/27 3.652458e-03 SMAD3;IFNG
123 MAPK cascade (GO:0000165) 4/303 3.746044e-03 PTGER4;LRRK2;IL2RA;JAK2
124 regulation of B cell activation (GO:0050864) 2/28 3.746044e-03 IL10;TNFAIP3
125 cellular response to interleukin-6 (GO:0071354) 2/28 3.746044e-03 JAK2;IL6ST
126 positive regulation of defense response to virus by host (GO:0002230) 2/28 3.746044e-03 IL23R;IL12B
127 positive regulation of nitric oxide metabolic process (GO:1904407) 2/28 3.746044e-03 SMAD3;IFNG
128 positive regulation of protein import into nucleus (GO:0042307) 2/28 3.746044e-03 SMAD3;IFNG
129 regulation of T cell mediated cytotoxicity (GO:0001914) 2/29 3.989152e-03 IL23R;IL12B
130 negative regulation of interleukin-1 production (GO:0032692) 2/30 4.205478e-03 IL10;TNFAIP3
131 positive regulation of protein import (GO:1904591) 2/30 4.205478e-03 SMAD3;IFNG
132 negative regulation of interleukin-6 production (GO:0032715) 2/31 4.457893e-03 IL10;TNFAIP3
133 negative regulation of gene expression (GO:0010629) 4/322 4.465449e-03 PTGER4;IL10;NOS2;IFNG
134 cellular response to oxygen-containing compound (GO:1901701) 4/323 4.483746e-03 IL10;NRIP1;RORC;TNFAIP3
135 positive regulation of response to external stimulus (GO:0032103) 3/139 4.485050e-03 PTGER4;IFNG;IL12B
136 B cell proliferation (GO:0042100) 2/32 4.577885e-03 IL10;CD40
137 superoxide metabolic process (GO:0006801) 2/32 4.577885e-03 NOS2;NCF4
138 positive regulation of receptor signaling pathway via JAK-STAT (GO:0046427) 2/33 4.834058e-03 IL10;JAK2
139 regulation of activated T cell proliferation (GO:0046006) 2/34 5.058772e-03 IL23R;IL12B
140 positive regulation of pri-miRNA transcription by RNA polymerase II (GO:1902895) 2/34 5.058772e-03 IL10;SMAD3
141 cellular response to organic cyclic compound (GO:0071407) 3/150 5.354212e-03 LRRK2;NRIP1;RORC
142 regulation of protein import into nucleus (GO:0042306) 2/36 5.513132e-03 SMAD3;IFNG
143 macrophage activation (GO:0042116) 2/36 5.513132e-03 IFNG;JAK2
144 positive regulation of T cell mediated immunity (GO:0002711) 2/36 5.513132e-03 IL23R;IL12B
145 positive regulation of Wnt signaling pathway (GO:0030177) 3/153 5.513132e-03 LRRK2;TNFAIP3;RSPO3
146 positive regulation of protein kinase activity (GO:0045860) 3/154 5.579350e-03 IFNG;IL23R;IL12B
147 negative regulation of epithelial cell apoptotic process (GO:1904036) 2/37 5.705937e-03 IL10;TNFAIP3
148 lymphocyte proliferation (GO:0046651) 2/38 5.977465e-03 IL10;CD40
149 positive regulation of protein phosphorylation (GO:0001934) 4/371 6.744077e-03 CD40;IFNG;LRRK2;JAK2
150 negative regulation of proteolysis (GO:0045861) 2/43 7.446194e-03 IL10;LRRK2
151 negative regulation of type I interferon production (GO:0032480) 2/43 7.446194e-03 IL10;TNFAIP3
152 positive regulation of T cell differentiation (GO:0045582) 2/43 7.446194e-03 IL23R;IL12B
153 defense response to bacterium (GO:0042742) 3/176 7.817542e-03 NOS2;IL23R;IL12B
154 negative regulation of immune response (GO:0050777) 3/178 7.941593e-03 PTGER4;IL10;TNFAIP3
155 regulation of pri-miRNA transcription by RNA polymerase II (GO:1902893) 2/45 7.941593e-03 IL10;SMAD3
156 regulation of receptor signaling pathway via JAK-STAT (GO:0046425) 2/45 7.941593e-03 IL10;JAK2
157 regulation of B cell proliferation (GO:0030888) 2/46 8.088553e-03 IL10;CD40
158 negative regulation of tumor necrosis factor production (GO:0032720) 2/46 8.088553e-03 IL10;TNFAIP3
159 erythrocyte differentiation (GO:0030218) 2/46 8.088553e-03 IKZF1;JAK2
160 positive regulation of nucleocytoplasmic transport (GO:0046824) 2/46 8.088553e-03 SMAD3;IFNG
161 negative regulation of mitotic cell cycle (GO:0045930) 2/48 8.639307e-03 IL10;SMAD3
162 negative regulation of tumor necrosis factor superfamily cytokine production (GO:1903556) 2/48 8.639307e-03 IL10;TNFAIP3
163 regulation of interleukin-10 production (GO:0032653) 2/48 8.639307e-03 IL23R;IL12B
164 cellular response to mechanical stimulus (GO:0071260) 2/49 8.944878e-03 PTGER4;CD40
165 cellular response to tumor necrosis factor (GO:0071356) 3/194 9.574500e-03 CD40;TNFSF15;JAK2
166 myeloid cell differentiation (GO:0030099) 2/52 9.940472e-03 IKZF1;JAK2
167 regulation of gene expression (GO:0010468) 6/1079 1.052304e-02 IL10;SMAD3;NOS2;IFNG;IL12B;PRDM1
168 positive regulation of interleukin-1 beta production (GO:0032731) 2/56 1.137097e-02 IFNG;JAK2
169 positive regulation of interferon-gamma production (GO:0032729) 2/57 1.170539e-02 IL23R;IL12B
170 negative regulation of inflammatory response (GO:0050728) 3/212 1.195664e-02 PTGER4;IL10;TNFAIP3
171 negative regulation of multicellular organismal process (GO:0051241) 3/214 1.213597e-02 PTGER4;IL10;IFNG
172 positive regulation of protein modification process (GO:0031401) 3/214 1.213597e-02 CD40;IFNG;LRRK2
173 negative regulation of autophagy (GO:0010507) 2/59 1.223914e-02 IL10;LRRK2
174 positive regulation of interleukin-1 production (GO:0032732) 2/62 1.341685e-02 IFNG;JAK2
175 apoptotic process (GO:0006915) 3/231 1.471611e-02 IFNG;IL2RA;JAK2
176 regulation of autophagy (GO:0010506) 3/231 1.471611e-02 IL10;IFNG;LRRK2
177 positive regulation of gene expression (GO:0010628) 4/482 1.472191e-02 PTGER4;IL10;SMAD3;IFNG
178 positive regulation of cell death (GO:0010942) 2/66 1.482965e-02 IFNG;LRRK2
179 positive regulation of transcription, DNA-templated (GO:0045893) 6/1183 1.546345e-02 IL10;CD40;SMAD3;HNF4A;NRIP1;RORC
180 interferon-gamma-mediated signaling pathway (GO:0060333) 2/68 1.546345e-02 IFNG;JAK2
181 positive regulation of protein localization to nucleus (GO:1900182) 2/68 1.546345e-02 SMAD3;IFNG
182 positive regulation of MAP kinase activity (GO:0043406) 2/69 1.582493e-02 CD40;LRRK2
183 positive regulation of DNA-binding transcription factor activity (GO:0051091) 3/246 1.688787e-02 IL10;CD40;SMAD3
184 positive regulation of nucleic acid-templated transcription (GO:1903508) 4/511 1.745108e-02 IL10;SMAD3;HNF4A;RORC
185 positive regulation of phosphorylation (GO:0042327) 3/253 1.806956e-02 CD40;IFNG;LRRK2
186 positive regulation of interleukin-6 production (GO:0032755) 2/76 1.870662e-02 NOS2;IFNG
187 negative regulation of NF-kappaB transcription factor activity (GO:0032088) 2/79 2.002929e-02 IL10;TNFAIP3
188 regulation of protein phosphorylation (GO:0001932) 3/266 2.002929e-02 CD40;IFNG;LRRK2
189 negative regulation of cell cycle (GO:0045786) 2/80 2.002929e-02 IL10;SMAD3
190 regulation of fat cell differentiation (GO:0045598) 2/80 2.002929e-02 SMAD3;RORC
191 positive regulation of cell adhesion (GO:0045785) 2/80 2.002929e-02 IL12B;JAK2
192 response to interferon-gamma (GO:0034341) 2/80 2.002929e-02 CD40;IL23R
193 regulation of interleukin-8 production (GO:0032677) 2/81 2.041370e-02 IL10;NOS2
194 lymphocyte differentiation (GO:0030098) 2/84 2.179828e-02 IL10;IKZF1
195 B cell activation (GO:0042113) 2/85 2.207817e-02 IL10;CD40
196 positive regulation of protein catabolic process (GO:0045732) 2/85 2.207817e-02 IFNG;TNFAIP3
197 regulation of interferon-gamma production (GO:0032649) 2/86 2.235771e-02 IL23R;IL12B
198 regulation of neuron death (GO:1901214) 2/86 2.235771e-02 IFNG;LRRK2
199 positive regulation of transcription by RNA polymerase II (GO:0045944) 5/908 2.247143e-02 IL10;CD40;SMAD3;HNF4A;NRIP1
200 negative regulation of cellular process (GO:0048523) 4/566 2.247143e-02 IL10;SMAD3;HNF4A;JAK2
201 negative regulation of binding (GO:0051100) 2/88 2.247143e-02 LRRK2;JAK2
202 positive regulation of programmed cell death (GO:0043068) 3/286 2.247143e-02 NOS2;IFNG;LRRK2
203 second-messenger-mediated signaling (GO:0019932) 2/89 2.247143e-02 NOS2;LRRK2
204 positive regulation of autophagy (GO:0010508) 2/90 2.247143e-02 IFNG;LRRK2
205 regulation of lipid metabolic process (GO:0019216) 2/92 2.247143e-02 HNF4A;RORC
206 positive regulation of cellular component biogenesis (GO:0044089) 2/92 2.247143e-02 SMAD3;IFNG
207 negative regulation of hydrogen peroxide-induced neuron death (GO:1903208) 1/5 2.247143e-02 IL10
208 negative regulation of interferon-alpha production (GO:0032687) 1/5 2.247143e-02 IL10
209 regulation of cell maturation (GO:1903429) 1/5 2.247143e-02 LRRK2
210 regulation of core promoter binding (GO:1904796) 1/5 2.247143e-02 IFNG
211 regulation of germinal center formation (GO:0002634) 1/5 2.247143e-02 TNFAIP3
212 regulation of growth hormone receptor signaling pathway (GO:0060398) 1/5 2.247143e-02 JAK2
213 regulation of guanylate cyclase activity (GO:0031282) 1/5 2.247143e-02 NOS2
214 neuroinflammatory response (GO:0150076) 1/5 2.247143e-02 IFNG
215 ciliary neurotrophic factor-mediated signaling pathway (GO:0070120) 1/5 2.247143e-02 IL6ST
216 regulation of mononuclear cell proliferation (GO:0032944) 1/5 2.247143e-02 IL12B
217 regulation of morphogenesis of a branching structure (GO:0060688) 1/5 2.247143e-02 LRRK2
218 regulation of natural killer cell differentiation (GO:0032823) 1/5 2.247143e-02 PRDM1
219 peptidyl-cysteine S-nitrosylation (GO:0018119) 1/5 2.247143e-02 NOS2
220 regulation of NK T cell proliferation (GO:0051140) 1/5 2.247143e-02 IL12B
221 endothelial cell apoptotic process (GO:0072577) 1/5 2.247143e-02 IL10
222 positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation (GO:0032831) 1/5 2.247143e-02 IFNG
223 positive regulation of dopamine receptor signaling pathway (GO:0060161) 1/5 2.247143e-02 LRRK2
224 positive regulation of leukocyte mediated immunity (GO:0002705) 1/5 2.247143e-02 NOS2
225 positive regulation of mononuclear cell proliferation (GO:0032946) 1/5 2.247143e-02 IL12B
226 intracellular distribution of mitochondria (GO:0048312) 1/5 2.247143e-02 LRRK2
227 kidney morphogenesis (GO:0060993) 1/5 2.247143e-02 LRRK2
228 prostaglandin transport (GO:0015732) 1/5 2.247143e-02 NOS2
229 protein K29-linked deubiquitination (GO:0035523) 1/5 2.247143e-02 TNFAIP3
230 regulation of cell cycle (GO:0051726) 3/296 2.247143e-02 SMAD3;IFNG;IL12B
231 innate immune response (GO:0045087) 3/302 2.364532e-02 CD40;NOS2;IL23R
232 regulation of MAP kinase activity (GO:0043405) 2/97 2.409510e-02 CD40;LRRK2
233 cellular response to chemical stress (GO:0062197) 2/101 2.468977e-02 LRRK2;NCF4
234 negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis (GO:1903588) 1/6 2.468977e-02 IL12B
235 activation of Janus kinase activity (GO:0042976) 1/6 2.468977e-02 JAK2
236 negative regulation of cellular extravasation (GO:0002692) 1/6 2.468977e-02 PTGER4
237 negative regulation of cytosolic calcium ion concentration (GO:0051481) 1/6 2.468977e-02 SMAD3
238 negative regulation of establishment of protein localization to mitochondrion (GO:1903748) 1/6 2.468977e-02 LRRK2
239 negative regulation of excitatory postsynaptic potential (GO:0090394) 1/6 2.468977e-02 LRRK2
240 negative regulation of toll-like receptor 2 signaling pathway (GO:0034136) 1/6 2.468977e-02 TNFAIP3
241 regulation of immunoglobulin mediated immune response (GO:0002889) 1/6 2.468977e-02 IL10
242 regulation of protein autoubiquitination (GO:1902498) 1/6 2.468977e-02 LRRK2
243 regulation of retrograde transport, endosome to Golgi (GO:1905279) 1/6 2.468977e-02 LRRK2
244 regulation of vitamin D biosynthetic process (GO:0060556) 1/6 2.468977e-02 IFNG
245 response to manganese ion (GO:0010042) 1/6 2.468977e-02 LRRK2
246 positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target (GO:0002860) 1/6 2.468977e-02 IL12B
247 positive regulation of natural killer cell mediated immune response to tumor cell (GO:0002857) 1/6 2.468977e-02 IL12B
248 telencephalon cell migration (GO:0022029) 1/6 2.468977e-02 LRRK2
249 positive regulation of Wnt signaling pathway, planar cell polarity pathway (GO:2000096) 1/6 2.468977e-02 RSPO3
250 protein deubiquitination involved in ubiquitin-dependent protein catabolic process (GO:0071947) 1/6 2.468977e-02 TNFAIP3
251 protein K29-linked ubiquitination (GO:0035519) 1/6 2.468977e-02 RNF186
252 regulation of protein ubiquitination (GO:0031396) 2/109 2.706172e-02 LRRK2;TNFAIP3
253 response to tumor necrosis factor (GO:0034612) 2/110 2.706172e-02 CD40;JAK2
254 arginine catabolic process (GO:0006527) 1/7 2.706172e-02 NOS2
255 regulation of calcidiol 1-monooxygenase activity (GO:0060558) 1/7 2.706172e-02 IFNG
256 negative regulation of membrane protein ectodomain proteolysis (GO:0051045) 1/7 2.706172e-02 IL10
257 regulation of extracellular matrix assembly (GO:1901201) 1/7 2.706172e-02 SMAD3
258 cellular response to prostaglandin E stimulus (GO:0071380) 1/7 2.706172e-02 PTGER4
259 nodal signaling pathway (GO:0038092) 1/7 2.706172e-02 SMAD3
260 regulation of natural killer cell mediated cytotoxicity directed against tumor cell target (GO:0002858) 1/7 2.706172e-02 IL12B
261 regulation of T cell tolerance induction (GO:0002664) 1/7 2.706172e-02 IL2RA
262 regulation of transforming growth factor beta2 production (GO:0032909) 1/7 2.706172e-02 SMAD3
263 positive regulation of interleukin-23 production (GO:0032747) 1/7 2.706172e-02 IFNG
264 positive regulation of lymphocyte apoptotic process (GO:0070230) 1/7 2.706172e-02 IL10
265 SMAD protein complex assembly (GO:0007183) 1/7 2.706172e-02 SMAD3
266 T-helper 17 cell differentiation (GO:0072539) 1/7 2.706172e-02 RORC
267 protein localization to endoplasmic reticulum exit site (GO:0070973) 1/7 2.706172e-02 LRRK2
268 positive regulation of metabolic process (GO:0009893) 2/113 2.799051e-02 IFNG;JAK2
269 activation of protein kinase activity (GO:0032147) 2/114 2.836188e-02 TNFSF15;JAK2
270 cellular response to molecule of bacterial origin (GO:0071219) 2/115 2.855549e-02 IL10;TNFAIP3
271 regulation of alpha-beta T cell differentiation (GO:0046637) 1/8 2.855549e-02 PRDM1
272 regulation of chemokine (C-C motif) ligand 5 production (GO:0071649) 1/8 2.855549e-02 IL10
273 regulation of dopamine receptor signaling pathway (GO:0060159) 1/8 2.855549e-02 LRRK2
274 negative regulation of response to reactive oxygen species (GO:1901032) 1/8 2.855549e-02 LRRK2
275 cellular response to interleukin-21 (GO:0098757) 1/8 2.855549e-02 STAT4
276 regulation of interleukin-18 production (GO:0032661) 1/8 2.855549e-02 IL10
277 cytoplasmic sequestering of NF-kappaB (GO:0007253) 1/8 2.855549e-02 IL10
278 regulation of nitric oxide metabolic process (GO:0080164) 1/8 2.855549e-02 JAK2
279 regulation of nitrogen compound metabolic process (GO:0051171) 1/8 2.855549e-02 IFNG
280 regulation of protein ADP-ribosylation (GO:0010835) 1/8 2.855549e-02 IFNG
281 positive regulation of adaptive immune response (GO:0002821) 1/8 2.855549e-02 IL6ST
282 regulation of response to wounding (GO:1903034) 1/8 2.855549e-02 IL10
283 regulation of toll-like receptor 3 signaling pathway (GO:0034139) 1/8 2.855549e-02 TNFAIP3
284 positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway (GO:2001269) 1/8 2.855549e-02 JAK2
285 regulation of vascular wound healing (GO:0061043) 1/8 2.855549e-02 TNFAIP3
286 interleukin-21-mediated signaling pathway (GO:0038114) 1/8 2.855549e-02 STAT4
287 positive regulation of steroid biosynthetic process (GO:0010893) 1/8 2.855549e-02 IFNG
288 tyrosine phosphorylation of STAT protein (GO:0007260) 1/8 2.855549e-02 JAK2
289 negative regulation of amyloid-beta clearance (GO:1900222) 1/8 2.855549e-02 IFNG
290 cellular response to lipopolysaccharide (GO:0071222) 2/120 2.902446e-02 IL10;TNFAIP3
291 cellular response to interferon-gamma (GO:0071346) 2/121 2.938750e-02 IFNG;JAK2
292 regulation of anatomical structure morphogenesis (GO:0022603) 2/123 2.954877e-02 LRRK2;TNFAIP3
293 cellular response to organic substance (GO:0071310) 2/123 2.954877e-02 SMAD3;LRRK2
294 cellular response to oxidative stress (GO:0034599) 2/125 2.954877e-02 LRRK2;NCF4
295 negative regulation of cell growth (GO:0030308) 2/126 2.954877e-02 SMAD3;HNF4A
296 negative regulation of growth (GO:0045926) 2/126 2.954877e-02 SMAD3;HNF4A
297 negative regulation of bone resorption (GO:0045779) 1/9 2.954877e-02 TNFAIP3
298 negative regulation of leukocyte proliferation (GO:0070664) 1/9 2.954877e-02 TNFAIP3
299 regulation of complement-dependent cytotoxicity (GO:1903659) 1/9 2.954877e-02 IL10
300 CD40 signaling pathway (GO:0023035) 1/9 2.954877e-02 CD40
301 negative regulation of production of molecular mediator of immune response (GO:0002701) 1/9 2.954877e-02 IL10
302 regulation of ER to Golgi vesicle-mediated transport (GO:0060628) 1/9 2.954877e-02 LRRK2
303 regulation of interleukin-23 production (GO:0032667) 1/9 2.954877e-02 IFNG
304 regulation of leukocyte mediated cytotoxicity (GO:0001910) 1/9 2.954877e-02 NOS2
305 ornithine metabolic process (GO:0006591) 1/9 2.954877e-02 HNF4A
306 peptidyl-cysteine modification (GO:0018198) 1/9 2.954877e-02 NOS2
307 regulation of protein deacetylation (GO:0090311) 1/9 2.954877e-02 IFNG
308 epithelial cell apoptotic process (GO:1904019) 1/9 2.954877e-02 IL10
309 positive regulation of B cell differentiation (GO:0045579) 1/9 2.954877e-02 IL10
310 regulation of Wnt signaling pathway, planar cell polarity pathway (GO:2000095) 1/9 2.954877e-02 RSPO3
311 positive regulation of extracellular matrix assembly (GO:1901203) 1/9 2.954877e-02 SMAD3
312 intestinal epithelial cell differentiation (GO:0060575) 1/9 2.954877e-02 IL6ST
313 positive regulation of protein kinase C signaling (GO:0090037) 1/9 2.954877e-02 CD40
314 T-helper 1 type immune response (GO:0042088) 1/9 2.954877e-02 IL12B
315 positive regulation of macromolecule biosynthetic process (GO:0010557) 2/129 3.067746e-02 IFNG;JAK2
316 arginine metabolic process (GO:0006525) 1/10 3.112631e-02 NOS2
317 negative regulation of heterotypic cell-cell adhesion (GO:0034115) 1/10 3.112631e-02 IL10
318 B cell homeostasis (GO:0001782) 1/10 3.112631e-02 TNFAIP3
319 regulation of cell-cell adhesion involved in gastrulation (GO:0070587) 1/10 3.112631e-02 IL10
320 negative regulation of receptor binding (GO:1900121) 1/10 3.112631e-02 IL10
321 cellular response to dopamine (GO:1903351) 1/10 3.112631e-02 LRRK2
322 negative regulation of toll-like receptor 4 signaling pathway (GO:0034144) 1/10 3.112631e-02 TNFAIP3
323 cellular response to prostaglandin stimulus (GO:0071379) 1/10 3.112631e-02 PTGER4
324 regulation of hydrogen peroxide-induced cell death (GO:1903205) 1/10 3.112631e-02 LRRK2
325 negative regulation of vascular endothelial growth factor signaling pathway (GO:1900747) 1/10 3.112631e-02 IL12B
326 regulation of tau-protein kinase activity (GO:1902947) 1/10 3.112631e-02 IFNG
327 positive regulation of circadian rhythm (GO:0042753) 1/10 3.112631e-02 RORC
328 response to dopamine (GO:1903350) 1/10 3.112631e-02 LRRK2
329 positive regulation of tissue remodeling (GO:0034105) 1/10 3.112631e-02 IL12B
330 primary miRNA processing (GO:0031053) 1/10 3.112631e-02 SMAD3
331 protein K11-linked deubiquitination (GO:0035871) 1/10 3.112631e-02 TNFAIP3
332 negative regulation of inflammatory response to antigenic stimulus (GO:0002862) 2/136 3.218490e-02 PTGER4;IL10
333 regulation of inflammatory response to antigenic stimulus (GO:0002861) 2/137 3.253781e-02 PTGER4;IL10
334 enzyme linked receptor protein signaling pathway (GO:0007167) 2/140 3.254603e-02 JAK2;IL6ST
335 transcription initiation from RNA polymerase II promoter (GO:0006367) 2/140 3.254603e-02 HNF4A;RORC
336 negative regulation of bone remodeling (GO:0046851) 1/11 3.254603e-02 TNFAIP3
337 regulation of cytoplasmic transport (GO:1903649) 1/11 3.254603e-02 LRRK2
338 negative regulation of oxidative stress-induced neuron death (GO:1903204) 1/11 3.254603e-02 IL10
339 negative regulation of protein maturation (GO:1903318) 1/11 3.254603e-02 LRRK2
340 cellular response to interleukin-2 (GO:0071352) 1/11 3.254603e-02 IL2RA
341 regulation of mitochondrial depolarization (GO:0051900) 1/11 3.254603e-02 LRRK2
342 positive regulation of cellular respiration (GO:1901857) 1/11 3.254603e-02 IFNG
343 regulation of toll-like receptor 2 signaling pathway (GO:0034135) 1/11 3.254603e-02 TNFAIP3
344 response to corticosteroid (GO:0031960) 1/11 3.254603e-02 IL10
345 response to prostaglandin E (GO:0034695) 1/11 3.254603e-02 PTGER4
346 response to sterol (GO:0036314) 1/11 3.254603e-02 RORC
347 interleukin-2-mediated signaling pathway (GO:0038110) 1/11 3.254603e-02 IL2RA
348 positive regulation of non-canonical Wnt signaling pathway (GO:2000052) 1/11 3.254603e-02 RSPO3
349 positive regulation of cellular catabolic process (GO:0031331) 2/141 3.278813e-02 IFNG;LRRK2
350 protein localization to organelle (GO:0033365) 2/142 3.313523e-02 LRRK2;RNF186
351 negative regulation of B cell proliferation (GO:0030889) 1/12 3.449149e-02 IL10
352 adipose tissue development (GO:0060612) 1/12 3.449149e-02 RORC
353 negative regulation of cellular response to vascular endothelial growth factor stimulus (GO:1902548) 1/12 3.449149e-02 IL12B
354 regulation of B cell apoptotic process (GO:0002902) 1/12 3.449149e-02 IL10
355 cellular response to growth hormone stimulus (GO:0071378) 1/12 3.449149e-02 JAK2
356 phospholipid homeostasis (GO:0055091) 1/12 3.449149e-02 HNF4A
357 positive regulation of leukocyte proliferation (GO:0070665) 1/12 3.449149e-02 IL12B
358 mitochondrion distribution (GO:0048311) 1/12 3.449149e-02 LRRK2
359 regulation of cytosolic calcium ion concentration (GO:0051480) 2/148 3.483861e-02 PTGER4;SMAD3
360 positive regulation of intracellular protein transport (GO:0090316) 2/148 3.483861e-02 SMAD3;IFNG
361 negative regulation of autophagosome assembly (GO:1902902) 1/13 3.603399e-02 LRRK2
362 purine ribonucleoside triphosphate metabolic process (GO:0009205) 1/13 3.603399e-02 LRRK2
363 regulation of actomyosin structure organization (GO:0110020) 1/13 3.603399e-02 PTGER4
364 negative regulation of macromolecule biosynthetic process (GO:0010558) 1/13 3.603399e-02 IL10
365 regulation of lysosomal lumen pH (GO:0035751) 1/13 3.603399e-02 LRRK2
366 regulation of receptor signaling pathway via STAT (GO:1904892) 1/13 3.603399e-02 JAK2
367 regulation of response to stress (GO:0080134) 1/13 3.603399e-02 IL10
368 response to UV-B (GO:0010224) 1/13 3.603399e-02 IL12B
369 icosanoid secretion (GO:0032309) 1/13 3.603399e-02 NOS2
370 macrophage activation involved in immune response (GO:0002281) 1/13 3.603399e-02 IFNG
371 positive regulation of purine nucleotide biosynthetic process (GO:1900373) 1/13 3.603399e-02 NOS2
372 protein autophosphorylation (GO:0046777) 2/159 3.776375e-02 LRRK2;JAK2
373 negative regulation of interleukin-12 production (GO:0032695) 1/14 3.776375e-02 IL10
374 cytoplasmic sequestering of transcription factor (GO:0042994) 1/14 3.776375e-02 IL10
375 defense response to protozoan (GO:0042832) 1/14 3.776375e-02 CD40
376 regulation of protein kinase C signaling (GO:0090036) 1/14 3.776375e-02 CD40
377 positive regulation of cyclase activity (GO:0031281) 1/14 3.776375e-02 NOS2
378 growth hormone receptor signaling pathway via JAK-STAT (GO:0060397) 1/14 3.776375e-02 JAK2
379 immune response-regulating cell surface receptor signaling pathway (GO:0002768) 1/14 3.776375e-02 CD40
380 positive regulation of protein deacetylation (GO:0090312) 1/14 3.776375e-02 IFNG
381 positive regulation of SMAD protein signal transduction (GO:0060391) 1/14 3.776375e-02 JAK2
382 negative regulation of blood pressure (GO:0045776) 1/15 3.890427e-02 NOS2
383 negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway (GO:1902236) 1/15 3.890427e-02 LRRK2
384 negative regulation of protein processing (GO:0010955) 1/15 3.890427e-02 LRRK2
385 regulation of integrin activation (GO:0033623) 1/15 3.890427e-02 PTGER4
386 nitric oxide biosynthetic process (GO:0006809) 1/15 3.890427e-02 NOS2
387 regulation of non-canonical Wnt signaling pathway (GO:2000050) 1/15 3.890427e-02 RSPO3
388 positive regulation of acute inflammatory response (GO:0002675) 1/15 3.890427e-02 IL6ST
389 positive regulation of exosomal secretion (GO:1903543) 1/15 3.890427e-02 IFNG
390 GTP metabolic process (GO:0046039) 1/15 3.890427e-02 LRRK2
391 positive regulation of heterotypic cell-cell adhesion (GO:0034116) 1/15 3.890427e-02 IL10
392 positive regulation of lyase activity (GO:0051349) 1/15 3.890427e-02 NOS2
393 positive regulation of membrane protein ectodomain proteolysis (GO:0051044) 1/15 3.890427e-02 IFNG
394 superoxide anion generation (GO:0042554) 1/15 3.890427e-02 NCF4
395 macrophage differentiation (GO:0030225) 1/15 3.890427e-02 IFNG
396 mammary gland epithelium development (GO:0061180) 1/15 3.890427e-02 JAK2
397 DNA-templated transcription, initiation (GO:0006352) 2/168 3.986146e-02 HNF4A;RORC
398 activation of NF-kappaB-inducing kinase activity (GO:0007250) 1/16 3.986146e-02 TNFSF15
399 negative regulation of epithelial cell differentiation (GO:0030857) 1/16 3.986146e-02 IFNG
400 regulation of amyloid-beta clearance (GO:1900221) 1/16 3.986146e-02 IFNG
401 regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis (GO:1903587) 1/16 3.986146e-02 IL12B
402 regulation of cellular carbohydrate metabolic process (GO:0010675) 1/16 3.986146e-02 RORC
403 regulation of defense response to virus (GO:0050688) 1/16 3.986146e-02 TNFAIP3
404 cellular response to sterol (GO:0036315) 1/16 3.986146e-02 RORC
405 regulation of interleukin-1 production (GO:0032652) 1/16 3.986146e-02 IL10
406 regulation of isotype switching (GO:0045191) 1/16 3.986146e-02 IL10
407 nitric oxide metabolic process (GO:0046209) 1/16 3.986146e-02 NOS2
408 positive regulation of alpha-beta T cell proliferation (GO:0046641) 1/16 3.986146e-02 IL12B
409 positive regulation of endothelial cell apoptotic process (GO:2000353) 1/16 3.986146e-02 CD40
410 glutamine family amino acid catabolic process (GO:0009065) 1/16 3.986146e-02 NOS2
411 positive regulation of isotype switching (GO:0045830) 1/16 3.986146e-02 CD40
412 protein localization to endoplasmic reticulum (GO:0070972) 1/16 3.986146e-02 LRRK2
413 negative regulation of nucleic acid-templated transcription (GO:1903507) 3/464 4.161928e-02 IFNG;HNF4A;IKZF1
414 negative regulation of chemokine production (GO:0032682) 1/17 4.161928e-02 IL10
415 negative regulation of cytokine production involved in immune response (GO:0002719) 1/17 4.161928e-02 IL10
416 regulation of cell killing (GO:0031341) 1/17 4.161928e-02 IL10
417 negative regulation of vascular associated smooth muscle cell proliferation (GO:1904706) 1/17 4.161928e-02 IL10
418 regulation of neuroblast proliferation (GO:1902692) 1/17 4.161928e-02 LRRK2
419 positive regulation of muscle hypertrophy (GO:0014742) 1/17 4.161928e-02 IL6ST
420 regulation of mitotic cell cycle (GO:0007346) 2/178 4.222887e-02 IL10;SMAD3
421 positive regulation of cellular biosynthetic process (GO:0031328) 2/180 4.291341e-02 SMAD3;IFNG
422 regulation of adaptive immune response (GO:0002819) 1/18 4.291341e-02 IL6ST
423 negative regulation of interleukin-8 production (GO:0032717) 1/18 4.291341e-02 IL10
424 nitric oxide mediated signal transduction (GO:0007263) 1/18 4.291341e-02 NOS2
425 regulation of membrane depolarization (GO:0003254) 1/18 4.291341e-02 LRRK2
426 positive regulation of amyloid-beta formation (GO:1902004) 1/18 4.291341e-02 IFNG
427 positive regulation of cardiac muscle hypertrophy (GO:0010613) 1/18 4.291341e-02 IL6ST
428 regulation of transforming growth factor beta production (GO:0071634) 1/18 4.291341e-02 SMAD3
429 positive regulation of extracellular matrix organization (GO:1903055) 1/18 4.291341e-02 SMAD3
430 positive regulation of peptidyl-serine phosphorylation of STAT protein (GO:0033141) 1/18 4.291341e-02 IFNG
431 activin receptor signaling pathway (GO:0032924) 1/19 4.424043e-02 SMAD3
432 regulation of epithelial cell differentiation (GO:0030856) 1/19 4.424043e-02 IFNG
433 regulation of exosomal secretion (GO:1903541) 1/19 4.424043e-02 IFNG
434 regulation of lymphocyte proliferation (GO:0050670) 1/19 4.424043e-02 IL12B
435 regulation of peptidyl-serine phosphorylation of STAT protein (GO:0033139) 1/19 4.424043e-02 IFNG
436 excitatory postsynaptic potential (GO:0060079) 1/19 4.424043e-02 LRRK2
437 regulation of synaptic vesicle exocytosis (GO:2000300) 1/19 4.424043e-02 LRRK2
438 regulation of vascular endothelial growth factor signaling pathway (GO:1900746) 1/19 4.424043e-02 IL12B
439 mitochondrion localization (GO:0051646) 1/19 4.424043e-02 LRRK2
440 myeloid cell activation involved in immune response (GO:0002275) 1/19 4.424043e-02 IFNG
441 regulation of GTPase activity (GO:0043087) 2/189 4.496435e-02 CD40;LRRK2
442 negative regulation of cell differentiation (GO:0045596) 2/191 4.560699e-02 SMAD3;IFNG
443 regulation of acute inflammatory response (GO:0002673) 1/20 4.560699e-02 IL6ST
444 regulation of lymphocyte differentiation (GO:0045619) 1/20 4.560699e-02 PRDM1
445 regulation of protein kinase A signaling (GO:0010738) 1/20 4.560699e-02 LRRK2
446 regulation of tissue remodeling (GO:0034103) 1/20 4.560699e-02 IL12B
447 growth hormone receptor signaling pathway (GO:0060396) 1/20 4.560699e-02 JAK2
448 positive regulation of natural killer cell mediated cytotoxicity (GO:0045954) 1/20 4.560699e-02 IL12B
449 positive regulation of nitrogen compound metabolic process (GO:0051173) 1/20 4.560699e-02 IFNG
450 negative regulation of macromolecule metabolic process (GO:0010605) 2/194 4.614804e-02 NOS2;IFNG
451 regulation of programmed cell death (GO:0043067) 2/194 4.614804e-02 LRRK2;JAK2
452 cellular response to estradiol stimulus (GO:0071392) 1/21 4.691709e-02 NRIP1
453 positive regulation of glycolytic process (GO:0045821) 1/21 4.691709e-02 IFNG
454 innate immune response in mucosa (GO:0002227) 1/21 4.691709e-02 NOS2
455 positive regulation of phosphatase activity (GO:0010922) 1/21 4.691709e-02 IFNG
456 positive regulation of purine nucleotide metabolic process (GO:1900544) 1/21 4.691709e-02 IFNG
457 positive regulation of transcription regulatory region DNA binding (GO:2000679) 1/21 4.691709e-02 IFNG
458 positive regulation of vascular associated smooth muscle cell proliferation (GO:1904707) 1/21 4.691709e-02 IL10
459 regulation of signal transduction (GO:0009966) 2/198 4.708871e-02 LRRK2;RSPO3
460 regulation of macromolecule metabolic process (GO:0060255) 2/200 4.786710e-02 IL10;PRDM1
461 negative regulation of interleukin-2 production (GO:0032703) 1/22 4.848545e-02 TNFAIP3
462 negative regulation of lymphocyte activation (GO:0051250) 1/22 4.848545e-02 TNFAIP3
463 regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway (GO:1902235) 1/22 4.848545e-02 LRRK2
464 positive regulation of amyloid precursor protein catabolic process (GO:1902993) 1/22 4.848545e-02 IFNG
465 regulation of angiogenesis (GO:0045765) 2/203 4.867177e-02 CD40;TNFAIP3
466 negative regulation of GTPase activity (GO:0034260) 1/23 4.969388e-02 LRRK2
467 negative regulation of intracellular protein transport (GO:0090317) 1/23 4.969388e-02 LRRK2
468 regulation of neural precursor cell proliferation (GO:2000177) 1/23 4.969388e-02 LRRK2
469 ERK1 and ERK2 cascade (GO:0070371) 1/23 4.969388e-02 PTGER4
470 regulation of SMAD protein signal transduction (GO:0060390) 1/23 4.969388e-02 JAK2
471 regulation of steroid metabolic process (GO:0019218) 1/23 4.969388e-02 RORC
472 regulation of toll-like receptor 4 signaling pathway (GO:0034143) 1/23 4.969388e-02 TNFAIP3
473 positive regulation of epithelial cell apoptotic process (GO:1904037) 1/23 4.969388e-02 CD40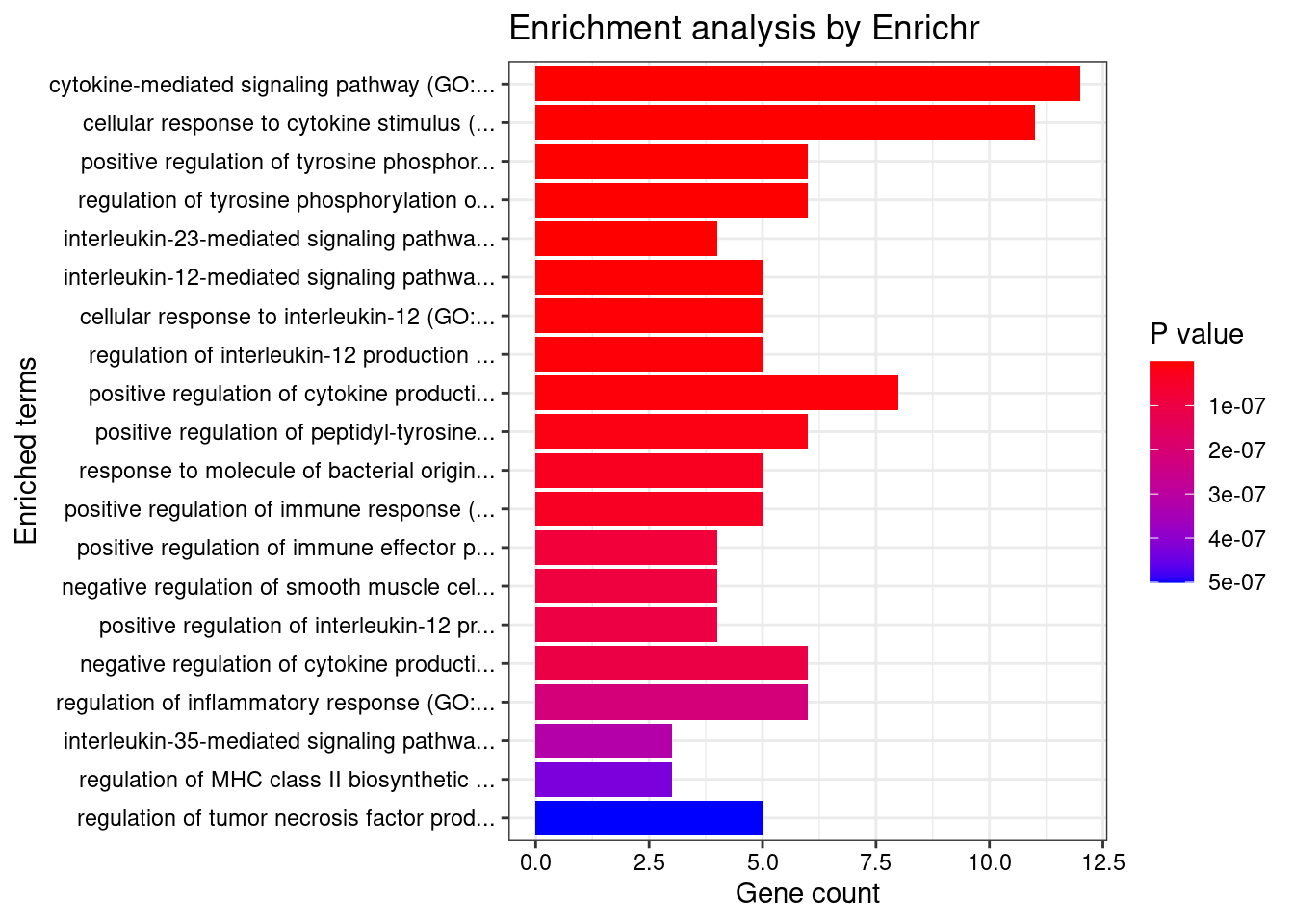
| Version | Author | Date |
|---|---|---|
| 177114d | wesleycrouse | 2022-05-04 |
save(enrich_results, file="ABC_IBD_genes_enrichment.RData")
write.csv(enrich_results, file="ABC_IBD_genes_enrichment.csv")Summary table for selected tissue groups
selected_groups <- c("Blood or Immune", "Digestive")
selected_genes <- unique(unlist(sapply(df_group[selected_groups], function(x){x$ctwas})))
weight_groups <- weight_groups[order(weight_groups$group),]
selected_weights <- weight_groups$weight[weight_groups$group %in% selected_groups]
gene_pips_by_weight <- gene_pips_by_weight_bkup
results_table <- as.data.frame(round(gene_pips_by_weight[selected_genes,selected_weights],3))
results_table$n_discovered <- apply(results_table>0.8,1,sum,na.rm=T)
results_table$n_imputed <- apply(results_table, 1, function(x){sum(!is.na(x))-1})
results_table <- cbind(results_table, G_list[sapply(rownames(results_table), match, table=G_list$hgnc_symbol),c("chromosome_name","start_position","end_position","nearby","nearest")])
results_table$known <- rownames(results_table) %in% known_genes
#load("group_enrichment_results.RData")
group_enrichment_results$group <- as.character(group_enrichment_results$group)
group_enrichment_results$db <- as.character(group_enrichment_results$db)
group_enrichment_results <- group_enrichment_results[group_enrichment_results$group %in% selected_groups,,drop=F]
results_table$enriched_terms <- sapply(rownames(results_table), function(x){paste(group_enrichment_results$Term[grep(x, group_enrichment_results$Genes)],collapse="; ")})
write.csv(results_table, file=paste0("summary_table_crohns.csv"))Overlap of enrichment analysis
#collect GO terms for selected genes
dbs <- c("GO_Biological_Process_2021")
GO_enrichment <- enrichr(selected_genes, dbs)Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Parsing results... Done.enrich_results_selected_genes <- GO_enrichment[[db]]
#enrich_results <- GO_enrichment[[db]]
#save(enrich_results, file="selected_genes_enrichment.RData")
#load("selected_genes_enrichment.RData")
#enrich_results_selected_genes <- enrich_results
load("ABC_IBD_genes_enrichment.RData")
enrich_results_known_genes <- enrich_results
overlap_table <- as.data.frame(matrix(F, nrow(enrich_results_known_genes), length(selected_genes)))
overlap_table <- cbind(enrich_results_known_genes$Term, overlap_table)
colnames(overlap_table) <- c("Term", selected_genes)
for (i in 1:nrow(overlap_table)){
Term <- overlap_table$Term[i]
if (Term %in% enrich_results_selected_genes$Term){
Term_genes <- enrich_results_selected_genes$Genes[enrich_results_selected_genes$Term==Term]
overlap_table[i, unlist(strsplit(Term_genes, ";"))] <- T
}
}
write.csv(overlap_table, file="GO_overlap_crohns.csv")
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.5 disgenet2r_0.99.2 WebGestaltR_0.4.4 enrichR_3.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.8 reshape2_1.4.3 purrr_0.3.4 lattice_0.20-38 colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 utf8_1.2.1 rlang_0.4.11 later_0.8.0 pillar_1.6.1 withr_2.4.1 glue_1.4.2 DBI_1.1.1 gdtools_0.1.9 rngtools_1.5 doRNG_1.8.2 plyr_1.8.4 foreach_1.5.1 lifecycle_1.0.0 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0 workflowr_1.6.2 codetools_0.2-16 evaluate_0.14 labeling_0.3 knitr_1.23 doParallel_1.0.16 httpuv_1.5.1 curl_3.3 parallel_3.6.1 fansi_0.5.0 Rcpp_1.0.6 readr_1.4.0 promises_1.0.1 scales_1.1.0 jsonlite_1.6 apcluster_1.4.8 farver_2.1.0 fs_1.3.1 hms_1.1.0 rjson_0.2.20 digest_0.6.20 stringi_1.4.3 dplyr_1.0.7 grid_3.6.1 rprojroot_2.0.2 tools_3.6.1 magrittr_2.0.1 tibble_3.1.2 crayon_1.4.1
[56] whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2 Matrix_1.2-18 data.table_1.14.0 svglite_1.2.2 rmarkdown_1.13 httr_1.4.1 iterators_1.0.13 R6_2.5.0 igraph_1.2.4.1 git2r_0.26.1 compiler_3.6.1